Abstract
Background
Ewing sarcoma (ES) is an aggressive childhood bone and soft tissue cancer. KIAA1429 is one type of N6-methyladenosine (m6A) writer that plays a tumor-progressive role in various cancers, but the role of KIAA1429 in ES remains to be elucidated. The aim of the study was to investigate the role of KIAA1429 in ES.
Methods
We performed a multi-omic screen including CRISPR-Cas9 functional genomic and transcriptomic approaches, and identified that KIAA1429 played a significant role in ES progression. Gene knockdown, quantitative real-time PCR (Q-RT-PCR), immunoblotting, CellTiter-Glo assays, clonogenic assays, a subcutaneous xenograft model and immunohistochemistry were used to assess the functional role of KIAA1429 in ES. We mainly conducted RNA sequencing (RNA-seq) in ES cells to analyze the downstream regulatory mechanism of KIAA1429. An integrative analysis of chromatin immunoprecipitation sequencing (ChIP-seq) and RNA-seq indicated the upstream regulatory mechanism of KIAA1429.
Results
In vitro and in vivo CRISPR-Cas9 knockout screening identified KIAA1429 as an ES-dependent gene. Genetic suppression of KIAA1429 inhibited ES cell proliferation and tumorigenicity both in vitro and in vivo. Further studies revealed that KIAA1429 promotes ES tumorigenesis by regulating the ribosome-associated cell cycle and cancer-related inflammation. Interestingly, we found that STAT3 was a target of KIAA1429 and that a STAT3 inhibitor reduced KIAA1429 transcript levels, indicating positive feedback between KIAA1429 and STAT3. Finally, we found that NKX2-2 bound to the KIAA1429 promoter and transactivated KIAA1429.
Conclusion
Our study systematically analyzed ES-dependent epigenetic/transcriptional regulatory genes and identified KIAA1429 as a biomarker of tumor progression in ES, providing a potential therapeutic target for treating ES.
Similar content being viewed by others
Introduction
Ewing sarcoma (ES) is generally recognized as an aggressive cancer of bone (approximately 70%) and soft tissue (approximately 30%) that affects children, adolescents and young adults [1, 2]. ES is molecularly characterized by a fusion of an FET gene and an ETS gene, and the most common fusion type is EWS-FLI1 [1, 2]. Despite the fact that therapeutic developments in ES over the past decade have improved the 5-year overall survival rate (~70%), many patients suffer from severe bone ache, and more than 25% of patients are diagnosed with distant metastases, impairing quality of life and finally reducing survival [1, 3]. Most patients with ES usually show relapse within 2 years of initial diagnosis and require complex therapeutic strategies, including chemotherapy, surgery and other therapies [2]. However, these approaches are sometimes limited and are followed by a variety of complications, such as loss of appetite, malnutrition and myelotoxicity in chemotherapy and shock, pain and amputation in surgery [4]. In general, ES is difficult to treat, and it is urgent to further explore the mechanism of ES to find new treatment alternatives.
Epigenetic/transcriptional regulation including DNA methylation, histone modification, nucleosome remodeling, and modulation of three-dimensional chromatin structure, is critical to physiologic and pathophysiologic control of cell fate and proliferation [5, 6]. Multiple previous studies have indicated that epigenetic control plays a pivotal role in ES progression [6]. In particular, previous pan-cancer genome studies have shown high-frequency epigenetic/transcriptional-associated mutations and low-frequency somatic mutations in childhood tumors, indicating that epigenetic/transcriptional-associated pathogenesis and targeted therapeutic strategies are worthy of further study [7, 8]. The N6-methyladenosine (m6A) modification is an epigenetic modification that refers to the addition or removal of a methyl group to/from the nitrogen the 6th carbon of adenine nucleotide, which is the most common epigenetic modification in RNA molecules [9]. The m6A network involves writers such as METTL3 and METTL14, readers such as IGF2BP3 and YTHDF2, and erasers such as ALKBH5 and FTO [10]. Previous studies showed that the m6A process is involved in cancer progression; however, few reports on this topic have been found for ES [11].
Based on CRISPR-Cas9 screening of whole genome and of epigenetic/transcriptional-targeted genes in vitro and in vivo, our study identified KIAA1429 (also called Vir Like M6A Methyltransferase Associated, VIRMA), a “writer” of m6A modification [9], as an ES-dependent gene. Furthermore, we demonstrated that KIAA1429 was characterized by high expression and tumorigenic dependence and promoted ES proliferation in in vitro and in vivo models. We also used transcriptomic and epigenomic approaches to further investigate the upstream and downstream mechanisms underlying KIAA1429 in ES.
