Abstract
Background
The Coronavirus disease 2019 (COVID-19) pandemic has underlined the urgent need for reliable, multicenter, and full-admission intensive care data to advance our understanding of the course of the disease and investigate potential treatment strategies. In this study, we present the Dutch Data Warehouse (DDW), the first multicenter electronic health record (EHR) database with full-admission data from critically ill COVID-19 patients.
Methods
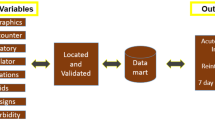
A nation-wide data sharing collaboration was launched at the beginning of the pandemic in March 2020. All hospitals in the Netherlands were asked to participate and share pseudonymized EHR data from adult critically ill COVID-19 patients. Data included patient demographics, clinical observations, administered medication, laboratory determinations, and data from vital sign monitors and life support devices. Data sharing agreements were signed with participating hospitals before any data transfers took place. Data were extracted from the local EHRs with prespecified queries and combined into a staging dataset through an extract–transform–load (ETL) pipeline. In the consecutive processing pipeline, data were mapped to a common concept vocabulary and enriched with derived concepts. Data validation was a continuous process throughout the project. All participating hospitals have access to the DDW. Within legal and ethical boundaries, data are available to clinicians and researchers.
Results
Out of the 81 intensive care units in the Netherlands, 66 participated in the collaboration, 47 have signed the data sharing agreement, and 35 have shared their data. Data from 25 hospitals have passed through the ETL and processing pipeline. Currently, 3464 patients are included in the DDW, both from wave 1 and wave 2 in the Netherlands. More than 200 million clinical data points are available. Overall ICU mortality was 24.4%. Respiratory and hemodynamic parameters were most frequently measured throughout a patient's stay. For each patient, all administered medication and their daily fluid balance were available. Missing data are reported for each descriptive.
Conclusions
In this study, we show that EHR data from critically ill COVID-19 patients may be lawfully collected and can be combined into a data warehouse. These initiatives are indispensable to advance medical data science in the field of intensive care medicine.
Similar content being viewed by others
Introduction
The Corona virus disease 2019 (COVID-19) pandemic has placed an unprecedented burden on intensive care units around the world. Many intensive care units still face high death rates, and the number of critically ill patients still exceeds available intensive care unit (ICU) beds in some areas [1]. More than ever before, COVID-19 has shown the need for concerted research efforts among the intensive care community to understand the course of severe COVID-19 disease, to identify potential treatment strategies and to guide resource allocation.
Research with routinely collected electronic health record (EHR) data has increasingly gained interest in the ICU over the last decade [2]. There has been a widespread transition toward EHR systems, enabling the routine capture of individual patient data throughout ICU admission [3]. Moreover, several individual hospitals have extracted these EHR data and converted them into critical care datasets available for research, including the Medical Information Mart for Intensive Care (MIMIC) [4], AmsterdamUMCdb [5], and HiRID [6]. These datasets have laid the groundwork for working with EHR data and have advanced medical data science in the field of critical care.
However, rather than single-center data alone, the COVID-19 pandemic has underlined the need for accurate and verifiable multicenter data [7, 8]. The novelty of COVID-19 and absence of treatment guidelines resulted in practice variation between centers, emphasizing the limits of single-center research and the need for multicenter research into effective treatment strategies [9]. Furthermore, medical transfers, different levels of care, and care practice differences between hospitals hamper the extrapolation of single-center data. Patient demographics, for example, have been shown to differ considerably between centers [10]. Multicenter data are therefore crucial, but assembling data from multiple centers yields major challenges.
We initiated a large-scale data sharing collaboration in the Netherlands that resulted in the Dutch Data Warehouse (DDW), a complete-admission and multicenter database with EHR data from critically ill COVID-19 patients. The DDW was designed with an interdisciplinary team of legal advisors, privacy officers, data engineers, IT-professionals, data scientists, statisticians, and clinicians. This paper presents a full report on the first stable version of the database and addresses the major challenges in the construction of the DDW. Given the crisis, a brief overview of the preliminary dataset was published as a letter [11]. In the present report, we expand on the methodology underlying the DDW and show the patient population currently included.
Methods
The data sharing collaborative was started at the beginning of the COVID-19 crisis in the Netherlands in March 2020. All hospitals in the Netherlands with an intensive care unit were approached to participate. Per hospital, an intensivist and IT-professional served as contacts for local study approval, data expertise, and data extraction. All hospitals that participated have access to the cumulative dataset for research purposes. The process of obtaining legal approval and the extract–transform–load (ETL) pipeline, as well as the data mapping, data enrichment, and data validation process are described in detail. An overview of the project can be found in Fig. 1.
