Abstract
Introduction
Increasing evidence supports a role for mitochondrial dysfunction in organ injury and immune dysregulation in sepsis. Although differential expression of mitochondrial genes in blood cells has been reported for several diseases in which bioenergetic failure is a postulated mechanism, there are no data about the blood cell mitochondrial transcriptome in pediatric sepsis.
Methods
We conducted a focused analysis using a multicenter genome-wide expression database of 180 children ≤10 years of age with septic shock and 53 healthy controls. Using total RNA isolated from whole blood within 24 hours of PICU admission for septic shock, we evaluated 296 nuclear-encoded mitochondrial genes using a false discovery rate of 1%. A series of bioinformatic approaches were applied to compare differentially expressed genes across previously validated gene expression-based subclasses (groups A, B, and C) of pediatric septic shock.
Results
In total, 118 genes were differentially regulated in subjects with septic shock compared to healthy controls, including 48 genes that were upregulated and 70 that were downregulated. The top scoring canonical pathway was oxidative phosphorylation, with general downregulation of the 51 genes corresponding to the electron transport system (ETS). The top two gene networks were composed primarily of mitochondrial ribosomal proteins highly connected to ETS complex I, and genes encoding for ETS complexes I, II, and IV that were highly connected to the peroxisome proliferator activated receptor (PPAR) family. There were 162 mitochondrial genes differentially regulated between groups A, B, and C. Group A, which had the highest maximum number of organ failures and mortality, exhibited a greater downregulation of mitochondrial genes compared to groups B and C.
Conclusions
Based on a focused analysis of a pediatric septic shock transcriptomic database, nuclear-encoded mitochondrial genes were differentially regulated early in pediatric septic shock compared to healthy controls, as well as across genotypic and phenotypic distinct pediatric septic shock subclasses. The nuclear genome may be an important mechanism contributing to alterations in mitochondrial bioenergetic function and outcomes in pediatric sepsis.
Similar content being viewed by others
Introduction
Septic shock is a leading cause of morbidity and mortality in the pediatric intensive care unit (PICU) [1],[2]. With improved therapies to reverse shock, progressive multi-organ failure and secondary infection from acquired immunoparalysis are now the main antecedents to sepsis-associated death [3],[4]. Increasing evidence supports a role for mitochondrial bioenergetic dysfunction in the pathobiology of organ injury and immune dysregulation in sepsis [5]-[7].
Circulating blood cells from critically ill patients with septic shock exhibit decreased oxidative respiration, electron chain complex activity, mitochondrial turnover, and mitochondrial membrane potential [8]-[12]. Blood is an easily accessible tissue that can be used to directly measure mitochondrial dysfunction in immune cells and may reflect a systemic process affecting other vital organs. Mitochondrial dysfunction in blood cells has been associated with severity of illness, organ dysfunction, mortality, and immunoparalysis in human sepsis [8]-[12], including children [13]. Differential expression of mitochondrial genes in blood cells has been reported for several diseases in which bioenergetic failure is a postulated mechanism [14]-[16], and injection of endotoxin has been shown to cause widespread suppression of genes encoding for mitochondrial ATP production and protein synthesis within human leukocytes [17]. However, there are no data about the blood cell mitochondrial transcriptome in pediatric sepsis. Identification of mitochondrial genomic changes within blood cells could provide clinically relevant biomarkers, offer insight into biological mechanisms, and inform therapeutic targets related to mitochondrial bioenergetic dysfunction for children with sepsis.
While mitochondria contain their own circular genome, the majority of the mitochondrial proteins comprising subunits of the electron transport system (ETS) are encoded by nuclear genes, including 38/45 for complex I, 4/4 for complex II, 10/11 for complex III, 10/13 for complex IV, and 17/19 for complex V (ATP synthase). In addition, all of the 79 known mitochondrial ribosomal proteins (MRPs) are encoded by the nuclear genome [18],[19]. These ETS and ribosomal proteins are synthesized within the cytoplasm and then imported into the mitochondria.
Over the last decade, we have generated an extensive genome-wide expression database of children with septic shock drawn from multiple centers in the U.S. [20]. The database has enabled the discovery of gene expression-based subclasses of pediatric septic shock [21]-[23], stratification biomarkers [24]-[31], diagnostic biomarkers [32]-[35], and novel therapeutic targets [36]-[42]. Here, we mined the database to test the hypothesis that expression of whole blood-derived nuclear-encoded mitochondrial genes will be differentially regulated between pediatric patients with septic shock and nonseptic controls within the first 24 hours of presentation to the PICU. We further hypothesized that nuclear-encoded mitochondrial genes would be differentially regulated across genotypic and phenotypic distinct subclasses of pediatric septic shock. We tested these hypotheses using a focused analytical approach in which we restricted the working gene list to 296 nuclear-encoded mitochondrial genes, as previously reported by Lunnon et al. [14].
Methods
Patients and data collection
The study protocol was approved by the Institutional Review Boards of each participating institution: Cincinnati Children's Hospital Medical Center, The Children's Hospital of Philadelphia, University of California Benioff Children's Hospital Oakland, Penn State Hershey Children's Hospital, Children's Mercy Hospital, Children's Hospital of Orange County, Akron Children's Hospital, Children's National Medical Center, Miami Children's Hospital, Texas Children's Hospital, and CS Mott Children's Hospital at the University of Michigan. Children ≤10 years of age admitted to the PICU who met pediatric-specific criteria for septic shock were eligible for enrollment [43]. Age-matched controls were recruited from the ambulatory departments of participating institutions using published inclusion and exclusion criteria [36]. All subjects and data collection methods have been previously reported in microarray-based studies addressing hypotheses entirely different from that of the current study and details of the study protocol were previously published [21]-[23],[32],[33],[36],[37],[44]-[48]. All microarray data have been deposited in the National Center for Biotechnology (NCBI) Gene Expression Omnibus (Accession numbers: GSE26440 and GSE26378).
RNA extraction and microarray hybridization
Written informed consent was obtained from the parents or legal guardians of all septic shock and control subjects to participate in this study. Blood samples were obtained within the first 24 hours of meeting criteria for septic shock. Total RNA was isolated from whole blood using the PaxGene™ Blood RNA System (PreAnalytiX, Qiagen/Becton Dickson, Valencia, CA, USA). Microarray hybridization was performed as previously described using the Human Genome U133 Plus 2.0 GeneChip (Affymetrix, Santa Clara, CA, USA) [36].