Materials and methods
CRISPR-Cas9 knockout screening
CRISPR-Cas9 screening was performed as described previously [12]. Briefly, human ES cells (A673 and SKNMC cells) were transduced with lentiCas9-Blast (Addgene, 52962) (MOI<0.7) to generate Cas9-expressing cells, and then the cells were transduced with an sgRNA library targeting epigenetic/transcriptional regulatory genes (MOI < 0.3). The sgRNA library contains 6,528 sgRNAs (1,103 epigenetic/transcriptional genes with 6,379 sgRNAs and 149 nontargeting control (NTC) sgRNAs) (Supplementary Table 1). Transduced cells were subsequently cultured in vitro or subcutaneously injected into Nu/Nu mice for viability screening in vitro or in vivo. Genomic DNA was extracted from cultured cells and subcutaneous tumors for sequencing and was further analyzed by the MAGECK algorithm [13] (Supplementary Table 2).
Cell culture
ES cell lines A673 and SKNMC were purchased from the Cell Bank of Chinese Academy of Sciences. A673 and virus-packaging HEK293T cells were cultured in Dulbecco’s modified Eagle medium/high glucose (DMEM) (BasalMedia, #L110KJ) supplemented with 10% fetal bovine serum (FBS; Sigma, #F2442) and a 1× penicillin streptomycin (P/S) solution (BasalMedia, #S110JV). SKNMC cells were cultured in Eagle's minimum essential medium (MEM) (BasalMedia, #L510KJ) supplemented with 10% FBS and 1×P/S. STR sequencing of ES cell lines was performed by BIOWING Biotech Co., Ltd (Shanghai, China) (Supplemental Table 3).
Virus packaging and transfection
Lentiviral shRNA plasmids were constructed by inserting target oligonucleotides into the pLKO.1 (Addgene, #10878) plasmid. Plasmid DNA was extracted using a DNA extraction kit (Vazyme, DC112-01). Lentivirus was packaged by transfecting the plasmids with packaging vectors (psPAX and pMD2.G) and PEI MAX solution (Polysciences, #24765) into HEK293T cells. The viral supernatant was then collected, filtered with a 0.45 μm strainer, concentrated with PEG6000 (Sigma, #81253), resolved in PBS and then aliquoted for further transfection. Cells were infected with viruses and selected with puromycin (1 μg/mL, Yeasen, 60210ES25) for 72 hours. The target oligonucleotides are listed in Supplemental Table 4.
Immunoblotting
Cell samples were lysed in RIPA buffer (Thermo Fisher Scientific, #89900) and protein concentration was quantified using the Pierce BCA kit (Thermo Fisher Scientific, #23225). Denatured proteins (10-20 μg/lane) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were then blocked with 5% fat-free milk (BD Biosciences, #232100) in Tris buffered saline containing Tween 20 (TBST), followed by incubation of rabbit anti-human KIAA1429 (1:500; Proteintech, #25712-1-AP) or rabbit anti-human β-tubulin (1:5000; Abcam, #ab6046) antibody overnight. Then HRP-conjugated goat anti-rabbit IgG (0.2 μg/ml; Pierce, #31460) antibody was used. A luminescence image analyzer (Fujifilm, LAS-4000) was used to visualize the bands after incubation with enhanced chemiluminescence reagents (Millipore, WBKLS0500).
Quantitative reverse transcription PCR (Q-RT-PCR)
Approximately 2.5x105 cells were lysed with TRIzol reagent (Thermo Fisher Scientific, #TR118) and total RNA was extracted. RNA was reverse transcribed to cDNA with a High-Capacity RNA-to-cDNA kit (Thermo Fisher Scientific, #4387406). Quantitative PCR was performed using an Applied Biosystems QuantStudio™ 5 Real-Time PCR System (Thermo Fisher Scientific, #A34322) with SYBR Green Master buffer (ROX) (Thermo Fisher Scientific, #A25742). GAPDH was used as an internal control, and mRNA levels were calculated using the 2^delta Ct method. The primer sequences are summarized in Supplemental Table 4.
Cell viability assay
Cells were seeded in triplicate in 96-well plates at a density of 2000 cells/well in 100 μL of culture medium. The CellTiter-Glo® luminescent cell viability assay (Promega, #G7573) was then performed to assess cell viability on days 0, 2 and 4 according to the manufacturer’s protocol.
Colony formation assay
A total of 500 cells/well were seeded in 6-well plates. Fresh medium was added every 5 days in the first 7 days. Half of the medium was removed and then the same amount of new medium was added every 4 days afterward. Generally, colonies were visible after 8-21 days, and the cells were washed with PBS, fixed with a 10% neutralized formaldehyde solution, and then stained with 0.5% crystal violet (Sigma, #C6158-100G) containing 25% methanol.