Legal and privacy
In close collaboration with data protection officers (DPO), health care lawyers, and intensivists, we drafted a data sharing agreement (DSA) and a multidisciplinary report on the lawful collection of EHR data during the COVID-19 crisis. Under the General Data Protection Regulation (GDPR) and Dutch law, data subjects are required to give explicit consent for the processing of their data. We argued, however, that during the COVID-19 crisis asking consent could not be reasonably expected from health care workers due to (a) the large number of expected patients and associated time burden in an already overstrained health care system, (b) the danger of spreading or contracting the virus upon contact with patients or their families, and (c) the poor clinical condition of many patients in the intensive care. Consent was therefore not only impractical, but often infeasible. In addition, alternative forms of data collection to construct a database of this size were unavailable and selection bias would have ensued in case of failed consents.
As under non-crisis circumstances, COVID-19 data necessary for scientific purposes may be gathered when researchers “provide for suitable and specific measures to safeguard the fundamental rights and interests of the data subject” (GDPR, Article 9, paragraph j) [12]. Therefore, we (a) pseudonymized data in the providing hospital, (b) informed patients through media and local hospital outlets about the possibility to opt out, and (c) signed data sharing agreements regulating privacy of patients. The study proposal and documentation were reviewed and approved by the institutional review board of Amsterdam UMC location VUmc prior to study onset. Data sharing agreements were approved locally in each hospital before data transfers took place. The DSA has been added to the Additional files 1 and 2. All institutional review board documentation is available upon request from the corresponding author.
Extract–transform–load pipeline
In collaboration with local IT-experts, template Structured Query Language (SQL) queries were written to automatically extract EHR data from each of the major EHR systems in the Netherlands: MetaVision (iMDsoft, Tel Aviv, Israel), HiX (ChipSoft, Amsterdam, The Netherlands), and Epic (Epic Systems, Verona, WA, United States). Intensive care COVID-19 patients were labelled locally by the participating hospitals. All adult patients with laboratory-confirmed COVID-19 or a Reporting and Data System (CO-RADS) score with clinical suspicion compatible with the diagnosis were labeled for inclusion (13).
The extracted data included demographics, clinical observations manually entered by the clinical team, administered medication, laboratory determinations, and data from vital sign monitors and life support devices such as mechanical ventilators, renal replacement devices and extracorporeal life support devices. Clinical notes, radiology reports and images, pathology and microbiology data were not extracted due to the additional complexity of these data and potential privacy implications. We included Dutch national registry data on patient comorbidities since these data are unsystematically recorded in the EHR and are frequently part of clinical notes [13].
IT experts from the participating hospitals adjusted the structured queries to local system configurations and performed the data extraction and pseudonymisation. Pseudonymisation was performed using a Secure Hash Algorithm (SHA-256). Data were stored in CSV format and shared with end-to-end encryption. Data extractions were performed upon request depending on the number of newly admitted patients. Upon receiving the data transfers, tables from the different EHR systems were restructured and data were combined into a staging database. A first data validation step was performed checking tables for completeness of columns, missing data, headers, and delimiters. This process was repeated per hospital to ensure completeness of data. After the staging database, data went through the data processing pipeline to be mapped, enriched, further validated and restructured to facilitate research.
Data mapping
One of the major challenges in combining multicenter EHR data is to find corresponding parameters between hospitals. No mandated set of recorded parameters exists for ICUs in the Netherlands, nor is there a standardized nomenclature for parameters, which results in between-hospital differences on several levels. First, parameter names may differ between hospitals and may include abbreviations, generating a plethora of unique parameters. In addition, certain parameters may be recorded in one hospital, but not in another. For example, not all hospitals record Richmond Agitation and Sedation Scales (RASS). Moreover, the level of parameter detail may differ between hospitals. One hospital may distinguish between alanine transferase (ALAT) measured in blood versus ALAT measured in other body fluids. Lastly, varying units between centers further hampers finding corresponding parameters. These between-hospital differences greatly complicate the combination of multicenter EHR data.
Through a process called mapping, parameters from different hospitals are linked to a concept from a predefined vocabulary. Although international vocabularies such as Logical Observation Identifiers Names and Codes (LOINC) and Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) exist [14,15,16], no widespread mapping tooling is available and existing vocabularies may not yet be complete for the intensive care unit [17]. Considering the urgency of the COVID-19 pandemic, we therefore created our own vocabulary of 942 clinically relevant parameters. We incorporated all 5.456 medications included in the Anatomical Therapeutic Chemical (ATC) classifications from the World Health Organization Collaborating Center for Drugs Statistics Methodology [18]. Most, but not all hospitals specified ATC codes for administered medication. Medications without an ATC code were mapped manually. Finally, we created a separate vocabulary of categories for 54 categorical concepts such as heart rhythm. These vocabularies included prespecified concepts for these categories, such as atrial fibrillation, ventricular tachycardia, and so on in the case of heart rhythm.
