Abstract
Introduction
Septic-shock-associated acute kidney injury (SSAKI) carries high morbidity in the pediatric population. Effective treatment strategies are lacking, in part due to poor detection and prediction. There is a need to identify novel candidate biomarkers of SSAKI. The objective of our study was to determine whether microarray data from children with septic shock could be used to derive a panel of candidate biomarkers for predicting SSAKI.
Methods
A retrospective cohort study compared microarray data representing the first 24 hours of admission for 179 children with septic shock with those of 53 age-matched normal controls. SSAKI was defined as a >200% increase of baseline serum creatinine, persistent to 7 days after admission.
Results
Patients with SSAKI (n = 31) and patients without SSAKI (n = 148) were clinically similar, but SSAKI carried a higher mortality (45% vs. 10%). Twenty-one unique gene probes were upregulated in SSAKI patients versus patients without SSAKI. Using leave-one-out cross-validation and class prediction modeling, these probes predicted SSAKI with a sensitivity of 98% (95% confidence interval (CI) = 81 to 100) and a specificity of 80% (95% CI = 72 to 86). Serum protein levels of two specific genes showed high sensitivity for predicting SSAKI: matrix metalloproteinase-8 (89%, 95% CI = 64 to 98) and elastase-2 (83%, 95% CI = 58 to 96). Both biomarkers carried a negative predictive value of 95%. When applied to a validation cohort, although both biomarkers carried low specificity (matrix metalloproteinase-8: 41%, 95% CI = 28 to 50; and elastase-2: 49%, 95% CI = 36 to 62), they carried high sensitivity (100%, 95% CI = 68 to 100 for both).
Conclusions
Gene probes upregulated in critically ill pediatric patients with septic shock may allow for the identification of novel candidate serum biomarkers for SSAKI prediction.
Similar content being viewed by others
Introduction
Septic shock leads to significant morbidity and mortality in critically ill adult and pediatric patients [1, 2]. Meanwhile, acute kidney injury (AKI) is also known to be independently associated with mortality and morbidity in critically ill patients. The treatment of sepsis costs the US population over $15 billion/year for adults and over $2 billion/year for children, while the costs for AKI approach $10 billion/year for adults alone [3, 4]. Sepsis is the most common precipitant for AKI in both populations, and the development of kidney injury in the context of sepsis is a poor prognostic sign. Together the two disease processes carry up to 75% mortality [5–9].
Effective therapies for septic-shock-associated acute kidney injury (SSAKI) are lacking. Detection schemes for SSAKI have been and still are dependent on serum creatinine, a flawed real-time marker of AKI [10, 11]. Diagnoses of SSAKI based upon changes in creatinine, therefore, are considerably varied and create heterogeneity between studies investigating AKI therapy. Biomarker research seeking to identify more robust markers of AKI has yielded promising results. Neutrophil gelatinase-associated lipocalin (NGAL), IL-18, and cystatin C have all demonstrated encouraging efficacy for predicting ischemic or nephrotoxic AKI and its severity [12–14]. Sepsis intrinsically induces increased expression of some AKI biomarkers (for example, NGAL), and studies of NGAL levels in patients with SSAKI often demonstrate high sensitivity with modest specificity [9, 14–17].
Importantly, the pathophysiology of SSAKI may be unique from that of ischemic or nephrotoxic AKI [18]. Therapies aiming to restore renal perfusion in ischemic AKI [19–21] have not been demonstrated to be particularly effective and may be even less effective in SSAKI, a process that may not be secondary to impaired glomerular preload. Persistent SSAKI may fall into the class of fluid-unresponsive AKI [22]. Renal replacement therapy has been used as therapy for AKI and data exist demonstrating that initiation prior to accumulation of excessive fluid overload may improve outcomes [23, 24]. The aggregate data, however, show that patients with SSAKI have consistently increased mortality, even with early renal replacement therapy initiation [25, 26]. The modest efficacy of biomarkers at identifying SSAKI also underscores the notion that the pathophysiology of SSAKI is unique from other etiologies of AKI. There is a need to identify novel candidate biomarkers of SSAKI, which would expedite early treatment aimed at preventing the effects of this highly morbid complication of sepsis.
We have generated an extensive genome-wide expression database from children with septic shock by way of microarray technology and have now leveraged this database to identify candidate biomarkers for SSAKI [27–32]. Herein we report the identification of 21 unique gene probes upregulated in patients with SSAKI, within the first 24 hours of admission to the pediatric intensive care unit (PICU), and their ability to robustly predict SSAKI. Two readily measurable gene products from this list, matrix metalloproteinase-8 (MMP-8) and neutrophil elastase-2, show high sensitivity for SSAKI in a cohort of patients with septic shock.
Materials and methods
Patients and data collection
The study protocol was approved by the Institutional Review Boards of each participating institution (n = 11). Children ≤10 years of age admitted to the PICU and meeting pediatric-specific criteria for septic shock were eligible for enrollment [33]. Controls, used to normalize the microarray data across the patients with septic shock and to conduct the three-group analysis of variance in the first derivation analysis, were recruited from the ambulatory departments of participating institutions using published inclusion and exclusion criteria [28–32]. These controls were required to reliably compare data across different batches of samples. All patients and controls in the derivation cohort were previously reported in microarray-based studies addressing hypotheses entirely different from that of the current report [28–32]. All microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO:GSE26440 and GEO:GSE26378). The patients in the validation cohort have not been previously reported in any manner.