FACS assay
Cell proliferation and apoptosis were measured with Click-iT™ Plus EdU Flow Cytometry Assay Kits (Invitrogen, C10632) and FITC Annexin V Apoptosis Detection Kit (MultiSciences, AT-101) according to the manufacturer’s instructions, respectively. Fluorescence activated cell sorting (FACS) analysis was performed on a CytoFLEX FACS instrument (Beckman Coulter) and the data were analyzed with FlowJo software (FlowJo).
Tetracycline-inducible VIRMA knockdown (Tet-on)
The shKIAA1429 sequences were cloned and inserted into the Tet-on puromycin-resistant plasmid (Addgene, #21915). Tet-on-shKIAA1429 plasmids were used for virus packaging in HEK293T cells and stably transfected ES cells were established by puromycin selection. KIAA1429 knockdown was induced in vitro using doxycycline (Dox) at a concentration of 200 ng/mL.
Animal experiments
The Medical Experimental Animal Administrative Committee in Shanghai approved all animal experiments. BALB/c nude female mice (4-6 weeks) were purchased from the Experimental Animal Center of the Chinese Academy of Sciences (Shanghai, China). For subcutaneous cell line xenografts, 2.5 x 106 ES Tet-on-shKIAA1429 cells were subcutaneously transplanted in the dorsal flanks of mice on each side. When the tumors reached a volume of approximately 100 mm3, the mice were randomly divided into two groups and given water containing 2 mg/mL Dox with 2% sucrose or 2% sucrose as a control. Tumor volume was measured with the formula 1/2 (long axis * short axis^2). Mice with tumors larger than 1,500 mm3 were euthanized.
Histological analysis
Histological analyses, including hematoxylin-eosin (HE) and immunohistochemical (IHC) staining, were performed by Servicebio Biotechnology Company (Shanghai, China). IHC staining was performed using a primary anti-Ki-67 antibody (Servicebio, #GB121499). The stained cells were counted as a percentage of the total positive cells in the five random fields of view using the IHC profiler plugin in ImageJ software (v1.52p, USA).
Public data acquisition
The transcriptomic datasets of ES tissues (GSE17679, GSE34620, GSE12102 and GSE142162), ES cells (GSE17679 and GSE36133), normal skeletal muscles (GSE17679 and GSE38718) and human mesenchymal stem cells (hMSC) (GSE7888), and chromatin immunoprecipitation sequencing (ChIP-seq) datasets (GSE141493, GSE176400) for Fig. 7D were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Data on expression and dependency in cancer cell lines were downloaded from the DepMap database (https://depmap.org/portal/; version 22Q2). ChIP-seq datasets in Fig. 6H were downloaded from the Cistrome database (http://cistrome.org/).
Analysis of RNA sequencing (RNA-seq) and public ChIP-seq data
The analytical approach was previously described [14, 15]. For RNA-seq analysis of our own samples, reads were mapped to the hg38 reference genome using HISAT2. Read counts were generated with HTSeq (version 0.11.1) and fragments per kilobase million (FPKM) values were calculated. ChIP-seq data were mapped and visualized using the Integrative Genome Viewer (IGV; version 2.16.0) software and WashU Epigenome Browse (http://epigenomegateway.wustl.edu/browser/) according to BigWig files on the hg19 genome track.
ChIP-qPCR analysis
ChIP-qPCR was performed as previously described [16]. Briefly, 1 × 107 cells were prepared for chromatin immunoprecipitation. Subsequently, 10 μg of chromatin was immunoprecipitated with 10 μL STAT3 (Cell Signaling Technology, 12640S) or 1 μL IgG antibody (Cell Signaling Technology, 2729) and 50 μL Pierce™ ChIP-grade Protein A/G magnetic beads. Primers for qPCR were designed according to the peak sequence of STAT3 at the promoter of KIAA1429 (Fig .6H) via online software (https://bioinfo.ut.ee/primer3-0.4.0/), and the sequences were listed in Supplementary Table 4.
Identification of differentially expressed genes (DEGs) and construction of a Venn diagram
For public microarray dataset analyses, DEGs were acquired using the Transcriptome Analysis Console (TAC, V4.0.1) software with default settings. For RNA-seq data, DEGs were analyzed using the false discovery rate (FDR) moderated limma test (package “DESeq2” in R). The cutoff was set for DEG selection based on the criterion of |log2 (fold change, FC)| > 0.6 with FDR < 0.05 in public datasets or |log2FC| > 0.3 with P < 0.05 in RNA-seq data. We constructed a Venn diagram using an available online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) to visualize the overlapping genes.