Data analysis
We analyzed existing, normalized microarray data. The original analyses were performed using one patient sample per chip. Image files were captured using an Affymetrix GeneChip Scanner 3000. Raw data files (.CEL) were subsequently preprocessed using robust multiple-array average (RMA) normalization and GeneSpring GX 7.3 software (Agilent Technologies, Palo Alto, CA, USA). All signal intensity-based data were used after RMA normalization, which specifically suppresses all but significant variation among lower intensity probe sets [49]. All chips representing septic shock samples were then normalized to the respective median values of controls on a per gene basis.
Differences in mRNA abundance between the study groups were determined using analysis of variance (ANOVA) and corrections for multiple comparisons using a Benjamini-Hochberg false discovery rate (FDR) of 1%. We did not include predetermined fold expression filters in the analysis because the biological implications of a specific threshold change in gene expression are not clear. Thus, in order to account for the possibility that even a modest change in expression within a set of genes from a common metabolic pathway could yield dramatic variability in flux through that pathway [50], we considered all statistical differences in gene expression using the 1% FDR to be significant in this analysis. For clarity, further details regarding microarray data analysis and gene list generation will be provided in the Results section.
Gene lists of differentially regulated genes were analyzed using the Ingenuity Pathways Analysis (IPA) application (Ingenuity Systems, Redwood City, CA, USA) to explore potential associations with specific domains of mitochondrial function [44],[47],[48]. IPA is a database generated from peer-reviewed scientific publications that provides a tool for discovery of signaling pathways and gene networks within the uploaded gene lists. Adjunct analyses of gene lists were conducted using the ToppGene application [51].
Gene expression mosaics representing the expression patterns of differentially regulated genes were generated using the Gene Expression Dynamics Inspector (GEDI) [22],[23],[52],[53]. The signature graphical outputs of GEDI are expression mosaics that give microarray data a `face' that is intuitively recognizable via human pattern recognition. The algorithm for creating the mosaics is a self-organizing map that enables use of human pattern recognition to perform a global analysis of complex genomic data [54].
Ordinal and continuous clinical variables not normally distributed were analyzed via ANOVA on Ranks. Dichotomous clinical variables were analyzed using a chi-square test (SigmaStat Software, Systat Software, Inc., San Jose, CA, USA).
Results
Differential regulation of nuclear-encoded mitochondrial genes in patients with septic shock versus healthy controls
One hundred and eighty pediatric subjects with septic shock and 53 healthy pediatric controls were available for analysis. Table 1 provides the demographic characteristics of the two study groups.
To determine which mitochondrial genes were differentially regulated between patients with septic shock and controls, we conducted an ANOVA starting with 296 nuclear-encoded mitochondrial genes. One hundred and eighteen of the 296 nuclear-encoded mitochondrial genes (40%) were differentially regulated between the two study groups, including 48 upregulated and 70 downregulated in subjects with septic shock. The list of all 118 genes is provided in Table S1 in Additional file 1.
Since limiting the starting gene list for this analysis to the 296 nuclear-encoded mitochondrial genes is a potential source of bias, we conducted an identical analysis starting with all available genes on the array (54,675). This analysis yielded 18,429 differentially regulated genes between septic shock and controls, indicating that the expected rate of differential gene expression is 34% when all genes on the array are considered. Thus, the observed rate of 40% differentially regulated genes when considering only the 296 nuclear-encoded mitochondrial genes is greater than the expected rate of 34% (P = 0.025, chi-square). This suggests that our results were not simply due to bias introduced by restricting the analysis to the mitochondrial genes.
To visualize the biological function of the 118 differentially regulated genes, we uploaded the gene list to the IPA platform and focused the data output on canonical pathways and gene networks. The top scoring canonical pathway was `oxidative phosphorylation'. Figure 1 illustrates the differential regulation of the 51 genes corresponding to the oxidative phosphorylation pathway, which were generally downregulated (that is the degree of green intensity relative to red intensity) in the subjects with septic shock relative to controls (see Table S2 in Additional file 2 for complete gene list).
Differential expression of genes corresponding to the oxidative phosphorylation pathway. Differential expression of individual nuclear-encoded genes corresponding to mitochondrial electron transport system complexes I to V based on false discovery rate of 1%. Red intensity correlates with increased gene expression and green intensity correlates with decreased gene expression. The 51 genes corresponding to the oxidative phosphorylation pathway were generally downregulated (that is greater degree of green relative to red intensity) in the subjects with septic shock relative to controls.
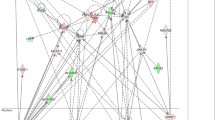
The top two gene networks from the IPA analysis are shown in Figures 2 and 3, with gene nodes being colored based on the degree of increased (red) or decreased (green) expression in subjects with septic shock relative to controls. The gene network shown in Figure 2 is composed primarily of MRPs highly connected to the mitochondrial ETS complex I (nicotinamide adenine dinucleotide (NADH) dehydrogenase), and most of these genes are downregulated in septic shock relative to controls (see Table S3 in Additional file 3 for complete gene list). Consistent with this finding, cross-referencing the network genes to the ToppGene platform returned `NADH dehydrogenase activity' as the top molecular function. The gene network shown in Figure 3 is composed of genes encoding for subunits of ETS complex I, II, and IV that are highly connected to gene nodes corresponding to the peroxisome proliferator-activated receptor (PPAR) family (see Table S4 in Additional file 4 for complete gene list). Cross-referencing the network genes to the ToppGene platform returned the nonspecific function of `oxidoreductase activity' as the top molecular function.
Differentially regulated genes corresponding to a gene network composed of mitochondrial ribosomal proteins highly connected to the mitochondrial electron transport system (ETS) complex I (nicotinamide adenine dinucleotide (NADH) dehydrogenase). The degree of green intensity in a gene node corresponds to decreased expression and the degree of red intensity in a given gene node corresponds to increased expression in the subjects with septic shock, relative to controls, respectively. The list of network genes is provided in Table S3 in Additional file 3.
Differentially regulated genes corresponding to a gene network having peroxisome proliferator-activated receptor (PPAR)-related genes as highly connected nodes. The degree of green intensity in a gene node corresponds to decreased expression and the degree of red intensity in a given gene node corresponds to increased expression in the subjects with septic shock, relative to controls, respectively. The list of network genes is provided in Table S4 in Additional file 4.
