Abstract
Data derived from the electronic health record (EHR) is frequently extracted using undefined approaches that may affect the accuracy of collected variables. Further, efforts to assess data accuracy often suffer from limited collaboration between clinicians and data analysts who perform the extraction. In this manuscript, we describe the methodology behind creation of a structured, rigorously derived intensive care unit (ICU) data mart based on data automatically and routinely derived from the EHR. This ICU data mart includes high-quality data elements commonly used for quality improvement and research purposes. These data elements were identified by physicians working closely with data analysts to iteratively develop and refine algorithmic definitions for complex outcomes and risk factors. We contend that this methodology can be reproduced and applied across other institution or to other clinical domains to create high quality data marts, inclusive of complex outcomes data.
Similar content being viewed by others
Background
The electronic health record (EHR) contains a vast quantity of data due to its observation nature, holding great promise as a valuable, efficient, and cost-effective tool. These data can inform quality improvement and research initiatives, especially those related to medical resources and patient outcomes [1,2,3]. In its initial implementation, however, the EHR rarely captures outcomes of interest to key stakeholders reliably and accurately due to frequent limitations resulting from disorganized, incorrect, or missing variables that lack vigorous extraction methodologies. Together this limits the data’s validity and utility [4]. In order to provide validated results for scientific interpretation, vigorous, reproducible, and validated techniques must be established for each EHR variable of interest.
Many institutions rely on a structured repository of data, drawn from the EHR, to facilitate ongoing access, a so-called data warehouse or data mart [5, 6]. This data repository is frequently created after the EHR has been created and, at many institutions, is created and maintained by data analysts working in isolation from front-line clinicians. The intensive care unit (ICU) is a particularly challenging area for creation of a data mart. Critically ill patients suffer life threatening organ pathology in at least one, if not many, organ systems. These patients are generally intensively monitored, with very high frequency of physiologic data capture. Laboratory data may be obtained multiple times per day. Multiple organ support modalities may be employed, with complex documentation and monitoring to quantify the degree of support. Once data are located though, they can support surveillance, decision support, and modeling of outcomes [7].
We describe a methodology for the creation of a structured, rigorously constructed intensive care unit (ICU) data mart based on data automatically and routinely derived from the EHR. This was performed through identification of data elements commonly collected as part of routine clinical care that also hold value for quality improvement and research purposes. We then present a methodology for collecting, structuring, and assessing accuracy of data elements through sequential collection cycles. Importantly, this work was completed with data analysts and clinicians working side-by-side throughout the process to ensure data and clinical accuracy.
Methods
Identification
A multidisciplinary project team including clinical intensivists and data analysts met daily (virtually or in person) throughout construction, assembling a priority list of physiologic, laboratory, demographic, and billing data. The presence of data availability in routine clinical practice was confirmed using extensive chart review and identification. Data analysts worked to identify the location of variables within the data architecture underlying the EHR (Epic, Verona, WI). All elements were investigated for extent of chart documentation, frequency, duplication in different sites within the medical record, and agreement with patient clinical course. As is common within our EHR, data have the ability to be entered under variations of variable names housed on different database tables. Methodologic screening across database tables using clinician feedback was performed to ensure capture of variables across electronic sources. Variable location was confirmed after location(s) of these key variables within the EHR were vetted across data analysts and clinician sources. Key team members involved in identification of variables include anesthesiology and internal medicine faculty and trainees, business intelligence analysts, and database administrators who together function as a team and are all involved in daily core meetings.
Extraction
We created algorithmic definitions for complex data elements, including most outcomes, leveraging existing literature, when available. Test patients were extracted and algorithms iteratively refined at least weekly based on results of below preliminary validation methods. The number of iterations required for each variable was variable dependent, influenced by fidelity of the variable within the EHR. The following preliminary data extraction methodology was used to assess data quality for each variable throughout subsequent extractions: range check (is the data collection within a physiologic range?), type/format check (is the data presented in the format expected for that value?), check digit/length check (has the data been entered or saved correctly within the EHR?), and finally lookup (within a random sample of patients, are data obtained an accurate reflection of the patients’ clinical course?).