After informed consent from legal guardians, blood samples were obtained within 24 hours of initial presentation to the PICU with septic shock. Clinical and laboratory data were collected daily while in the PICU, and were stored using a web-based database. Mortality was tracked for 28 days after enrollment.
SSAKI was defined as a >200% rise of serum creatinine relative to the median normal value for age [34, 35], persistent to day 7 of PICU admission. For example, if a patient had a >200% rise of serum creatinine upon admission to the PICU but returned to <200% by day 7 (without renal replacement therapy), then that patient was classified as not having SSAKI. Although stringent, this definition allowed us to identify those patients with persistent fluid-unresponsive AKI. No patient in this study had known pre-existing chronic kidney disease. Patients who died before day 7 with a creatinine rise >200% from baseline were included.
RNA extraction and microarray hybridization
Total RNA was isolated from whole blood using the PaxGene™ Blood RNA System (PreAnalytiX, Qiagen/Becton Dickson, Valencia, CA, USA). Microarray hybridization was performed as previously described using the Human Genome U133 Plus 2.0 GeneChip (Affymetrix, Santa Clara, CA, USA) [27–32].
Serum protein biomarker measurements
Measurements of serum MMP-8 and elastase-2 protein levels were performed using a Luminex 200 multi-plex instrument (Luminex Corporation, Austin, TX, USA) and antibody-coated magnetic beads (Millipore, Billerica, MA, USA) following the manufacturer's specifications.
Data analysis
Analyses were performed using one patient sample per chip. Image files were captured using an Affymetrix GeneChip Scanner 3000. CEL files were subsequently preprocessed using robust multiple-array average normalization and GeneSpring GX 7.3 software (Agilent Technologies, Palo Alto, CA, USA). All signal intensity-based data were used after robust multiple-array average normalization, which specifically suppresses all but significant variation among lower intensity probe sets [36]. All chips representing patient samples were then normalized to the respective median values of controls.
Differences in mRNA abundance between the study categories were measured using GeneSpring GX 7.3 software, as were the class prediction procedures. Receiver operating characteristic (ROC) curves and calculations of biomarker performance were performed using SPSS (IBM Corporation, Somers, NY, USA). Ordinal and continuous clinical variables not normally distributed were analyzed via analysis of variance on ranks. Dichotomous clinical variables were analyzed using a chi-square test (SigmaStat software; Systat Software, Inc., San Jose, CA, USA).
Results
Initial derivation of candidate biomarkers for SSAKI
Based on complete microarray data from 179 children with septic shock, we used a multistage approach to begin deriving a panel of candidate biomarkers to predict SSAKI. All of the microarray data in the derivation process represent the first 24 hours of admission to the PICU with septic shock. Table 1 provides the clinical characteristics of the derivation cohort, consisting of 148 patients without SSAKI and 31 patients with SSAKI. The patients with SSAKI had a higher Pediatric Risk of Mortality score and a higher mortality rate, compared with the patients without SSAKI. All other variables shown in Table 1 were not significantly different between the two groups.
In the first derivation stage we conducted a two-step statistical test to determine which gene probes on the array (>80,000 gene probes) were differentially regulated between patients with and without SSAKI. In step one we conducted a three-group analysis of variance using normal controls (n = 53), patients without SSAKI, and patients with SSAKI as the comparison groups, and corrections for multiple comparisons (Benjamini-Hochberg false discovery rate = 5%). This was followed by a post hoc test (Tukey) to isolate the gene probes differentially regulated between patients with and without SSAKI (100 gene probes, see Additional file 1).
The 100-gene probe list presented in Additional file 1 corresponds to 61 unique and well-annotated genes. Twenty-one of the gene probes were upregulated in the patients with SSAKI, relative to the patients without SSAKI (Table 2). These 21 gene probes were subsequently used in a leave-one-out cross-validation procedure (Support Vector Machines algorithm) to predict 'SSAKI' and 'no SSAKI' classes in the derivation cohort. The leave-one-out cross-validation procedure removes a single observation from the original sample as validation - and analyzes the remaining observations as comparators. The procedure is repeated for each observation in the sample so that each is used once as validation. Figure 1 provides the 2 × 2 contingency table demonstrating the results of the leave-one-out cross validation procedures, and the associated performance calculations. Figure 1 demonstrates that the expression patterns of these 21 upregulated gene probes can predict SSAKI with a high degree of sensitivity and modestly high specificity in the derivation cohort. In addition, the expression patterns of these 21 upregulated gene probes have a high negative predictive value for SSAKI in the derivation cohort. Accordingly these 21 gene probes represent potential candidate biomarkers for predicting SSAKI.