Differential expression of nuclear-encoded mitochondrial genes across gene expression-based subclasses of pediatric septic shock
We next determined if the 296 nuclear-encoded mitochondrial genes were differentially regulated (ANOVA; 1% FDR) between septic shock survivors (n = 151) and nonsurvivors (n = 29). Two of the 296 nuclear-encoded mitochondrial genes (cytochrome c oxidase subunit VIIb and NADH dehydrogenase flavoprotein 2, 24 kiloDaltons (kDa)) were differentially regulated between survivors and nonsurvivors. Given the relatively few patients that did not survive, we conducted an alternative analysis in which we compared patients with a `complicated course' (n = 52; defined as patients who either died by 28 days or had persistence of two or more organ failures on day 7 of septic shock) to patients without a complicated course (n = 128) [29],[31],[55]. Three of the 296 genes (cytochrome c oxidase subunit VIIb; NADH dehydrogenase flavoprotein 2, 24 kDa; and NADH dehydrogenase 1β subcomplex, 6, 17 kDA) were differentially regulated between patients with and without a complicated course.
We previously reported and validated three gene expression-based subclasses (groups A, B, and C) of pediatric septic shock having clinically significant phenotypic differences [21]-[23]. The subclass-defining genes correspond to adaptive immunity, glucocorticoid receptor signaling, and PPARβ signaling. Because the gene network shown in Figure 3 contained highly connected gene nodes corresponding to the PPAR family, we next determined if the 296 nuclear-encoded mitochondrial genes were differentially regulated across groups A, B, and C.
The clinical and demographic data for the patients in septic shock groups A (n = 54), B (n = 73), C (n = 53) are shown in Table 2. Patients in group A had a higher mortality rate, pediatric risk of mortality (PRISM) score, pediatric sepsis biomarker risk model (PERSEVERE)-based mortality risk, and maximum number of organ failures compared to patients in groups B and C. Among the patients with available serum lactate data, group A patients had higher median lactate concentrations at study entry compared to groups B and C. There were also some intergroup differences with respect to age, white blood cell counts, and exposure to corticosteroids.
Using a three-group ANOVA with a false discovery rate of 1%, 162 of the 296 (55%) nuclear-encoded mitochondrial genes were differentially regulated between groups A, B, and C (see Table S5 in Additional file 5 for complete gene list). As a control for this analysis, we generated a random list of 296 genes and conducted an identical three-group ANOVA to determine how many of the randomly selected genes were differentially regulated between groups A, B, and C. We conducted 10 iterations of this process and determined that on average, 118 (± 10 standard deviations) randomly selected genes were differentially regulated between the three groups. Thus, based on random sampling, the expected rate of differentially regulated genes between the three groups is approximately 40%, which is significantly lower than the observed rate of 55% when we focused the analysis to the 296 nuclear-encoded genes (P <0.001, chi-square).
We next uploaded the values of these 162 genes to the GEDI platform to construct gene expression mosaics for the three septic shock subgroups. Figure 4 shows the respective gene expression mosaics for each subgroup and provides a global representation of how the 162 nuclear-encoded mitochondrial genes were differentially expressed across these three groups. Overall, group A exhibited a greater repression of nuclear-encoded mitochondrial genes, compared to subgroups B and C, as evidenced by the greater proportion of blue color intensity.
Gene Expression Dynamics Inspector-generated mosaics of differentially expressed mitochondrial genes for the three septic shock subgroups. The 162 genes are depicted along the same coordinates across the three expression mosaics. Red intensity correlates with increased gene expression and blue intensity correlates with decreased gene expression. Clear differences in color patterns illustrate differential expression of mitochondrial genes across patient subgroups A, B, and C, with general downregulation in group A. Group A subjects have higher illness severity, higher mortality, and higher organ failure burden.
We next uploaded the 162 mitochondrial gene list to the IPA platform to visualize biological function. This analysis yielded `oxidative phosphorylation' as the most significant canonical pathway rather than a specific domain of mitochondrial function. Figure 5 illustrates the expression of these 47 genes (relative to controls) corresponding to oxidative phosphorylation in each septic shock subclass (see Table S6 in Additional file 6 for complete gene list). Group A exhibited downregulation (that is the degree of green intensity relative to red intensity) of genes for all five ETS complexes. In contrast, group B exhibited upregulation of genes for ETS complex II, with concomitant downregulation of genes for ETS complexes I, III, IV, and V; group C exhibited upregulation of genes for ETS complex III, with concomitant downregulation of genes for ETS complexes I, II, IV, and V.
Differential regulation of oxidative phosphorylation genes for the three septic shock subgroups. Differential expression of individual nuclear-encoded genes across patient subgroups A, B, and C that correspond to mitochondrial electron transport system (ETS) complexes I to V based on a false discovery rate of 1%. Red intensity correlates with increased gene expression and green intensity correlates with decreased gene expression. Group A exhibited downregulation (that is greater degree of green intensity relative to red intensity) of genes for all ETS complexes, whereas group B exhibited upregulation of genes for complex II and group C exhibited upregulation of genes for complex III.
Discussion
In this focused analysis of a comprehensive genomic expression database, we found that nuclear-encoded mitochondrial genes are differentially regulated early in pediatric septic shock compared to healthy controls. We also compared expression of the nuclear-encoded mitochondrial genes across previously defined and validated subclasses of pediatric septic shock with distinct phenotypic characteristics. Although these subclasses were defined primarily by differential expression of genes corresponding to adaptive immunity, glucocorticoid receptor signaling, and PPARβ signaling, we found that the nuclear-encoded mitochondrial genes were also differentially regulated across these subclasses, with a greater degree of repression in the subclass of patients with the most organ dysfunction and highest mortality.
Fifty-one nuclear genes encoding subunits of the mitochondrial ETS complexes were differentially regulated in blood samples from children with septic shock, with a greater degree of downregulation overall. These findings parallel the decrease in leukocyte gene expression for subunits of the ETS complexes I to V that occurs four to six hours after endotoxin injection in healthy human volunteers [17]. The mitochondrial ETS involves over 100 proteins derived from the nuclear and mitochondrial genomes assembled into five complexes. Although it is difficult to predict how the sum of the changes we observed might alter total function of the respiratory chain, our data raise the notion that genomic influences could affect mitochondrial oxidative respiration and therefore cellular bioenergetic and organ function in sepsis. In particular, the prominence of NADH dehydrogenase activity in the top gene networks is consistent with decreased complex I gene expression [56] and activity noted in skeletal muscle in human adult sepsis [57],[58]. Prior studies using blood samples from patients with sepsis have also demonstrated altered respiratory chain activity in peripheral blood mononuclear cells and platelets [9],[11],[13],[59],[60].