The received parameters were manually mapped per hospital to the predefined concept vocabulary. In order to facilitate the mapping process, the median, interquartile ranges, number of measurements, min, max, number and percentage of unique patients with the parameter, unit, and the most frequent value were calculated per parameter and exported to Google sheets for the mapping. Consequently, the concepts were aggregated into higher level concepts by the clinical team. For example, temperatures measured in the bladder and esophagus were both aggregated into the higher-level temperature concept. Both the detailed as well as the aggregated mappings are available in the DDW. Next, units were checked for each parameter and adjusted where necessary. Lastly, all mappings were independently reviewed by an intensive care clinician and discussed with the original hospital in case of uncertainty about the mapping. An overview of the most frequent concepts in the DDW can be found in Table 1.
Data enrichment
Because several medical concepts are insufficiently stored in the EHR, we added derived concepts to the DDW based on clinical expertise. These concepts included the conversion of recorded concepts, the addition of novel clinical concepts, and the calculation of clinical scores. The conversion of concepts ensured that concepts were added to the database when they could be derived from other available concepts. For example, respiratory system compliance can be calculated when tidal volume and driving pressure are available [19]. Secondly, clinical concepts that have been described in the literature were added to the DDW and included ventilatory ratio [20], physiologic dead space [21], and mechanical power [22]. These derived concepts can be found in Table 2 and included specific algorithms per concept to ensure the correct selection of underlying parameters. Lastly, clinical scores such as the Sequential Organ Failure Assessment (SOFA) score [23] and the Acute Physiology and Chronic Health Evaluation II (APACHE II) score [24] were calculated from the data per calendar day for each patient and can be found in Additional file 3: Table S1.
In addition to the derived concepts, some concepts required more complex derivation algorithms. Notably, patient in- and extubation times may not be easily or reliably available in EHR data, or result from multiple data columns. Therefore, we developed an algorithm that determines the start and end of intubation episodes based on other concepts. The overview of this algorithm has been published previously [11].
Data validation
Data validation and quality control were integrated throughout the project. The internal validity of the data was safeguarded by incorporating data that were validated by the clinical team during routine care, comparing calculated clinical scores against the manually recorded benchmarking scores from national registry data, and by data verification checks with the original hospital. In addition, several checkpoints ensured accurate processing of the data throughout the ETL and data processing pipeline. First, patient tables, headers, and column data were checked for completeness in the ETL pipeline. Secondly, parameter mappings were checked by an intensive care clinician and were therefore independently performed by two clinicians. Next, value distribution plots were continuously generated as part of the processing pipeline. These plots show the distribution of all parameters from all hospitals that were mapped to a certain concept and easily identify aberrant mappings. For all concepts, medically impossible cutoff values were determined by the clinical domain experts. Finally, demographics and any inconsistencies in the distributions or mapping were validated with their original hospital.
Data and code availability
The pipelines were constructed in Python 3 (Python Software Foundation). The resulting DDW is stored on a remote server. An application programming interface (API) was developed to facilitate data access. Access to the server is regulated to comply with the data sharing agreements. All hospitals have access to the data. External researchers can get access to all data in collaboration with any of the participating hospitals. The list of collaborators is available in the co-author list and in the declarations section. The collaborators may be contacted directly, through the corresponding author, or through the contact information on Amsterdammedicaldatascience.nl [25]. Research questions have to be in the line with the reason for data collection as outlined in the DSA; the investigation of the ICU course of COVID-19 or its potential treatments. In addition, researchers have to sign a code of conduct before getting access to the data. Data access is granted by Amsterdam UMC; compliance with the DSA is the responsibility of the researcher and hospital accessing the data. A repository to process the data warehouse, including more information on table structures and data content, is available on Gitlab. Anyone can get access to the repository by contacting the corresponding author.
Results
The data sharing collaboration was initiated in March 2020. Out of 81 hospitals with an intensive care unit in the Netherlands, 66 hospitals currently participate in the project (7 hospitals did not have the IT infrastructure or resources to carry out the data extraction, 1 hospital did not treat COVID-19 patients, and 7 did not want to participate or did not respond), 47 have signed the data sharing agreement and 35 have shared their data. The time to get approval and extract data ranged between less than 1 month and 6 months between hospitals. So far, data from 25 hospitals have passed through the ETL and data processing pipelines and are currently included in the DDW. These hospitals amount to a total of 3463 patients, both from wave 1 and wave 2 in the Netherlands. From these patients, more than 200 million clinical data points are available.
Parameter mapping
The mapping process of the received parameters resulted in a large mapping structure between all hospitals and EHR systems. From the staging database, 67,236 parameters (32,570 parameters from EPIC, 19,492 from Hix, and 15,174 parameters from MetaVision) were mapped to the common vocabulary. Next, 14,656 text parameters were mapped to categorical concepts. Part of these mappings were aggregated into 289 higher level concept names. The final list of the most frequent concepts and their clinical categories can be found in Table 1.