Results of the leave-one-out cross-validation procedure involving 21 gene probes. The procedure was based on a Support Vector Machines algorithm and was targeted at prediction of 'SSAKI' and 'no SSAKI' classes. Performance calculations provided as the percentage with 95% confidence intervals (CIs). LR, likelihood ratio; NPV, negative predictive value; PPV, positive predictive value; SSAKI, sepsis-shock-associated acute kidney injury.
Matrix metalloproteinase-8 and elastase-2 serum levels as predictors of SSAKI
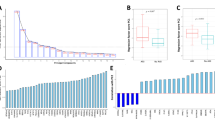
To further test the ability of the gene probes to predict SSAKI, we conducted initial measurements of selected gene probe products. Two gene probes corresponding to MMP-8 and one gene probe corresponding to elastase-2 were included in the 21 gene probes used in the class prediction modeling procedure described above. Since MMP-8 and elastase-2 protein levels can be readily measured in the serum compartment, we tested the ability of MMP-8 and elastase-2 serum protein levels to predict SSAKI in the derivation cohort. There were 150 parallel serum samples from the derivation cohort available for this analysis (132 patients without SSAKI and 18 patients with SSAKI). Figure 2 demonstrates that both MMP-8 and elastase-2 serum protein levels were higher in patients with SSAKI, compared with patients without kidney injury, thus corroborating the microarray data.
Serum matrix metalloproteinase-8 and elastase-2 levels in patients with and without sepsis-shock-associated acute kidney injury. Serum matrix metalloproteinase-8 (MMP-8) and elastase-2 levels are higher in patients with sepsis-shock-associated acute kidney injury (SSAKI) than patients without SSAKI. Patients with SSAKI (n = 132) were compared with patients without SSAKI (n = 18). Serum samples were obtained within 24 hours of admission to the pediatric ICU with septic shock and were drawn in parallel with RNA samples. Medians and interquartile ranges for the targets of interest for no SSAKI versus SSAKI: MMP-8, 26 (9 to 49) versus 40 (20 to 98), P = 0.029; and neutrophil elastase-2, 340 (9 to 571) and 574 (444 to 911), P = 0.01 (rank-sum test).
We next constructed ROC curves to determine the ability of serum protein levels of MMP-8 and elastase-2, respectively, to predict SSAKI in the derivation cohort (Figure 3). From these ROC curves we empirically selected cut-off values of 11 ng/ml (MMP-8) and 235 ng/ml (elastase-2) with an a priori goal of achieving a high sensitivity at the expense of specificity. The performance characteristics of the respective cut-off values are shown in Table 3. For both MMP-8 and elastase-2, the respective cut-off values were able to predict SSAKI with a relatively high sensitivity. In addition, both cut-off values had a high negative predictive value for the development of SSAKI. When we incorporated both MMP-8 and elastase-2 into a multiple regression analysis, we were unable to substantially improve the ability to predict SSAKI compared with the ability of each candidate biomarker alone (data not shown).
Matrix metalloproteinase-8 and elastase-2 as candidate biomarkers for predicting sepsis-shock-associated acute kidney injury. Receiver operating characteristic curves for matrix metalloproteinase-8 (MMP-8) and elastase-2 as candidate biomarkers for predicting sepsis-shock-associated acute kidney injury. Area under the curve = 0.659 (95% confidence interval = 0.520 to 0.798) and area under the curve = 0.688 (95% confidence interval = 0.549 to 0.826) for matrix MMP-8 and elastase-2, respectively.
Validation of MMP-8 and elastase-2 performance
Having demonstrated the individual performances of MMP-8 and elastase-2 as candidate biomarkers for predicting SSAKI in the derivation cohort, we next applied the same respective cut-off values to a separate validation cohort of patients. The validation cohort consisted of 59 patients without SSAKI and 11 patients with SSAKI. The clinical characteristics of the validation cohort were not significantly different from those of the derivation cohort, except that in the validation cohort the patients with SSAKI had a higher proportion of males and a lower proportion of patients with negative cultures compared with the patients without SSAKI (Table 4).
Table 5 provides the respective performance calculations for the two candidate biomarkers in the validation cohort. Both candidate biomarkers were able to predict SSAKI with 100% sensitivity in the validation cohort. However, the markers carried limited positive predictive value. In the validation cohort, 35 patients had an MMP-8 level >11 ng/ml without sustained SSAKI. In these patients, eight (23%) met criteria for at least mild AKI (pediatric Risk, Injury, Failure, Loss, End-Stage kidney injury (RIFLE) Risk) for at least 1 day. Also in the validation cohort, 30 patients had an elastase-2 level >235 ng/ml without sustained SSAKI. OK Nine (30%) of these patients met at least pediatric RIFLE Risk criteria for at least 1 day. Both biomarkers had negative predictive values for SSAKI of 100%, an important finding that adds clinical utility for practitioners treating patients who carry risks for developing severe AKI. Collectively, our results indicate that admission microarray data can be used to identify novel candidate biomarkers for SSAKI.
Discussion
We have demonstrated that microarray data can be leveraged to identify gene expression patterns common in SSAKI. Although mRNA levels do not necessarily correlate with protein expression, analysis of MMP-8 and elastase-2 protein levels in the serum compartment indicates that our gene expression data can identify novel serum protein biomarkers for SSAKI.