A similar genome-wide analysis using skeletal muscle from 17 adult patients with sepsis-induced multi-organ failure found that 82 mitochondrial genes were differentially regulated compared to healthy controls (74 upregulated, 8 downregulated) [61]. Decreased ETS enzyme activity was also observed and attributed to a loss of mitochondrial content. The authors concluded that deficits in ETS activity could not be explained by decreased mitochondrial gene expression because of the overall absence of repression of mitochondrial genes. However, these patients were studied later in their septic course when a decrease in mitochondrial content seems to be most prominent, as opposed to our study in which patients were studied in the acute phase when decreased ETS function predominates. More recently, Carre et al. performed muscle biopsies early after onset (one to two days) of critical septic illness in adult patients and observed that an overall decrease in ETS gene expression was associated with decreased protein content and activity of ETS complexes I and IV [56]. Concurrent measurements of mitochondrial gene expression and function are needed to determine if the changes we observed in mitochondrial gene expression are sufficient to alter mitochondrial function in blood cells (and other tissues) in pediatric sepsis. We note, however, that serum lactate concentrations were highest in group A. While this suggests a potential association between downregulation of mitochondrial genes and mitochondrial dysfunction, these data should be interpreted cautiously because they are incomplete and represent an indirect, nonspecific measure of mitochondrial function.
Genes encoding the MRPs were also predominately downregulated in pediatric septic shock. This finding should not be construed as specific to mitochondria, as global downregulation of ribosomal gene transcription may occur nonspecifically during extreme biological conditions such as septic shock [17],[62]. Similar downregulation of MRPs has been previously described in blood and brain tissue from patients with Alzheimer's disease [14]. Like bacteria and eukaryotic cell cytoplasm, mitochondria contain their own ribosomes. The MRPs are encoded in nuclear genes, synthesized in the cytoplasm, and then imported into mitochondria. MRPs assemble with mitochondrial-transcribed rRNAs to form two ribosomes that are responsible for translating the 37 mitochondrial-encoded genes, including 13 protein subunits critical to the function of ETS complexes I, III, IV, and V (ATP synthase). Thus, MRPs play a critical role in mitochondrial protein synthesis and bioenergetic function [18],[19]. Known mutations in MRPs are associated with lactic acidosis, organ dysfunction, and early death [18], and several MRPs (MRPS29, MRPS30) have been implicated in apoptosis [19]. Moreover, changes in mitochondrial turnover (including biogenesis, mitophagy, and fusion/fission) have been associated with clinical outcomes in sepsis [5],[63]. The significant repression of MRPs in our study points to changes in the mitochondrial protein synthesis machinery as one potential mechanism that could lead to mitochondrial dysfunction and diminished mitochondrial content in sepsis.
The differential regulation of mitochondrial genes across subclasses of pediatric septic shock supports a link between mitochondrial gene expression and clinical outcomes, including organ failure and mortality. These subclasses were previously identified based on hierarchical clustering, with patients in group A exhibiting the greatest repression of genes corresponding to key signaling pathways of the adaptive immune system, glucocorticoid receptor signaling, and PPARα signaling [21]. In the current study, patients in group A also exhibited a greater repression of nuclear-encoded mitochondrial genes than groups B and C, especially in ETS complexes II and III. However, we caution that since we observed minimal differential regulation of mitochondrial genes when directly comparing survivors and nonsurvivors, or patients with and without a complicated course, we cannot rule out a `coupling effect' in which changes in mitochondrial gene expression are enhanced by other biologic pathways that differ between genomically defined subclasses. Mitochondria are involved in a variety of cell signaling pathways underlying the immune response, including cytokine release, inflammasome formation, and formation of reactive oxygen species [64]. It will be important to establish the extent to which mitochondrial gene expression may truly affect phenotypic differences in septic shock through its role in the immune system and other cell signaling pathways versus more direct effects on cellular bioenergetics.
We note the limitations of our study. First, this was a post hoc, focused analysis using a limited set of 296 nuclear-encoded mitochondrial genes. To reduce the likelihood of false-positive results, we used a relatively stringent FDR of 1% and conducted control analyses to determine expected rates of differential gene expression based on either all available genes on the array, or 10 iterations of analyses based on randomly selected genes. In both cases, we found that the observed rate of differential gene expression in mitochondrial genes was greater than the expected random rate. Nonetheless, our focused analytical approach does not allow us to conclude that the pediatric septic shock transcriptome is specifically enriched for genes corresponding to mitochondrial function. We can only conclude that if the analytical approach is limited to nuclear-encoded mitochondrial genes, we find differential regulation of these genes in children with septic shock, and across subgroups of patients with septic shock.
Second, sufficient data were available from only one time point. Although blood sampling within 24 hours of initial presentation to the PICU with septic shock was likely to capture maximum clinical acuity, we were unable to test temporal changes in the mitochondrial transcriptome with evolution of the septic course. Third, the data are based on whole blood-derived RNA, which carries the potential for confounding by differential white blood cell counts. Although platelet counts did not differ between the three septic shock subclasses, group A had a lower total leukocyte count with a greater percentage of lymphocytes and fewer neutrophils. Lymphocytes have a relatively lower mitochondrial content than neutrophils, though how this effects nuclear-encoded mitochondrial gene expression is not clear [65],[66], and have been shown to have slightly less gene upregulation in sepsis [67]. However, we have previously shown that whole blood-derived RNA can yield biologically meaningful data, gene expression profiles have revealed similar themes in leukocyte subsets and whole blood [62], and our current data are consistent with mitochondrial gene expression profiles from previous laboratory- and clinical-based studies [17],[56],[68]-[70]. Fourth, since concomitant measures of mitochondrial function were not available we cannot determine how the observed changes might alter the total function of respiratory chain and ATP production. Although the fold change in gene expression was modest in most cases, our findings were similar to magnitude of changes in mitochondrial gene expression profiles observed in prior studies [56],[68],[71]. However, it also possible that primary mitochondrial bioenergetic dysfunction itself leads to changes in nuclear gene expression [72]. Finally, because we used a nuclear gene array platform, changes in mitochondrial-encoded genes were not included in this study. These genes are critical to the function of the ETS and mitochondrial protein synthesis and should be considered in future studies.