Data tables
Figure 2 gives an overview of the included data in the DDW. Table 1 lists the most frequent concepts found in the DDW with the number of total measurements, and the number of patients and number of hospitals with at least one measurement available for that concept. The data are available in separate tables and include a patients table with demographics and admission details; a single-timestamp table with all observations and measurements recorded at a single point in time; a range measurements table that contains parameters with a start and an end timestamp such as urine output, fluid output, and body position; a medications table with start times, end times, and dosing information; a diagnosis table with ICD-10 codes when available; a parameters table with the summary of all parameters currently included in the DDW; an intubations table with the start and end of invasive mechanical ventilation; a comorbidities table; and an outcomes table.
Overview of the Dutch Data Warehouse content. Overview of the data domains in the Dutch Data Warehouse. Examples of data are given per domain. EHR electronic health record, BMI body mass index, GCS Glasgow Coma Scale, RASS Richmond agitation and sedation scale, CAM-ICU confusion assessment method for the ICU, PEEP positive end-expiratory pressure, ECMO extracorporeal membrane oxygenation, IV intravenous
Clinical characteristics of patients
Table 3 describes the COVID-19 patients currently included in the DDW. The first patient was admitted on February 20, 2020, while the last patient was admitted on March 2, 2021. The median age was 64.0 (IQR 56.0, 72.0), and the majority of patients were male with a median BMI of 27.3 (IQR 24.3, 30.7). Overall ICU mortality was 24.4%.
Importantly, the DDW includes data throughout the ICU admission. The most common parameters were respiratory parameters, notably the fraction of inspired oxygen, the ventilation mode, and the positive end expiratory pressure. These parameters are measured and stored directly by the mechanical ventilator. Similarly, hemodynamic parameters that are automatically recorded and stored are most prevalent, including heart rate and blood pressure. Lastly, fluid balance and all administered medications are available for each patient. Missing data are reported in a separate column for each descriptive.
Discussion
In this study, we present the Dutch Data Warehouse, a large multicenter database with electronic health record data collected throughout the ICU admission of critically ill COVID-19 patients in the Netherlands. Currently, the DDW contains 3463 patients with over 200 million data points. The first stable version has been released and is available to researchers within ethical and legal boundaries.
The intensive care unit is a natural habitat for large data sharing collaboratives, as much data are collected through routine monitoring, life support devices, and by the clinical team. Although many publicly available single-center datasets have advanced our understanding of electronic health record data [4,5,6], multicenter data are crucial to enhance generalizability of results and account for between-center differences. The most important aspects of multicenter EHR data sharing include the legal framework, between-hospital concept mapping, and data preparation. Despite the complexity and volume of parameters received, we describe the legal basis for collecting these data under European privacy laws and show that these data can technically be combined into a data warehouse suitable for research.
The DDW has been used both as a research database and to create reports per hospital to compare local practices. The high granularity of the data, the wide variety of clinical parameters, and the availability of the data throughout the ICU stay make the database especially suitable for research. Clinical questions in a wide variety of areas relating to COVID-19 may be answered with the data, such as ventilation strategies, the timing and effects of proning, and the occurrence of superinfections. Apart from hard clinical endpoints such as mortality or length of stay, the DDW also allows for the investigation of intermediate clinical endpoints, such as line infections or improvements in P/F ratios. In addition to research, the dataset was used to create reports for hospitals to discuss and learn from treatment variation. These reports were created upon request and discussed confidentially with the participating hospitals.
For any medical data science project, and in particular projects throughout the COVID-19 pandemic, understanding and verifying the underlying data is crucial to interpret results. Reports have expressed worries about the quality of research conducted throughout the pandemic [26, 27]. The call for accurate, timely and reliable research data is larger than ever before. Only then, research can be replicated and checked by the scientific community. Undoubtedly, there will be mistakes and missing data in the Dutch Data Warehouse. Despite rigorous data preparation and validation, we believe that transparency of data and data sharing is key to continuously and collaboratively improve the dataset. Importantly, knowledge of intensive care medicine is indispensable when reviewing and evaluating the data, and thus, the involvement of critical care clinicians is paramount. With this report, we hope to encourage clinicians and researchers to get involved in data sharing collaborations. Moreover, we aim for this work to have laid out a roadmap for multicenter data sharing. Lastly, we have initiated ICUdata as a follow-up project. In this collaboration, we aim to collect and combine data from all ICU patients from as many ICUs as possible in the Netherlands. More information can be found on ICUdata.nl.
The DDW also comes with limitations. First of all, patient transfers could introduce bias since outcomes or prior admission data may not be available for these patients. However, whenever data were available from the receiving hospital, their admissions were connected in the DDW. Moreover, transfers show similar patient characteristics compared to non-transfers upon admission. Therefore, we believe the bias in these data will be limited. Secondly, since ICUs were operating at full capacity at times, it cannot be excluded that some patients that would have been admitted pre-COVID-19 are not currently in this dataset. Thirdly, like any EHR dataset, there will be missing data. We believe that transparency is essential to gauge potential limitations in specific research questions. More importantly, we aspire transparency to lead to changes in clinical practice to improve EHR datasets. Comorbidity data, for example, are frequently not structurally stored in EHRs. We included comorbidity data form Dutch national registry data, which may not be available in other countries. We encourage the community to think about minimally required datasets to be recorded and standardization of EHR parameters. This way, the field of medical data science can advance for the benefit of critically ill patients.