Although sepsis is the most important predictor of AKI in critically ill patients [37, 38], few clinical studies are dedicated to identifying the early phenotype of patients with SSAKI. Interrogation of several large databases of critically ill adult patients has yielded variables that may be predictive of SSAKI but are not consistently reliable (Table 6). The conclusions of these studies simply state that elevated proinflammatory markers portend adverse outcomes in acute kidney disease and death in patients with septic shock [39, 40].
NGAL has received considerable attention as a potential biomarker for AKI. NGAL derivation and validation studies were primarily performed in ischemic or nephrotoxic AKI with great efficacy [13], while investigations of NGAL biomarker utility in SSAKI have demonstrated variable results. In the general ICU population, serum NGAL levels have not been reliably demonstrated to be specific for SSAKI (Table 6). We previously published that while day 1 serum NGAL levels were elevated, with acceptable sensitivity, in children who develop SSAKI, the specificity was quite poor (39%) [41]. A recent prospective cohort study of 632 adult ICU patients demonstrated that the likelihood ratio for AKI (RIFLE Failure) was 1.71 for NGAL, and that elevated NGAL alone conferred an ROC area under curve of 0.77 for RIFLE Failure but when included in a 'most efficient clinical model' NGAL improved the ROC area under curve for RIFLE Failure to 0.96 [42]. OK In this study, specificity was 50% for a plasma NGAL cut-off value of 168 ng/ml. Emergency room serum NGAL levels >150 ng/ml were highly sensitive for AKI within 72 hours (96%), but specificity was only 51% [43]. NGAL is known to be released by activated neutrophils and appears to be intrinsically elevated in sepsis, which may explain its limitation as a biomarker specific for SSAKI.
A variable that may complicate delineation of biomarkers for SSAKI is that the pathophysiology of SSAKI may be different from that of ischemic or nephrotoxic AKI. Although acute tubular necrosis has traditionally been considered the etiology of AKI in sepsis, no conclusive pathologic evidence has demonstrated that this is true [18]. Animal models of SSAKI demonstrate increased, rather than decreased, renal blood flow and a state of hyperdynamic AKI [44]. The mechanisms mediating disease progression are multifactorial, and AKI during sepsis may be a result of acute tubular apoptosis rather than acute tubular necrosis [18]. Nonhemodynamic-dependent mechanisms of injury during sepsis, inflammatory and immunologic, may trigger this apoptotic response. This pathophysiology has probably decreased the utility of traditional tests based on the functionality of proximal tubular filtration [45].
Our study demonstrates that microarray-derived gene expression data can provide a tool for discovery of candidate biomarkers for SSAKI. Outside the neonatal population, we are the first group to attempt to characterize biomarkers specific to SSAKI (and not all-cause AKI) in children. Our data represent the first 24 hours of presentation to medical care, a timeframe that can be considered a therapeutic window for acute intervention. It is important to note that our patients were restricted to having septic shock, an exclusion criterion not previously used in pediatric studies investigating AKI biomarkers. Additionally, our definition of SSAKI identifies patients with severe, persistent kidney injury up to day 7. These candidate biomarkers thus identify patients with resuscitation unresponsive AKI (that is, patients having high serum creatinine levels at admission who subsequently did not normalize with standard resuscitation). The expression patterns of the 21 upregulated gene probes demonstrate excellent sensitivity and good specificity for SSAKI (Figure 1). The negative predictive value of nearly 100% carries obvious import to the bedside practitioner, but should be tempered by the fact that the prevalence of AKI in our cohort was relatively low (31/179, 17%). Individual performance calculations of two of these gene products, MMP-8 and elastase-2, demonstrate reasonable sensitivity in the derivation cohort (Table 3), but even higher sensitivity in the validation cohort (Table 5). Additionally, in the validation cohort, the specificity and negative predictive values of MMP-8 and elastase-2 increased further. Although MMP-8 and elastase-2 will require further prospective validation studies for confirmation, the results are a relevant demonstration that our gene expression methodology can successfully be leveraged against the tremendous heterogeneity in patients with SSAKI.
The biological links between MMP-8, elastase-2, and SSAKI are unclear at the present time. MMP-8 is a collagen-cleaving endopeptidase, secreted by neutrophils, involved in extracellular matrix remodeling. MMP-8 is implicated in the pathogenesis of numerous rheumatologic diseases and has been demonstrated as a biomarker for disease processes ranging from odontogenic inflammation, to left ventricular remodeling after myocardial infarction, to preterm labor [46–48]. We have recently identified that MMP-8 may be important in the pathogenesis of septic shock [49], and Hu and colleagues demonstrated that the use of an MMP-8 inhibitor protects mice against endotoxic shock [50]. There are no previous reports of MMP-8 involvement in the progression of kidney injury. Neutrophil elastase (elastase-2) is a serine protease secreted by neutrophils in response to bacterial infection, is a marker of inflammation, and has been documented as a biomarker of neonatal sepsis [51]. Interestingly, inhibition of neutrophil elastase reduced progressive renal injury seen in experimental direct endotoxin-induced AKI [52].