Transformation and loading
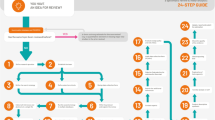
Once shown to be reproducible and appropriately valid based on above methodologies in a broad cohort of patients, structured query language (SQL) was used to extract, transform, and load data from the EHR for use into a relational database housed on a departmental server. This same departmental server houses intraoperative variables within a perioperative data warehouse (PDW) that were obtained through similar methodologies [8].The accuracy of loaded databases were again formally assessed using lookup strategies on large representative cohorts of ICU patients. For those variables with insufficient accuracy, investigators either returned to identification or extraction steps to improve data quality, or if no improvement strategies could be identified, removed data element from final inclusion within the data mart. After formal lookup assessments, patient outcome results were cataloged in appropriate outcomes tables for presentation and dissemination. This process is illustrated in Fig. 1. Total timeline for identification, extraction, and loading was approximately 3–6 months.
Progression Through Variable Identification. Figure 1 Variables presented have been identified by clinicians as high-yield variables for quality improvement and research purposes. As these variables are located and validated within the electronic health record, they are added within the data mart. These data are then analyzable to be able to draw conclusive findings regarding outcomes including acute kidney injury, reintubation rates, and 7-day mortality, to name a few. Directionality is dependent on quality of variable and accuracy across study stages, represented by the double-sided arrows
Results
A total of 459,465 ICU patient encounters were identified and included in the ICU data mart, accessible and maintained through the Division of Informatics within the Anesthesiology department. These patients include over 460,000,000 individual laboratory results and 4,610,776 vital signs (with 1-min fidelity in the first 24-h of admission). Using the above methodologies, a total of 26 outcomes were compiled (Table 1). These data have been structured within 19 tables, all of which have a sensitivity and specificity of greater than 95%. The iterative construction of our processes allows for continual structured updates and assessment of variables to maintain accuracy. When variables were not able to reach sufficient accuracy despite iterations, data analysts formed collaborative meetings to review, check, and improve techniques. If it was not possible to increase accuracy, variables were not advanced or included within final tables or projects due to lack of cogency.
Variables can be accessed or combined on request by data analysts. Reports are generated using data dictionaries to identify relevant variables dependent on the research or quality improvement project aims. Following institutional ethics approval, relevant data can be extracted and accessed in the desired format for clinicians or researchers.
Confirmed data are used to interpret patient outcomes for all patients within the ICU and ICU data mart. Additionally, these data can be joined to the 120 tables including more than 1900 unique variable columns within the existing perioperative data warehouse. While this division resides under the Department of Anesthesiology, it included data from ICU patients under Medicine and Surgery departments as well and is accessible through request. The division is responsible for support of administrators, clinicians, and researchers aiming to utilize ICU data for quality improvement or research endeavors following appropriate institutional approval.
Discussion
We present a methodology for building a robust and highly granular ICU data mart, leveraging the synergistic expertise of clinicians and data analysts. Optimizing the quality of data obtained from large databases will improve accuracy, results, and confidence within informatics research for quality improvement and research purposes.
These processes can be adapted to new variables as they present to provide real-time clinical data on large populations of patients within our ever-changing clinical environment. Several hospital systems involved in data informatics research have already established similar organizational methodologies to ensure quality of data obtained within their data warehouses [6, 9, 10]. As is often the case, it is accepted that data is available through the EHR, but the difficulty comes from the disconnects prevalent with extracting and utilizing that data [11]. The methodology presented provides a structure under which these data can be collected, incorporating key stakeholders requests and expertise, to draw results across institutional interfaces and patient locations. Together, these structured and validated methodologies strengthen the results obtained and the validity and trust within our research community. Even after establishing these methodologies, there is a need for consistent upkeep and maintenance of systems. Continual data maintenance and validation is not included within these methods but are equally essential to ensuring continuation of useable data collection. The major value for this established methodology lies in the additional variables and patient markers that are added to the EHR and identified as priority for inclusion within the data mart. These same processes are adapted to ensure quality data collection and trust of information obtained. Within workgroups, collection has been underway to extract variables that will identify positive coronavirus 2019 (COVID-19) test results into our variable lists using the presented methodology to confirm accuracy within results obtained across a variety of available laboratory data, as an example of the evolving needs addressed through adaptation of this same methodological structure.