Conclusions
In summary, our focused analysis demonstrated that nuclear-encoded mitochondrial genes were differentially regulated early in pediatric septic shock compared to healthy controls, as well as across genotypic and phenotypic distinct pediatric septic shock subclasses. Although no pathophysiologic consequences can be derived directly from these results, the findings nonetheless provide support for the hypothesis that differential regulation of nuclear-encoded mitochondrial genes may be an important mechanism contributing to alterations in mitochondrial bioenergetic function in pediatric sepsis.
Key messages
Nuclear-encoded mitochondrial genes corresponding to protein subunits of the mitochondrial ETS and mitochondrial ribosomes were differentially expressed in children with septic shock compared to healthy controls.
Patterns of nuclear-encoded mitochondrial gene expression differed across three previously validated gene expression-based subclasses of pediatric septic shock, with the greatest degree of repression in patients with the most organ dysfunction and highest mortality.
These findings provide the first evidence that the nuclear genome may be an important mechanism contributing to alterations in mitochondrial bioenergetic function within blood cells in pediatric sepsis.
Additional files
Abbreviations
- ANOVA:
-
analysis of variance
- ETS:
-
electron transport system
- FDR:
-
false discovery rate
- GEDI:
-
Gene Expression Dynamics Inspector
- IPA:
-
Ingenuity Pathways Analysis
- kDa:
-
kiloDaltons
- MRP:
-
mitochondrial ribosomal protein
- NADH:
-
nicotinamide adenine dinucleotide
- NCBI:
-
National Center for Biotechnology
- PERSEVERE:
-
pediatric sepsis biomarker risk model
- PICU:
-
pediatric intensive care unit
- PPAR:
-
peroxisome proliferator-activated receptor
- PRISM:
-
pediatric risk of mortality score
- RMA:
-
robust multiple-array average
References
Hartman ME, Linde-Zwirble WT, Angus DC, Watson RS: Trends in the epidemiology of pediatric severe sepsis. Pediatr Crit Care Med. 2013, 14: 686-693. 10.1097/PCC.0b013e3182917fad.
Weiss SL, Parker B, Bullock ME, Swartz S, Price C, Wainwright MS, Goodman DM: Defining pediatric sepsis by different criteria: Discrepancies in populations and implications for clinical practice. Pediatr Crit Care Med. 2012, 13: e219-e226. 10.1097/PCC.0b013e31823c98da.
Hall MW, Knatz NL, Vetterly C, Tomarello S, Wewers MD, Volk HD, Carcillo JA: Immunoparalysis and nosocomial infection in children with multiple organ dysfunction syndrome. Intensive Care Med. 2011, 37: 525-532. 10.1007/s00134-010-2088-x.
Proulx F, Joyal JS, Mariscalco MM, Leteurtre S, Leclerc F, Lacroix J: The pediatric multiple organ dysfunction syndrome. Pediatr Crit Care Med. 2009, 10: 12-22. 10.1097/PCC.0b013e31819370a9.
Crouser ED: Mitochondrial dysfunction in septic shock and multiple organ dysfunction syndrome. Mitochondrion. 2004, 4: 729-741. 10.1016/j.mito.2004.07.023.
Fink MP: Bench-to-bedside review: Cytopathic hypoxia. Crit Care. 2002, 6: 491-499. 10.1186/cc1824.
Singer M: Mitochondrial function in sepsis: acute phase versus multiple organ failure. Crit Care Med. 2007, 35: S441-S448. 10.1097/01.CCM.0000278049.48333.78.
Adrie C, Bachelet M, Vayssier-Taussat M, Russo-Marie F, Bouchaert I, Adib-Conquy M, Cavaillon JM, Pinsky MR, Dhainaut JF, Polla BS: Mitochondrial membrane potential and apoptosis peripheral blood monocytes in severe human sepsis. Am J Respir Crit Care Med. 2001, 164: 389-395. 10.1164/ajrccm.164.3.2009088.
Belikova I, Lukaszewicz AC, Faivre V, Damoisel C, Singer M, Payen D: Oxygen consumption of human peripheral blood mononuclear cells in severe human sepsis. Crit Care Med. 2007, 35: 2702-2708. 10.1097/01.CCM.0000295593.25106.C4.
Garrabou G, Moren C, Lopez S, Tobias E, Cardellach F, Miro O, Casademont J: The effects of sepsis on mitochondria. J Infect Dis. 2012, 205: 392-400. 10.1093/infdis/jir764.
Japiassu AM, Santiago AP, d'Avila JC, Garcia-Souza LF, Galina A, Castro Faria-Neto HC, Bozza FA, Oliveira MF: Bioenergetic failure of human peripheral blood monocytes in patients with septic shock is mediated by reduced F1Fo adenosine-5'-triphosphate synthase activity. Crit Care Med. 2011, 39: 1056-1063. 10.1097/CCM.0b013e31820eda5c.
Yamakawa K, Ogura H, Koh T, Ogawa Y, Matsumoto N, Kuwagata Y, Shimazu T: Platelet mitochondrial membrane potential correlates with severity in patients with systemic inflammatory response syndrome. J Trauma Acute Care Surg. 2013, 74: 411-417. 10.1097/TA.0b013e31827a34cf. discussion 418
Weiss SL, Selak MA, Tuluc F, Villarroel JP, Nadkarni VM, Deutschman CS, Becker LB: Mitochondrial dysfunction in peripheral mononuclear cells in pediatric septic shock. Pediatr Crit Care Med 2014. In press.,
Lunnon K, Ibrahim Z, Proitsi P, Lourdusamy A, Newhouse S, Sattlecker M, Furney S, Saleem M, Soininen H, Kloszewska I, Mecocci P, Tsolaki M, Vellas B, Coppola G, Geschwind D, Simmons A, Lovestone S, Dobson R, Hodges A: Mitochondrial dysfunction and immune activation are detectable in early Alzheimer's disease blood. J Alzheimers Dis. 2012, 30: 685-710.
Napoli E, Wong S, Hertz-Picciotto I, Giulivi C: Deficits in bioenergetics and impaired immune response in granulocytes from children with autism. Pediatrics. 2014, 133: e1405-1410-10.1542/peds.2013-1545.
Salehi MH, Kamalidehghan B, Houshmand M, Yong Meng G, Sadeghizadeh M, Aryani O, Nafissi S: Gene expression profiling of mitochondrial Oxidative Phosphorylation (OXPHOS) Complex I in Friedreich Ataxia (FRDA) patients. PLoS One. 2014, 9: e94069-10.1371/journal.pone.0094069.
Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF: A network-based analysis of systemic inflammation in humans. Nature. 2005, 437: 1032-1037. 10.1038/nature03985.
Christian BE, Spremulli LL: Mechanism of protein biosynthesis in mammalian mitochondria. Biochim Biophys Acta. 2012, 1819: 1035-1054. 10.1016/j.bbagrm.2011.11.009.
O'Brien TW: Properties of human mitochondrial ribosomes. IUBMB Life. 2003, 55: 505-513. 10.1080/15216540310001626610.
Wong HR: Genome-wide expression profiling in pediatric septic shock. Pediatr Res. 2013, 73: 564-569. 10.1038/pr.2013.11.
Wong HR, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Willson DF, Freishtat RJ, Anas N, Meyer K, Checchia PA, Monaco M, Odom K, Shanley TP: Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med. 2009, 7: 34-10.1186/1741-7015-7-34.
Wong HR, Wheeler DS, Tegtmeyer K, Poynter SE, Kaplan JM, Chima RS, Stalets E, Basu RK, Doughty LA: Toward a clinically feasible gene expression-based subclassification strategy for septic shock: proof of concept. Crit Care Med. 2010, 38: 1955-1961. 10.1097/CCM.0b013e3181b427e8.
Wong HR, Cvijanovich NZ, Allen GL, Thomas NJ, Freishtat RJ, Anas N, Meyer K, Checchia PA, Lin R, Shanley TP, Bigham MT, Wheeler DS, Doughty LA, Tegtmeyer K, Poynter SE, Kaplan JM, Chima RS, Stalets E, Basu RK, Varisco BM, Barr FE: Validation of a gene expression-based subclassification strategy for pediatric septic shock. Crit Care Med. 2011, 39: 2511-2517. 10.1097/CCM.0b013e3182257675.
Wong HR, Cvijanovich N, Wheeler DS, Bigham MT, Monaco M, Odoms K, Macias WL, Williams MD: Interleukin-8 as a stratification tool for interventional trials involving pediatric septic shock. Am J Respir Crit Care Med. 2008, 178: 276-282. 10.1164/rccm.200801-131OC.
Nowak JE, Wheeler DS, Harmon KK, Wong HR: Admission chemokine (C-C motif) ligand 4 levels predict survival in pediatric septic shock. Pediatr Crit Care Med. 2010, 11: 213-216. 10.1097/PCC.0b013e3181b8076c.
Kaplan JM, Wong HR: Biomarker discovery and development in pediatric critical care medicine. Pediatr Crit Care Med. 2011, 12: 165-173. 10.1097/PCC.0b013e3181e28876.
Wong HR, Salisbury S, Xiao Q, Cvijanovich NZ, Hall M, Allen GL, Thomas NJ, Freishtat RJ, Anas N, Meyer K, Checchia PA, Lin R, Shanley TP, Bigham MT, Sen A, Nowak J, Quasney M, Henricksen JW, Chopra A, Banschbach S, Beckman E, Harmon K, Lahni P, Lindsell CJ: The pediatric sepsis biomarker risk model. Crit Care. 2012, 16: R174-10.1186/cc11652.
Wong HR, Weiss SL, Giuliano JS, Wainwright MS, Cvijanovich NZ, Thomas NJ, Allen GL, Anas N, Bigham MT, Hall M, Freishtat RJ, Sen A, Meyer K, Checchia PA, Shanley TP, Nowak J, Quasney M, Chopra A, Fitzgerald JC, Gedeit R, Banschbach S, Beckman E, Lahni P, Hart K, Lindsell CJ: Testing the prognostic accuracy of the updated pediatric sepsis biomarker risk model. PloS one. 2014, 9: e86242-10.1371/journal.pone.0086242.
Wong HR, Weiss SL, Giuliano JS, Wainwright MS, Cvijanovich NZ, Thomas NJ, Allen GL, Anas N, Bigham MT, Hall M, Freishtat RJ, Sen A, Meyer K, Checchia PA, Shanley TP, Nowak J, Quasney M, Chopra A, Fitzgerald JC, Gedeit R, Banschbach S, Beckman E, Harmon K, Lahni P, Lindsell CJ: The temporal version of the pediatric sepsis biomarker risk model. PloS one. 2014, 9: e92121-10.1371/journal.pone.0092121.
Wong HR, Lindsell CJ, Pettila V, Meyer NJ, Thair SA, Karlsson S, Russell JA, Fjell CD, Boyd JH, Ruokonen E, Shashaty MG, Christie JD, Hart KW, Lahni P, Walley KR: A multibiomarker-based outcome risk stratification model for adult septic shock. Crit Care Med. 2014, 42: 781-789. 10.1097/CCM.0000000000000106.
Mickiewicz B, Vogel HJ, Wong HR, Winston BW: Metabolomics as a novel approach for early diagnosis of pediatric septic shock and its mortality. Am J Respir Crit Care Med. 2013, 187: 967-976. 10.1164/rccm.201209-1726OC.
Basu RK, Standage SW, Cvijanovich NZ, Allen GL, Thomas NJ, Freishtat RJ, Anas N, Meyer K, Checchia PA, Lin R, Shanley TP, Bigham MT, Wheeler DS, Devarajan P, Goldstein SL, Wong HR: Identification of candidate serum biomarkers for severe septic shock-associated kidney injury via microarray. Crit Care. 2011, 15: R273-10.1186/cc10554.
Wong HR, Cvijanovich NZ, Hall M, Allen GL, Thomas NJ, Freishtat RJ, Anas N, Meyer K, Checchia PA, Lin R, Bigham MT, Sen A, Nowak J, Quasney M, Henricksen JW, Chopra A, Banschbach S, Beckman E, Harmon K, Lahni P, Shanley TP: Interleukin-27 is a novel candidate diagnostic biomarker for bacterial infection in critically ill children. Crit Care. 2012, 16: R213-10.1186/cc11847.
Wong HR, Lindsell CJ, Lahni P, Hart KW, Gibot S: Interleukin 27 as a sepsis diagnostic biomarker in critically ill adults. Shock. 2013, 40: 382-386. 10.1097/SHK.0b013e3182a67632.
Basu RK, Wang Y, Wong HR, Chawla LS, Wheeler DS, Goldstein SL: Incorporation of biomarkers with the renal angina index for prediction of severe AKI in critically ill children. Clin J Am Soc Nephrol. 2014, 9: 654-662. 10.2215/CJN.09720913.