Our study has several limitations. Since in many cases neither a baseline creatinine nor a height was available, the definition of SSAKI in our population was based on a referenced median-for-age creatinine [34, 35]. Importantly, the initial pediatric RIFLE study also identified AKI in patients based on assumed estimated creatinine clearance when baseline creatinine levels were unavailable [53]. Second, some of the performance characteristics (negative and positive predictive values) are influenced by the relatively low prevalence of AKI in our cohort. This is probably secondary to the stringent a priori definition used for SSAKI. The intentional use of such a definition allowed us to identify the most severely affected patients with fluid-unresponsive AKI [22]. Third, the data for MMP-8 and elastase-2 continue to demonstrate the relatively low specificity that many candidate biomarkers carry for SSAKI. Still, when all 21 upregulated gene probes are analyzed using the leave-one-out procedure, the specificity increases to 80%, a level not yet seen for SSAKI biomarkers. Fourth, neither MMP-8 nor elastase-2 has been studied formally in AKI disease pathogenesis, from sepsis or other causes. Since MMP-8 and elastase-2 are neutrophil-derived proteases, it is tempting to speculate that they contribute to neutrophil tissue invasion and end-organ damage. In support of such speculation, our microarray data indicate that septic shock is associated with increased expression of several basement membrane-related genes (for example, laminin-family genes) that contribute to neutrophil diapedesis. Finally, our approach relies on extrapolation of mRNA expression data for the derivation of serum protein-based biomarkers. This can be problematic in that mRNA expression does not always correlate with protein expression. What will be required in the future is exploration into the feasibility of measuring the other candidate biomarkers, such as IFNα-inducible protein 27, which demonstrated a more than twofold mRNA expression level between patients with and without SSAKI.
Conclusions
In summary, we present a genomic-based methodology that can be used to identify novel candidate biomarkers for SSAKI. Additional validation studies and multicenter studies are required to investigate the potential of SSAKI biomarkers identified by gene array.
Key messages
-
We have used a microarray-based gene expression database to derive a panel of candidate biomarkers for early detection of SSAKI in critically ill children with septic shock.
-
The expression patterns of 21 gene probes were able to predict the development of SSAKI with a high degree of sensitivity and specificity.
-
The serum protein levels corresponding to two of these gene probes (MMP-8 and neutrophil elastase-2) were able to predict SSAKI with a high degree of sensitivity and a high negative predictive value in both a derivation cohort and a validation cohort.
-
The identified candidate genes could provide a foundation to robustly predict SSAKI.
Abbreviations
- AKI:
-
acute kidney injury
- CI:
-
confidence interval
- IFN:
-
interferon
- IL:
-
interleukin
- MMP-8:
-
matrix metalloproteinase-8
- NGAL:
-
neutrophil gelatinase-associated lipocalin
- PICU:
-
pediatric intensive care unit
- RIFLE:
-
Risk: Injury: Failure: Loss: End-Stage kidney injury
- ROC:
-
receiver operating characteristic
- SSAKI:
-
septic-shock-associated acute kidney injury.
References
Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR: Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001, 29: 1303-1310. 10.1097/00003246-200107000-00002.
Levy MM, Dellinger RP, Townsend SR, Linde-Zwirble WT, Marshall JC, Bion J, Schorr C, Artigas A, Ramsay G, Beale R, Parker MM, Gerlach H, Reinhart K, Silva E, Harvey M, Regan S, Angus DC: The Surviving Sepsis Campaign: results of an international guideline-based performance improvement program targeting severe sepsis. Crit Care Med. 2010, 38: 367-374. 10.1097/CCM.0b013e3181cb0cdc.
Watson RS, Carcillo JA, Linde-Zwirble WT, Clermont G, Lidicker J, Angus DC: The epidemiology of severe sepsis in children in the United States. Am J Respir Crit Care Med. 2003, 167: 695-701. 10.1164/rccm.200207-682OC.
Clermont G, Acker CG, Angus DC, Sirio CA, Pinsky MR, Johnson JP: Renal failure in the ICU: comparison of the impact of acute renal failure and end-stage renal disease on ICU outcomes. Kidney Int. 2002, 62: 986-996. 10.1046/j.1523-1755.2002.00509.x.
Uchino S: The epidemiology of acute renal failure in the world. Curr Opin Crit Care. 2006, 12: 538-543. 10.1097/01.ccx.0000247448.94252.5a.
Bagshaw SM, George C, Bellomo R: Early acute kidney injury and sepsis: a multicentre evaluation. Crit Care. 2008, 12: R47-
Schneider J, Khemani R, Grushkin C, Bart R: Serum creatinine as stratified in the RIFLE score for acute kidney injury is associated with mortality and length of stay for children in the pediatric intensive care unit. Crit Care Med. 2010, 38: 933-939. 10.1097/CCM.0b013e3181cd12e1.
Uchino S, Kellum JA, Bellomo R, Doig GS, Morimatsu H, Morgera S, Schetz M, Tan I, Bouman C, Macedo E, Gibney N, Tolwani A, Ronco C: Acute renal failure in critically ill patients: a multinational, multicenter study. JAMA. 2005, 294: 813-818. 10.1001/jama.294.7.813.