Conclusion
To the best of our knowledge, the Dutch Data Warehouse is the first dedicated multicenter and full-admission electronic health record database with highly granular clinical data from critically ill COVID-19 patients. We describe solutions for the legal aspects, ETL pipeline, data mapping, data enrichment, and data validation. Currently, 3463 patients are included in the DDW with over 200 million data points from patient demographics, clinical observations, administered medication, laboratory determinations, and vital sign monitors and life support devices. The resulting data warehouse is available to clinicians and researchers within ethical and legal boundaries. We expect this work will encourage clinicians and researchers to be involved in EHR data sharing collaborations to advance the field of medical data science.
Availability of data and materials
All participating hospitals have access to the data. External researchers can get access to all data in collaboration with any of the participating hospitals. The list of collaborators is available in the co-author list and in the declarations section, through the corresponding author, and through the contact details on amsterdammedicaldatascience.nl. Research questions have to be in line with the DSA; to investigate the course of COVID-19 in the ICU and to research potential treatments. Researchers have sign a code of conduct before accessing the data.
Abbreviations
- APACHE II:
-
Acute Physiology and Chronic Health Evaluation II
- ALAT:
-
Alanine transferase
- API:
-
Application programming interface
- ATC:
-
Anatomical Therapeutic Chemical
- BMI:
-
Body mass index
- DDW:
-
Dutch Data Warehouse
- DPO:
-
Data protection officers
- DSA:
-
Data sharing agreement
- EHR:
-
Electronic health record
- ETL:
-
Extract–transform–load
- GDPR:
-
General Data Protection Regulation
- ICD-10:
-
International Classification of Diseases
- IQR:
-
Interquartile range
- LOINC:
-
Logical Observation Identifiers Names and Codes
- RASS:
-
Richmond Agitation and Sedation Scales
- SNOMED-CT:
-
Systematized Nomenclature of Medicine Clinical Terms
- SOFA:
-
Sequential Organ Failure Assessment
- SQL:
-
Structured Query Language
References
Home [Internet]. Johns Hopkins Coronavirus Resour. Cent. [cited 2021 Jan 19]. https://coronavirus.jhu.edu/.
Gutierrez G. Artificial intelligence in the intensive care unit. Crit Care [Internet]. 2020 [cited 2021 Apr 22];24. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7092485/.
Oderkirk J. Readiness of electronic health record systems to contribute to national health information and research. OECD; 2017 [cited 2021 Apr 22]; https://www.oecd-ilibrary.org/social-issues-migration-health/readiness-of-electronic-health-record-systems-to-contribute-to-national-health-information-and-research_9e296bf3-en;jsessionid=YkvP_QGn-BoJb0Q_PXR-GhZF.ip-10-240-5-152.
Johnson AEW, Pollard TJ, Shen L, Lehman LH, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3:160035.
Thoral PJ, Peppink JM, Driessen RH, Sijbrands EJG, Kompanje EJO, Kaplan L, et al. Sharing ICU Patient Data Responsibly Under the Society of Critical Care Medicine/European Society of Intensive Care Medicine Joint Data Science Collaboration: The Amsterdam University Medical Centers Database (AmsterdamUMCdb) Example. Crit Care Med [Internet]. 2021 [cited 2021 Apr 22];Latest Articles. https://journals.lww.com/ccmjournal/Abstract/9000/Sharing_ICU_Patient_Data_Responsibly_Under_the.95320.aspx.
Hyland SL, Faltys M, Hüser M, Lyu X, Gumbsch T, Esteban C, et al. Early prediction of circulatory failure in the intensive care unit using machine learning. Nat Med. 2020;26:364–73.
Trias-Llimós S, Alustiza A, Prats C, Tobias A, Riffe T. The need for detailed COVID-19 data in Spain. Lancet Public Health. 2020;5:576.
Baker MG, Wilson N. The covid-19 elimination debate needs correct data. BMJ. 2020;371:m3883.
Azoulay E, de Waele J, Ferrer R, Staudinger T, Borkowska M, Povoa P, et al. International variation in the management of severe COVID-19 patients. Crit Care. 2020;24:486.
Qian Z, Alaa AM, van der Schaar M, Ercole A. Between-centre differences for COVID-19 ICU mortality from early data in England. Intensive Care Med. 2020;1–2.
Fleuren LM, de Bruin DP, Tonutti M, Lalisang RCA, Elbers PWG, Gommers D, et al. Large-scale ICU data sharing for global collaboration: the first 1633 critically ill COVID-19 patients in the Dutch Data Warehouse. Intensive Care Med. 2021. https://doi.org/10.1007/s00134-021-06361-x.