Wong HR, Shanley TP, Sakthivel B, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, Kalyanaraman M, Tofil NM, Penfil S, Monaco M, Tagavilla MA, Odoms K, Dunsmore K, Barnes M, Aronow BJ: Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol Genomics. 2007, 30: 146-155. 10.1152/physiolgenomics.00024.2007.
Cvijanovich N, Shanley TP, Lin R, Allen GL, Thomas NJ, Checchia P, Anas N, Freishtat RJ, Monaco M, Odoms K, Sakthivel B, Wong HR: Validating the genomic signature of pediatric septic shock. Physiol Genomics. 2008, 34: 127-134. 10.1152/physiolgenomics.00025.2008.
Cvijanovich NZ, King JC, Flori HR, Gildengorin G, Wong HR: Zinc homeostasis in pediatric critical illness. Pediatr Crit Care Med. 2009, 10: 29-34. 10.1097/PCC.0b013e31819371ce.
Solan PD, Dunsmore KE, Denenberg AG, Odoms K, Zingarelli B, Wong HR: A novel role for matrix metalloproteinase-8 in sepsis. Crit Care Med. 2012, 40: 379-387. 10.1097/CCM.0b013e318232e404.
Standage SW, Caldwell CC, Zingarelli B, Wong HR: Reduced peroxisome proliferator-activated receptor alpha expression is associated with decreased survival and increased tissue bacterial load in sepsis. Shock. 2012, 37: 164-169. 10.1097/SHK.0b013e31823f1a00.
Nowak JE, Harmon K, Caldwell CC, Wong HR: Prophylactic zinc supplementation reduces bacterial load and improves survival in a murine model of sepsis. Pediatr Crit Care Med. 2012, 13: e323-e329. 10.1097/PCC.0b013e31824fbd90.
Slinko S, Piraino G, Hake PW, Ledford JR, O'Connor M, Lahni P, Solan PD, Wong HR, Zingarelli B: Combined zinc supplementation with proinsulin C-peptide treatment decreases the inflammatory response and mortality in murine polymicrobial sepsis. Shock. 2014, 41: 292-300. 10.1097/SHK.0000000000000127.
Goldstein B, Giroir B, Randolph A: International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med. 2005, 6: 2-8. 10.1097/01.PCC.0000149131.72248.E6.
Shanley TP, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, Kalyanaraman M, Tofil NM, Penfil S, Monaco M, Odoms K, Barnes M, Sakthivel B, Aronow BJ, Wong HR: Genome-level longitudinal expression of signaling pathways and gene networks in pediatric septic shock. Mol Med. 2007, 13: 495-508. 10.2119/2007-00065.Shanley.
Wong HR, Cvijanovich N, Allen GL, Lin R, Anas N, Meyer K, Freishtat RJ, Monaco M, Odoms K, Sakthivel B, Shanley TP: Genomic expression profiling across the pediatric systemic inflammatory response syndrome, sepsis, and septic shock spectrum. Crit Care Med. 2009, 37: 1558-1566. 10.1097/CCM.0b013e31819fcc08.
Wong HR, Freishtat RJ, Monaco M, Odoms K, Shanley TP: Leukocyte subset-derived genomewide expression profiles in pediatric septic shock. Pediatr Crit Care Med. 2010, 11: 349-355.
Wynn JL, Cvijanovich NZ, Allen GL, Thomas NJ, Freishtat RJ, Anas N, Meyer K, Checchia PA, Lin R, Shanley TP, Bigham MT, Banschbach S, Beckman E, Wong HR: The influence of developmental age on the early transcriptomic response of children with septic shock. Mol Med. 2011, 17: 1146-1156. 10.2119/molmed.2011.00169.
Wong HR, Cvijanovich NZ, Allen GL, Thomas NJ, Freishtat RJ, Anas N, Meyer K, Checchia PA, Weiss SL, Shanley TP, Bigham MT, Banschbach S, Beckman E, Harmon K, Zimmerman JJ: Corticosteroids are associated with repression of adaptive immunity gene programs in pediatric septic shock. Am J Respir Crit Care Med. 2014, 189: 940-946. 10.1164/rccm.201401-0171OC.
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP: Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003, 4: 249-264. 10.1093/biostatistics/4.2.249.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP: Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005, 102: 15545-15550. 10.1073/pnas.0506580102.
Chen J, Bardes EE, Aronow BJ, Jegga AG: ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37: W305-W311. 10.1093/nar/gkp427.
Eichler GS, Huang S, Ingber DE: Gene Expression Dynamics Inspector (GEDI): for integrative analysis of expression profiles. Bioinformatics. 2003, 19: 2321-2322. 10.1093/bioinformatics/btg307.
Guo Y, Eichler GS, Feng Y, Ingber DE, Huang S: Towards a holistic, yet gene-centered analysis of gene expression profiles: a case study of human lung cancers. J Biomed Biotechnol. 2006, 2006: 69141-10.1155/JBB/2006/69141.
Gene Expression Dynamics Inspector [], [http://apps.childrenshospital.org/clinical/research/ingber/GEDI/gedihome.htm]
Abulebda K, Cvijanovich NZ, Thomas NJ, Allen GL, Anas N, Bigham MT, Hall M, Freishtat RJ, Sen A, Meyer K, Checchia PA, Shanley TP, Nowak J, Quasney M, Weiss SL, Chopra A, Banschbach S, Beckman E, Lindsell CJ, Wong HR: Post-ICU admission fluid balance and pediatric septic shock outcomes: a risk-stratified analysis. Crit Care Med. 2014, 42: 397-403. 10.1097/CCM.0b013e3182a64607.
Carre JE, Orban JC, Re L, Felsmann K, Iffert W, Bauer M, Suliman HB, Piantadosi CA, Mayhew TM, Breen P, Stotz M, Singer M: Survival in critical illness is associated with early activation of mitochondrial biogenesis. Am J Respir Crit Care Med. 2010, 182: 745-751. 10.1164/rccm.201003-0326OC.
Brealey D, Brand M, Hargreaves I, Heales S, Land J, Smolenski R, Davies NA, Cooper CE, Singer M: Association between mitochondrial dysfunction and severity and outcome of septic shock. Lancet. 2002, 360: 219-223. 10.1016/S0140-6736(02)09459-X.
Fredriksson K, Hammarqvist F, Strigard K, Hultenby K, Ljungqvist O, Wernerman J, Rooyackers O: Derangements in mitochondrial metabolism in intercostal and leg muscle of critically ill patients with sepsis-induced multiple organ failure. Am J Physiol Endocrinol Metab. 2006, 291: E1044-E1050. 10.1152/ajpendo.00218.2006.