Bagshaw SM, Uchino S, Bellomo R, Morimatsu H, Morgera S, Schetz M, Tan I, Bouman C, Macedo E, Gibney N, Tolwani A, Oudemans-van Straaten HM, Ronco C, Kellum JA: Septic acute kidney injury in critically ill patients: clinical characteristics and outcomes. Clin J Am Soc Nephrol. 2007, 2: 431-439. 10.2215/CJN.03681106.
Moran SM, Myers BD: Course of acute renal failure studied by a model of creatinine kinetics. Kidney Int. 1985, 27: 928-937. 10.1038/ki.1985.101.
Coca SG, Yalavarthy R, Concato J, Parikh CR: Biomarkers for the diagnosis and risk stratification of acute kidney injury: a systematic review. Kidney Int. 2008, 73: 1008-1016. 10.1038/sj.ki.5002729.
Parikh CR, Lu JC, Coca SG, Devarajan P: Tubular proteinuria in acute kidney injury: a critical evaluation of current status and future promise. Ann Clin Biochem. 2010, 47 (Pt 4): 301-312.
Mishra J, Dent C, Tarabishi R, Mitsnefes MM, Ma Q, Kelly C, Ruff SM, Zahedi K, Shao M, Bean J, Mori K, Barasch J, Deverajan P: Neutrophil gelatinase-associated lipocalin (NGAL) as a biomarker for acute renal injury after cardiac surgery. Lancet. 2005, 365: 1231-1238. 10.1016/S0140-6736(05)74811-X.
Zappitelli M, Washburn KK, Arikan AA, Loftis L, Ma Q, Devarajan P, Parikh CR, Goldstein SL: Urine neutrophil gelatinase-associated lipocalin is an early marker of acute kidney injury in critically ill children: a prospective cohort study. Crit Care. 2007, 11: R84-10.1186/cc6089.
Bagshaw SM, Langenberg C, Haase M, Wan L, May CN, Bellomo R: Urinary biomarkers in septic acute kidney injury. Intensive Care Med. 2007, 33: 1285-1296. 10.1007/s00134-007-0656-5.
Bagshaw SM, Bellomo R: Urine abnormalities in acute kidney injury and sepsis. Contrib Nephrol. 2010, 165: 274-283.
Bagshaw SM, Bennett M, Haase M, Haase-Fielitz A, Egi M, Morimatsu H, D'Amico G, Goldsmith D, Devarajan P, Bellomo R: Plasma and urine neutrophil gelatinase-associated lipocalin in septic versus non-septic acute kidney injury in critical illness. Intensive Care Med. 2010, 36: 452-461. 10.1007/s00134-009-1724-9.
Wan L, Bagshaw SM, Langenberg C, Saotome T, May C, Bellomo R: Pathophysiology of septic acute kidney injury: what do we really know?. Crit Care Med. 2008, 36 (4 Suppl): S198-S203.
Basu RK, Devarajan P, Wong H, Wheeler DS: An update and review of acute kidney injury in pediatrics. Pediatr Crit Care Med. 2011, 12: 339-347. 10.1097/PCC.0b013e3181fe2e0b.
Kellum JA, Ronco C, Mehta RL: Fluid management in acute kidney injury. Int J Artif Organs. 2008, 31: 94-95.
Venkataraman R: Can we prevent acute kidney injury?. Crit Care Med. 2008, 36 (4 Suppl): S166-S171.
Himmelfarb J, Joannidis M, Molitoris B, Schietz M, Okusa MD, Warnock D, Laghi F, Goldstein SL, Prielipp R, Parikh CR, Pannu N, Lobo SM, Shah S, D'Intini V, Kellum : Evaluation and initial management of acute kidney injury. Clin J Am Soc Nephrol. 2008, 3: 962-967. 10.2215/CJN.04971107.
Basu RK, Devarajan P, Wong H, Wheeler DS: An update and review of acute kidney injury in pediatrics. Pediatr Crit Care Med. 2011, 12: 339-347. 10.1097/PCC.0b013e3181fe2e0b.
Karvellas CJ, Farhat MR, Sajjad I, Mogensen SS, Leung AA, Wald R, Bagshaw SM: A comparison of early versus late initiation of renal replacement therapy in critically ill patients with acute kidney injury: a systematic review and meta-analysis. Crit Care. 2011, 15: R72-10.1186/cc10061.
Cho KC, Himmelfarb J, Paganini E, Ikizler TA, Soroko SH, Mehta RL, Chertow GM: Survival by dialysis modality in critically ill patients with acute kidney injury. J Am Soc Nephrol. 2006, 17: 3132-3138. 10.1681/ASN.2006030268.
Bellomo R, Prowle JR, Echeverri JE, Ligabo V, Ronco C: Fluid management in septic acute kidney injury and cardiorenal syndromes. Contrib Nephrol. 2010, 165: 206-218.