Similar to much of informatics research, our results are limited by the quality of data entered into the EHR. Missingness and inaccurate data elements can be screened and eliminated when detected, but such errors are difficult to prevent entirely. The ability of our algorithms and methods to identify accurate data with high fidelity on repeated queries is evidence of the rigor of our data extraction method. We recognize, however, this number is not validated and level of inaccuracies cannot be eliminated. As our systems change and update within our underlying EHR architecture, aspects of our data extraction may need to be updated as well to ensure continued legitimacy.
Our methodology and accuracy provides a strong foundation for the results obtained through our large ICU data mart. As we plan to add patient data throughout the hospitalization and perioperative periods, we will continue to establish structured methodologies to ensure data accuracy. Future uses of this work will aim to rigorously validate our results and those variables within our institution and across multiple health care centers to create multicenter perioperative data warehouses with rigorously validated patient variables for quality improvement and research purposes.
References
Brundin-Mather R, Soo A, Zuege DJ, Niven DJ, Fiest K, Doig CJ, et al. (2018) Secondary EMR data for quality improvement and research: A comparison of manual and electronic data collection from an integrated critical care electronic medical record system. J Crit Care. 47:295-301.
Chen LM, Kennedy EH, Sales A, Hofer TP. (2013) Use of health IT for higher-value critical care. N Engl J Med. 368:594-597.
King J, Patel V, Jamoom EW, Furukawa MF. (2014) Clinical benefits of electronic health record use: national findings. Health Serv Res. 49:392-404.
Docherty AB, Lone NI. (2015) Exploiting big data for critical care research. Curr Opin Crit Care. 21:467-472.
Kimball R, Ross M, Thorthwaite W, Becker B, Mundy J. The data warehouse lifecycle toolkit: John Wiley & Sons; 2008.
de Mul M, Alons P, van der Velde P, Konings I, Bakker J, Hazelzet J. (2012) Development of a clinical data warehouse from an intensive care clinical information system. Comput Methods Programs Biomed. 105:22-30.
Herasevich V, Pickering BW, Dong Y, Peters SG, Gajic O. (2010) Informatics infrastructure for syndrome surveillance, decision support, reporting, and modeling of critical illness. Mayo Clin Proc. 85:247-254.
Hofer IS, Gabel E, Pfeffer M, Mahbouba M, Mahajan A. (2016) A systematic approach to creation of a perioperative data warehouse. Anesth Analg. 122:1880-4.
Dewitt JG, Hampton PM. (2005) Development of a data warehouse at an academic health system: knowing a place for the first time. Acad Med. 80:1019-1025.
Weir CR, Hicken BL, Rappaport HS, Nebeker JR. (2006) Crossing the quality chasm: the role of information technology departments. Am J Med Qual. 21:382-393.
Freundlich RE, Lindsell CJ. (2022) We know what we want, it's just not there. J Clin Transl Sci. 6(1):e9.
Funding
Dr. Boncyk receives support from the National Institute on Aging (NIA) Administrative Supplement (R01AG053582), the Foundation for Anesthesia Education and Research (FAER), and the Society of Academic Associations of Anesthesiology & Perioperative Medicine (SAAAPM). Dr. Freundlich is supported by two grants from the National Institutes of Health (NIH): National Heart, Lung, and Blood Institute (NHLBI) grant K23HL148640 and National Center for Advancing Translational Sciences (NCATS) grant UL1TR002243. There was no direct funding towards this project.
Author information
Authors and Affiliations
Contributions
CB contributed to project design, data interpretation, and construction of manuscript. PB contributed to data collection and construction of manuscript. KM contributed to project design, data collection, data interpretation, and construction of manuscript. RF contributed to project design, data collection, data interpretation, and construction of manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This project received local Institutional Review Board (IRB) approval (#220897).
Informed consent
A waiver of informed consent was obtained with IRB approval.
Conflict of interest statement
Dr. Boncyk is a consultant for Sedana Medical. This manuscript does not reference any activities related to this consultancy and Dr. Boncyk declares no conflict of interest. Dr. Freundlich declares no conflict of interest. Pamela Butler declares no conflict of interest. Karen McCarthy declares no conflict of interest.
Clinical trial number
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Boncyk, C., Butler, P., McCarthy, K. et al. Validation of an Intensive Care Unit Data Mart for Research and Quality Improvement. J Med Syst 46, 81 (2022). https://doi.org/10.1007/s10916-022-01873-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-022-01873-5