Art. 9 GDPR – Processing of special categories of personal data [Internet]. Gen. Data Prot. Regul. GDPR. [cited 2021 Apr 24]. https://gdpr-info.eu/art-9-gdpr/.
Covid-19 op de IC [Internet]. [cited 2021 Apr 24]. https://www.stichting-nice.nl/.
Cornet R, de Keizer N. Forty years of SNOMED: a literature review. BMC Med Inform Decis Mak. 2008;8:S2.
Côté RA, Robboy S. Progress in Medical Information Management: Systematized Nomenclature of Medicine (SNOMED). JAMA. 1980;243:756–62.
Forrey AW, McDonald CJ, DeMoor G, Huff SM, Leavelle D, Leland D, et al. Logical observation identifier names and codes (LOINC) database: a public use set of codes and names for electronic reporting of clinical laboratory test results. Clin Chem. 1996;42:81–90.
Shahpori R, Doig C. Systematized Nomenclature of Medicine-Clinical Terms direction and its implications on critical care. J Crit Care. 2010;25(364):e1-9.
WHOCC - Structure and principles [Internet]. [cited 2021 Apr 25]. https://www.whocc.no/atc/structure_and_principles/.
Amato MBP, Meade MO, Slutsky AS, Brochard L, Costa ELV, Schoenfeld DA, et al. Driving pressure and survival in the acute respiratory distress syndrome. N Engl J Med. 2015;372:747–55.
Sinha P, Calfee CS, Beitler JR, Soni N, Ho K, Matthay MA, et al. Physiologic analysis and clinical performance of the ventilatory ratio in acute respiratory distress syndrome. Am J Respir Crit Care Med. 2019;199:333–41.
Intagliata S, Rizzo A, Gossman WG. Physiology, Lung Dead Space. StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2021 [cited 2021 Apr 25]. http://www.ncbi.nlm.nih.gov/books/NBK482501/.
Gattinoni L, Tonetti T, Cressoni M, Cadringher P, Herrmann P, Moerer O, et al. Ventilator-related causes of lung injury: the mechanical power. Intensive Care Med. 2016;42:1567–75.
Vincent JL, Moreno R, Takala J, Willatts S, De Mendonça A, Bruining H, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996;22:707–10.
Knaus WA, Draper EA, Wagner DP, Zimmerman JE. APACHE II: a severity of disease classification system. Crit Care Med. 1985;13:818–29.
Amsterdam Medical Data Science [Internet]. [cited 2020 Nov 20]. https://www.amsterdammedicaldatascience.nl/.
Quinn TJ, Burton JK, Carter B, Cooper N, Dwan K, Field R, et al. Following the science? Comparison of methodological and reporting quality of covid-19 and other research from the first wave of the pandemic. BMC Med. 2021;19:46.
Jung RG, Di Santo P, Clifford C, Prosperi-Porta G, Skanes S, Hung A, et al. Methodological quality of COVID-19 clinical research. Nat Commun. 2021;12:943.
Acknowledgements
The Dutch ICU Data Sharing Against COVID-19 Collaborators: From collaborating hospitals having shared data: Julia Koeter, MD, Intensive Care, Canisius Wilhelmina Ziekenhuis, Nijmegen, The Netherlands. Roger van Rietschote, Business Intelligence, Haaglanden MC, Den Haag,The Netherlands. M.C. Reuland, MD, Department of Intensive Care Medicine, Amsterdam UMC, Universiteit van Amsterdam, Amsterdam, The Netherlands. Laura van Manen, MD, Department of Intensive Care, BovenIJ Ziekenhuis, Amsterdam, The Netherlands. Leon Montenij, MD, PhD, Department of Anesthesiology, Pain Management and Intensive Care, Catharina Ziekenhuis Eindhoven, Eindhoven, The Netherlands. Jasper van Bommel, MD, PhD, Department of Intensive Care, Erasmus Medical Center, Rotterdam, The Netherlands. Roy van den Berg, Department of Intensive Care, ETZ Tilburg, Tilburg, The Netherlands. Ellen van Geest, Department of ICMT, Haga Ziekenhuis, Den Haag, The Netherlands. Anisa Hana, MD, PhD, Intensive Care, Laurentius Ziekenhuis, Roermond, The Netherlands. B. van den Bogaard, MD, PhD, ICU, OLVG, Amsterdam, The Netherlands. Prof. Peter Pickkers, Department of Intensive Care Medicine, Radboud University Medical Centre, Nijmegen, The Netherlands. Pim van der Heiden, MD, PhD, Intensive Care, Reinier de Graaf Gasthuis, Delft, The Netherlands. Claudia (C.W.) van Gemeren, MD, Intensive Care, Spaarne Gasthuis, Haarlem en Hoofddorp, The Netherlands. Arend Jan Meinders, MD, Department of Internal Medicine and Intensive Care, St Antonius Hospital, Nieuwegein, The Netherlands. Martha de Bruin, MD, Department of Intensive Care, Franciscus Gasthuis & Vlietland, Rotterdam, The