Wong HR, Wheeler DS, Tegtmeyer K, Poynter SE, Kaplan JM, Chima RS, Stalets E, Basu RK, Doughty LA: Toward a clinically feasible gene expression-based subclassification strategy for septic shock: proof of concept. Crit Care Med. 2010, 38: 1955-1961.
Cvijanovich N, Shanley TP, Lin R, Allen GL, Thomas NJ, Checchia P, Anas N, Freishtat RJ, Monaco M, Odoms K, Sakthivel B, Wong HR: Validating the genomic signature of pediatric septic shock. Physiol Genomics. 2008, 34: 127-134. 10.1152/physiolgenomics.00025.2008.
Shanley TP, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, Kalyanaraman M, Tofil NM, Penfil S, Monaco M, Odoms K, Barnes M, Sakthivel B, Aronow BJ, Wong HR: Genome-level longitudinal expression of signaling pathways and gene networks in pediatric septic shock. Mol Med. 2007, 13: 495-508.
Wong HR, Cvijanovich N, Allen GL, Lin R, Anas N, Meyer K, Freishtat RJ, Monaco M, Odoms K, Sakthivel B, Shanley TP: Genomic expression profiling across the pediatric systemic inflammatory response syndrome, sepsis, and septic shock spectrum. Crit Care Med. 2009, 37: 1558-1566. 10.1097/CCM.0b013e31819fcc08.
Wong HR, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Willson DF, Freishtat RJ, Anas N, Meyer K, Checchia PA, Monaco M, Odoms K, Shanley TP: Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med. 2009, 7: 34-10.1186/1741-7015-7-34.
Wong HR, Shanley TP, Sakthivel B, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, Kalyanaraman M, Tofil NM, Penfil S, Monaco M, Tagavilla MA, Odoms K, Dunsmore K, Barnes M, Aronow BJ: Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol Genomics. 2007, 30: 146-155. 10.1152/physiolgenomics.00024.2007.
Goldstein B, Giroir B, Randolph A: International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med. 2005, 6: 2-8. 10.1097/01.PCC.0000149131.72248.E6.
Finney H, Newman DJ, Thakkar H, Fell JM, Price CP: Reference ranges for plasma cystatin C and creatinine measurements in premature infants, neonates, and older children. Arch Dis Child. 2000, 82: 71-75. 10.1136/adc.82.1.71.
Bailey D, Phan V, Litalien C, Ducruet T, Merouani A, Lacroix J, Gauvin F: Risk factors of acute renal failure in critically ill children: a prospective descriptive epidemiological study. Pediatr Crit Care Med. 2007, 8: 29-35. 10.1097/01.pcc.0000256612.40265.67.
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP: Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003, 4: 249-264. 10.1093/biostatistics/4.2.249.
Neveu H, Kleinknecht D, Brivet F, Loirat P, Landais P: Prognostic factors in acute renal failure due to sepsis. Results of a prospective multicentre study. The French Study Group on Acute Renal Failure. Nephrol Dial Transplant. 1996, 11: 293-299.
Hoste EA, De Corte W: Epidemiology of AKI in the ICU. Acta Clin Belg Suppl. 2007, 2: 314-317.
Chawla LS, Seneff MG, Nelson DR, Williams M, Levy H, Kimmel PL, Macias WL: Elevated plasma concentrations of IL-6 and elevated APACHE II score predict acute kidney injury in patients with severe sepsis. Clin J Am Soc Nephrol. 2007, 2: 22-30.
Simmons EM, Himmelfarb J, Sezer MT, Chertow GM, Mehta RL, Paganini EP, Soroko S, Freedman S, Becker K, Spratt D, Shyr Y, Ikizler TA: Plasma cytokine levels predict mortality in patients with acute renal failure. Kidney Int. 2004, 65: 1357-1365. 10.1111/j.1523-1755.2004.00512.x.
Wheeler DS, Devarajan P, Ma Q, Harmon K, Monaco M, Cvijanovich N, Wong HR: Serum neutrophil gelatinase-associated lipocalin (NGAL) as a marker of acute kidney injury in critically ill children with septic shock. Crit Care Med. 2008, 36: 1297-1303. 10.1097/CCM.0b013e318169245a.
de Geus HR, Bakker J, Lesaffre EM, le Noble JL: Neutrophil gelatinase-associated lipocalin at ICU admission predicts for acute kidney injury in adult patients. Am J Respir Crit Care Med. 2011, 183: 907-914. 10.1164/rccm.200908-1214OC.
Shapiro NI, Trzeciak S, Hollander JE, Birkhahn R, Otero R, Osborn TM, Moretti E, Nguyen HB, Gunnerson K, Milzman D, Galeski DF, Goyal M, Cairns CB, Kupfer K, Lee SW, Rivers EP: The diagnostic accuracy of plasma neutrophil gelatinase-associated lipocalin in the prediction of acute kidney injury in emergency department patients with suspected sepsis. Ann Emerg Med. 2010, 56: 52-59. 10.1016/j.annemergmed.2010.02.010. e51
Bellomo R, Wan L, Langenberg C, May C: Septic acute kidney injury: new concepts. Nephron Exp Nephrol. 2008, 109: e95-e100. 10.1159/000142933.