Sjovall F, Morota S, Hansson MJ, Friberg H, Gnaiger E, Elmer E: Temporal increase of platelet mitochondrial respiration is negatively associated with clinical outcome in patients with sepsis. Crit Care. 2010, 14: R214-10.1186/cc9337.
Sjovall F, Morota S, Persson J, Hansson MJ, Elmer E: Patients with sepsis exhibit increased mitochondrial respiratory capacity in peripheral blood immune cells. Crit Care. 2013, 17: R152-10.1186/cc12831.
Fredriksson K, Tjader I, Keller P, Petrovic N, Ahlman B, Scheele C, Wernerman J, Timmons JA, Rooyackers O: Dysregulation of mitochondrial dynamics and the muscle transcriptome in ICU patients suffering from sepsis induced multiple organ failure. PLoS One. 2008, 3: e3686-10.1371/journal.pone.0003686.
Talwar S, Munson PJ, Barb J, Fiuza C, Cintron AP, Logun C, Tropea M, Khan S, Reda D, Shelhamer JH, Danner RL, Suffredini AF: Gene expression profiles of peripheral blood leukocytes after endotoxin challenge in humans. Physiol Genomics. 2006, 25: 203-215. 10.1152/physiolgenomics.00192.2005.
Gunst J, Derese I, Aertgeerts A, Ververs EJ, Wauters A, Van den Berghe G, Vanhorebeek I: Insufficient autophagy contributes to mitochondrial dysfunction, organ failure, and adverse outcome in an animal model of critical illness. Crit Care Med. 2013, 41: 182-194. 10.1097/CCM.0b013e3182676657.
Tait SW, Green DR: Mitochondria and cell signalling. J Cell Sci. 2012, 125: 807-815. 10.1242/jcs.099234.
Pyle A, Burn DJ, Gordon C, Swan C, Chinnery PF, Baudouin SV: Fall in circulating mononuclear cell mitochondrial DNA content in human sepsis. Intensive Care Med. 2010, 36: 956-962. 10.1007/s00134-010-1823-7.
Selak MA, Lyver E, Micklow E, Deutsch EC, Onder O, Selamoglu N, Yager C, Knight S, Carroll M, Daldal F, Dancis A, Lynch DR, Sarry JE: Blood cells from Friedreich ataxia patients harbor frataxin deficiency without a loss of mitochondrial function. Mitochondrion. 2011, 11: 342-350. 10.1016/j.mito.2010.12.003.
Salomao R, Brunialti MK, Gomes NE, Mendes ME, Diaz RS, Komninakis S, Machado FR, da Silva ID, Rigato O: Toll-like receptor pathway signaling is differently regulated in neutrophils and peripheral mononuclear cells of patients with sepsis, severe sepsis, and septic shock. Crit Care Med. 2009, 37: 132-139. 10.1097/CCM.0b013e318192fbaf.
Callahan LA, Supinski GS: Downregulation of diaphragm electron transport chain and glycolytic enzyme gene expression in sepsis. J Appl Physiol (1985). 2005, 99: 1120-1126. 10.1152/japplphysiol.01157.2004.
dos Santos CC, Murthy S, Hu P, Shan Y, Haitsma JJ, Mei SH, Stewart DJ, Liles WC: Network analysis of transcriptional responses induced by mesenchymal stem cell treatment of experimental sepsis. Am J Pathol. 2012, 181: 1681-1692. 10.1016/j.ajpath.2012.08.009.
Severino P, Silva E, Baggio-Zappia GL, Brunialti MK, Nucci LA, Rigato O, da Silva ID, Machado FR, Salomao R: Patterns of gene expression in peripheral blood mononuclear cells and outcomes from patients with sepsis secondary to community acquired pneumonia. PLoS One. 2014, 9: e91886-10.1371/journal.pone.0091886.
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC: PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003, 34: 267-273. 10.1038/ng1180.
Wallace DC, Fan W: Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2010, 10: 12-31. 10.1016/j.mito.2009.09.006.
Acknowledgements
Financial support was provided by National Institutes of Health Grants RC1HL100474, RO1GM064619, and RO1GM099773. Dr. Weiss is also supported by NIH K12HD047349-10. The NIH had no role in the design, collection, analysis, and interpretation of data, writing of the manuscript, or the decision to submit this manuscript for publication.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SLW and HRW conceived and developed the study, obtained funding for the study, conducted the analyses, and wrote the manuscript. NZC, GLA, NJT, RJF, NA, KM, PAC, TPS, MTB, and JF enrolled subjects at the participating institutions, provided clinical data and biological samples, and edited the manuscript. SB, EB, K Howard, and EF maintained the clinical database and coordinated all inter-institutional research activity. K Harmon maintained the biological repository and processed all biological samples. All authors read and approved the manuscript.
Electronic supplementary material
13054_2014_623_MOESM1_ESM.doc
Additional file 1: Table S1.: Mitochondrial genes (n = 118) differentially regulated between subjects with septic shock and normal controls. (DOC 140 KB)
13054_2014_623_MOESM2_ESM.doc
Additional file 2: Table S2.: List of genes (n = 51) corresponding to the oxidative phosphorylation pathway (Figure 1). (DOC 64 KB)
13054_2014_623_MOESM3_ESM.doc
Additional file 3: Table S3.: List of gene nodes in the gene network composed of mitochondrial ribosomal proteins (Figure 2). (DOC 48 KB)
13054_2014_623_MOESM4_ESM.doc
Additional file 4: Table S4.: List of gene nodes in the gene network having peroxisome proliferater-activated receptor (PPAR)-related genes as highly connected nodes (Figure 3). (DOC 47 KB)
13054_2014_623_MOESM5_ESM.doc
Additional file 5: Table S5.: Mitochondrial genes (n = 162) differentially regulated between subjects with septic shock groups A, B, and C. (DOC 156 KB)
13054_2014_623_MOESM6_ESM.doc
Additional file 6: Table S6.: List of genes (n = 47) corresponding to the oxidative phosphorylation pathway and differentially regulated between groups A, B, and C (Figure 5). (DOC 60 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Weiss, S.L., Cvijanovich, N.Z., Allen, G.L. et al. Differential expression of the nuclear-encoded mitochondrial transcriptome in pediatric septic shock. Crit Care 18, 623 (2014). https://doi.org/10.1186/s13054-014-0623-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13054-014-0623-9