Netherlands. Emma Rademaker, MD, MSc, Department of Intensive Care, UMC Utrecht, Utrecht, The Netherlands. Frits H.M. van Osch, PhD, Department of Clinical Epidemiology, VieCuri Medisch Centrum, Venlo, The Netherlands. Martijn de Kruif, MD, PhD, Department of Pulmonology, Zuyderland MC, Heerlen, The Netherlands. Nicolas Schroten, MD, Intensive Care, Albert Schweitzerziekenhuis, Dordrecht, The Netherlands. Klaas Sierk Arnold, MD, Anesthesiology, Antonius Ziekenhuis Sneek, Sneek, The Netherlands. J.W. Fijen, MD, PhD, Department of Intensive Care, Diakonessenhuis Hospital, Utrecht, The Netherland. Jacomar J.M. van Koesveld, MD, ICU, IJsselland Ziekenhuis, Capelle aan den IJssel, The Netherlands. Koen S. Simons, MD, PhD, Department of Intensive Care, Jeroen Bosch Ziekenhuis, Den Bosch, The Netherlands. Joost Labout, MD, PhD, ICU, Maasstad Ziekenhuis Rotterdam, The Netherlands. Bart van de Gaauw, MD, Martiniziekenhuis, Groningen, The Netherlands. Michael Kuiper, Intensive Care, Medisch Centrum Leeuwarden, Leeuwarden, The Netherlands. Albertus Beishuizen, MD, PhD, Department of Intensive Care, Medisch Spectrum Twente, Enschede, The Netherlands. Dennis Geutjes, Department of Information Technology, Slingeland Ziekenhuis, Doetinchem, The Netherlands. Johan Lutisan, MD, ICU, WZA, Assen, The Netherlands. Bart P. Grady, MD, PhD, Department of Intensive Care, Ziekenhuisgroep Twente, Almelo, The Netherlands. Remko van den Akker, Intensive Care, Adrz, Goes, The Netherlands. Tom A. Rijpstra, MD, Department of Intensive Care, Amphia Ziekenhuis, Breda, The Netherlands. Suat Simsek, MD PhD, Department of Internal Medicine/Endocrinology, Northwest Clinics, Alkmaar, the Netherlands. From collaborating hospitals having signed the data sharing agreement: Daniel Pretorius, MD, Department of Intensive Care Medicine, Hospital St Jansdal, Harderwijk, The Netherlands. Menno Beukema, MD, Department of Intensive Care, Streekziekenhuis Koningin Beatrix, Winterswijk, The Netherlands. Bram Simons, MD, Intensive Care, Bravis Ziekenhuis, Bergen op Zoom en Roosendaal, The Netherlands. A.A. Rijkeboer, MD, ICU, Flevoziekenhuis, Almere, The Netherlands. Marcel Aries, MD, PhD, MUMC+, University Maastricht, Maastricht, The Netherlands. Niels C. Gritters van den Oever, MD, Intensive Care, Treant Zorggroep, Emmen, The Netherlands. Martijn van Tellingen, MD, EDIC, Department of Intensive Care Medicine, afdeling Intensive Care, ziekenhuis Tjongerschans, Heerenveen, The Netherlands. Annemieke Dijkstra, MD, Department of Intensive Care Medicine, Het Van Weel-Bethesda Ziekenhuis, Dirksland, The Netherlands. Rutger van Raalte, Department of Intensive Care, Tergooi hospital, Hilversum, The Netherlands. From the Laboratory for Critical Care Computational Intelligence: Martin E. Haan, MD, Department of Intensive Care Medicine, Laboratory for Critical Care Computational Intelligence, Amsterdam Medical Data Science, Amsterdam UMC, Vrije Universiteit, Amsterdam, The Netherlands. Luca Roggeveen, MD, Department of Intensive Care Medicine, Laboratory for Critical Care Computational Intelligence, Amsterdam Medical Data Science, Amsterdam UMC, Vrije Universiteit, Amsterdam, The Netherlands. Fuda van Diggelen, MSc, Quantitative Data Analytics Group, Department of Computer Sciences, Faculty of Science, VU University, Amsterdam, The Netherlands. Ali el Hassouni, PhD, Quantitative Data Analytics Group, Department of Computer Sciences, Faculty of Science, VU University, Amsterdam, The Netherlands. David Romero Guzman, PhD, Quantitative Data Analytics Group, Department of Computer Sciences, Faculty of Science, VU University, Amsterdam, The Netherlands. Sandjai Bhulai, PhD, Analytics and Optimization Group, Department of Mathematics, Faculty of Science, Vrije Universiteit, Amsterdam, The Netherlands. Dagmar M. Ouweneel, PhD, Department of Intensive Care Medicine, Laboratory for Critical Care Computational Intelligence, Amsterdam Medical Data Science, Amsterdam UMC, Vrije Universiteit, Amsterdam, The Netherlands. Ronald Driessen, Department of Intensive Care Medicine, Laboratory for Critical Care Computational Intelligence, Amsterdam Medical Data Science, Amsterdam UMC, Vrije