Bagshaw SM, Langenberg C, Bellomo R: Urinary biochemistry and microscopy in septic acute renal failure: a systematic review. Am J Kidney Dis. 2006, 48: 695-705. 10.1053/j.ajkd.2006.07.017.
Rahkonen L, Rutanen EM, Nuutila M, Sainio S, Sorsa T, Paavonen J: Matrix metalloproteinase-8 in cervical fluid in early and mid pregnancy: relation to spontaneous preterm delivery. Prenat Diagn. 2010, 30: 1079-1085. 10.1002/pd.2614.
Sorsa T, Tervahartiala T, Leppilahti J, Hernandez M, Gamonal J, Tuomainen AM, Lauhio A, Pussinen PJ, Mantyla P: Collagenase-2 (MMP-8) as a point-of-care biomarker in periodontitis and cardiovascular diseases. Therapeutic response to non-antimicrobial properties of tetracyclines. Pharmacol Res. 2011, 63: 108-113. 10.1016/j.phrs.2010.10.005.
Zile MR, Desantis SM, Baicu CF, Stroud RE, Thompson SB, McClure CD, Mehurg SM, Spinale FG: Plasma biomarkers that reflect determinants of matrix composition identify the presence of left ventricular hypertrophy and diastolic heart failure. Circ Heart Fail. 2011, 4: 246-256. 10.1161/CIRCHEARTFAILURE.110.958199.
Solan PD, Dunsmore K, Denenberg AG, Odoms K, Zingarelli B, Wong HR: A novel role for matrix metalloproteinase-8 in sepsis. Crit Care Med. 2011, http://www.ncbi.nlm.nih.gov/pubmed/22020238, ,
Hu J, Van den Steen PE, Dillen C, Opdenakker G: Targeting neutrophil collagenase/matrix metalloproteinase-8 and gelatinase B/matrix metalloproteinase-9 with a peptidomimetic inhibitor protects against endotoxin shock. Biochem Pharmacol. 2005, 70: 535-544. 10.1016/j.bcp.2005.04.047.
Kingsmore SF, Kennedy N, Halliday HL, Van Velkinburgh JC, Zhong S, Gabriel V, Grant J, Beavis WD, Tchernev VT, Perlee L, Leinine S, Grimwade B, Sorette M, Edgar JD: Identification of diagnostic biomarkers for infection in premature neonates. Mol Cell Proteomics. 2008, 7: 1863-1875. 10.1074/mcp.M800175-MCP200.
Linas SL, Whittenburg D, Repine JE: Role of neutrophil derived oxidants and elastase in lipopolysaccharide-mediated renal injury. Kidney Int. 1991, 39: 618-623. 10.1038/ki.1991.73.
Akcan-Arikan A, Zappitelli M, Loftis LL, Washburn KK, Jefferson LS, Goldstein SL: Modified RIFLE criteria in critically ill children with acute kidney injury. Kidney Int. 2007, 71: 1028-1035. 10.1038/sj.ki.5002231.
Iglesias J, Marik PE, Levine JS: Elevated serum levels of the type I and type II receptors for tumor necrosis factor-alpha as predictive factors for ARF in patients with septic shock. Am J Kidney Dis. 2003, 41: 62-75. 10.1053/ajkd.2003.50024.
Hoste EA, Lameire NH, Vanholder RC, Benoit DD, Decruyenaere JM, Colardyn FA: Acute renal failure in patients with sepsis in a surgical ICU: predictive factors, incidence, comorbidity, and outcome. J Am Soc Nephrol. 2003, 14: 1022-1030. 10.1097/01.ASN.0000059863.48590.E9.
Martensson J, Bell M, Oldner A, Xu S, Venge P, Martling CR: Neutrophil gelatinase-associated lipocalin in adult septic patients with and without acute kidney injury. Intensive Care Med. 2010, 36: 1333-1340. 10.1007/s00134-010-1887-4.
Acknowledgements
The present work was supported by grants from the National Institutes of Health (R01GM064619 and RC1HL100474 to HRW).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
RKB assisted with data analysis and wrote the manuscript. SWS assisted with data analysis and database mining. NZC, GLA, NJT, RJF, NA, KM, PAC, RL, TPS, and MTB contributed patient samples and clinical data to the database. They also reviewed the manuscript prior to submission. DSW, PD, and SLG assisted in the initial study design, assisted with the data analysis, and edited the manuscript. HRW conceived the study, performed the majority of the analyses, and assisted with the writing of the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
13054_2011_9795_MOESM1_ESM.DOC
Additional file 1: Gene probes differentially regulated between patients with and without acute kidney injury. Additional File 1 is a table listing the gene probes identified from mRNA of patient whole blood samples that are differentially regulated (up or down) in patients with acute kidney injury versus those without kidney injury. (DOC 158 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Basu, R.K., Standage, S.W., Cvijanovich, N.Z. et al. Identification of candidate serum biomarkers for severe septic shock-associated kidney injury via microarray. Crit Care 15, R273 (2011). https://doi.org/10.1186/cc10554
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/cc10554