Universiteit, Amsterdam, The Netherlands. Jan Peppink, Department of Intensive Care Medicine, Laboratory for Critical Care Computational Intelligence, Amsterdam Medical Data Science, Amsterdam UMC, Vrije Universiteit, Amsterdam, The Netherlands. H.J. de Grooth, MD, PhD, Department of Intensive Care Medicine, Laboratory for Critical Care Computational Intelligence, Amsterdam Medical Data Science, Amsterdam UMC, Vrije Universiteit, Amsterdam, The Netherlands. G.J. Zijlstra, MD, PhD, Department of Intensive Care Medicine, Laboratory for Critical Care Computational Intelligence, Amsterdam Medical Data Science, Amsterdam UMC, Vrije Universiteit, Amsterdam, The Netherlands. A.J. van Tienhoven, MD, Department of Intensive Care Medicine, Laboratory for Critical Care Computational Intelligence, Amsterdam Medical Data Science, Amsterdam UMC, Vrije Universiteit, Amsterdam, The Netherlands. Evelien van der Heiden, MD, Department of Intensive Care Medicine, Amsterdam Medical Data Science, Amsterdam UMC, Vrije Universiteit, Amsterdam, The Netherlands. Jan Jaap Spijkstra, MD, PhD, Department of Intensive Care Medicine, Amsterdam Medical Data Science, Amsterdam UMC, Vrije Universiteit, Amsterdam, The Netherlands. Hans van der Spoel, MD, Department of Intensive Care Medicine, Amsterdam Medical Data Science, Amsterdam UMC, Vrije Universiteit, Amsterdam, The Netherlands. Angelique de Man, MD, PhD, Department of Intensive Care Medicine, Amsterdam Medical Data Science, Amsterdam UMC, Vrije Universiteit, Amsterdam, The Netherlands. Thomas Klausch, PhD, Department of Clinical Epidemiology, Laboratory for Critical Care Computational Intelligence, Amsterdam Medical Data Science, Amsterdam UMC, Vrije Universiteit, Amsterdam, The Netherlands. Heder J. de Vries, MD, Department of Intensive Care Medicine, Laboratory for Critical Care Computational Intelligence, Amsterdam Medical Data Science, Amsterdam UMC, Vrije Universiteit, Amsterdam, The Netherlands. From Pacmed: Adam Izdebski, Pacmed, Amsterdam, The Netherlands. Michael de Neree tot Babberich, MD, PhD, Pacmed, Amsterdam, The Netherlands. Olivier Thijssens, MSc, Pacmed, Amsterdam, The Netherlands. Lot Wagemakers, Pacmed, Amsterdam, The Netherlands. Hilde G.A. van der Pol, Pacmed, Amsterdam, The Netherlands. Tom Hendriks, Pacmed, Amsterdam, The Netherlands. Julie Berend, Pacmed, Amsterdam, The Netherlands. Virginia Ceni Silva, Pacmed, Amsterdam, The Netherlands. Robert F.J. Kullberg, MD, Pacmed, Amsterdam, The Netherlands. From RCCnet: Leo Heunks, MD, PhD, Department of Intensive Care Medicine, Amsterdam Medical Data Science, Amsterdam UMC, Vrije Universiteit, Amsterdam, The Netherlands. Nicole Juffermans, MD, PhD, ICU, OLVG, Amsterdam, The Netherlands. Arjen J.C. Slooter, MD, PhD, Department of Intensive Care Medicine, UMC Utrecht, Utrecht University, Utrecht, the Netherlands.
Funding
Partially funded by grants from ZonMw (Project 10430012010003, file 50-55700-98-908), Zorgverzekeraars Nederland and the Corona Research Fund. The sponsors had no role in any part of the study.
Author information
Authors and Affiliations
Contributions
LF drafted the manuscript. TD, DB, RL, MF, TM, MS, SV, AB, DQ, RN, TH, PT, WH and PE were involved in data processing and analytics. All authors contributed to data collection and critically reviewed the manuscript. All authors have full access to the data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The Medical Ethics Committee at Amsterdam UMC, location VUmc waived the need for patient informed consent and approved of an opt-out procedure for the collection of COVID-19 patient data during the COVID-19 crisis.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Data sharing agreement.
Additional file 2.
Institutional review board documentation.
Additional file 3: Table S1.
Overview of derived clinical score.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Fleuren, L.M., Dam, T.A., Tonutti, M. et al. The Dutch Data Warehouse, a multicenter and full-admission electronic health records database for critically ill COVID-19 patients. Crit Care 25, 304 (2021). https://doi.org/10.1186/s13054-021-03733-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13054-021-03733-z