Gene set enrichment analysis (GSEA)
GSEA was performed using GSEA 4.1.0 software according to the online instructions (http://www.broadinstitute.org/gsea/index.jsp). All cancer-related gene sets were downloaded from the official GSEA website, which includes gene sets (v7.5) of Hallmark (n=50), KEGG (n=186), Reactome (n=1,654) and Gene Ontology (GO; n=10,532). In addition, we extracted the top 500 genes (ranked by P value) after FLI1 knockdown in ES cells from GSE61950, GSE94277 and GSE27524, in combination with the top 500 genes upon the dependency score of A673 and SKNMC cells in the DepMap database, and built the gene set “Core ES signature” and other ES-associated gene sets. A NOM p value <0.05 or FDR q value < 0.25 was considered statistically significant.
Protein–protein interaction (PPI) network, Cytoscape and volcano plot
The STRING database (http://string-db.org/) was applied to determine the PPI network of downregulated genes with an interaction value of 0.4, after which Cytoscape combined with the CytoHubba plugin was used to visualize the PPI networks and hub genes. Volcano plots were visualized using ggplot2 package in R.
Statistical analyses
GraphPad Prism v9.5.1 or R v4.2.1 software was used for statistical analysis. Comparisons between two groups were performed using unpaired two-tailed Student’s t test, and comparisons among more than two groups were performed using one-way ANOVA. In vivo tumorigenicity was compared between the two groups using two-way ANOVA. P < 0.05 was considered to indicate statistical significance.
Results
CRISPR-Cas9 knockout screening identifies KIAA1429 as a critical tumor-dependent gene in ES
To systematically analyze tumor-dependent genes in ES, we first analyzed the public data of a genome-scale CRISPR knockout screen from DepMap. A total of 943 essential genes were identified in 13 ES cells with EWS-FLI1 fusion (Fig. 1A and D). Previous pan-cancer analysis demonstrated that mutation frequencies in childhood cancers were much lower than those in adult cancers, and genes linked to epigenetic modification emerged as the most common [7, 8], suggesting that epigenetic regulation plays a crucial role in pediatric tumors. Thus, we focused on tumor-dependent epigenetic/transcriptional regulatory factors in ES. We performed CRISPR knockout screening targeting epigenetic/transcriptional regulatory factors in two most commonly used ES cells A673 and SKNMC in vitro, and identified 369 and 297 tumor-dependent genes in A673 and SKNMC, respectively (Fig. 1B and E). To further narrow down the tumor-dependent genes of ES in vivo, we performed in vivo CRISPR knockout screening. A total of 319 and 85 tumor-dependent genes were identified in A673 and SKNMC cells, respectively (Fig. 1C and F). Comprehensive analysis of the above conditions identified 42 ES-dependent epigenetic/transcriptional regulatory factors (Fig. 1G, Supplementary Table 5). Among them, several known ES-dependent genes including AURKB, CDK9, MYC, RPA2, SSRP1, SUPT16H were found [17,18,19,20,21]. Functional enrichment analysis showed that RNA processing was the most significantly enriched term (Fig. 1G).
CRISPR-Cas9 knockout screening identifies ES-dependent genes. A Schematic diagram illustrating DepMap CRISPR-Cas9 knockout screening in vitro. B, C Schematic diagram illustrating CRISPR-Cas9 knockout screening targeting epigenetic/transcriptional regulatory genes in vitro (B) and in vivo (C). D Identification of ES-dependent genes in DepMap dataset (13 ES cells with EWS-FLI1 fusion, as shown in Fig. 3A). E, F Identification of ES-dependent epigenetic/transcriptional regulatory genes in ES cells in vitro (E) and vivo (F). G An integrated analysis of DepMap dataset and CRISPR-Cas9 screen targeting epigenetic/transcriptional regulatory genes. ES: Ewing sarcoma; FDR: false discovery rate
Then, we focused on analyzing a total of 17 ES-dependent RNA processing associated epigenetic/transcriptional genes based on the following criteria: (1) significantly upregulated in ES tumor tissues (GSE17679, GSE34620, GSE12102 and GSE142162) versus normal control tissues (GSE17679 and GSE38718) in GEO datasets (GPL570 platform); (2) significantly upregulated in 18 ES cell lines (GSE17679 and GSE36133) versus normal control human mesenchymal stem cell (hMSC) cells in a GEO dataset (GSE7888; GPL570 platform); and (3) the highest expression in ES among cancers in pan-cancer analysis from the CCLE dataset. Based on the above conditions, 4 candidate genes (KIAA1429, RBM14, SNRPD1, SNRPF) were found (Fig. 2A). However, KIAA1429 was the only gene with an effect score in the top 10% for the CRISPR screening of both A673 and SKNMC cells in vitro and in vivo (Fig. 2A).
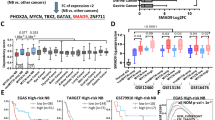
KIAA1429 is identified as an ES-dependent biomarker. A Identification of the essential epigenetic/transcriptional regulatory genes in ES. The yellow section indicated ES-dependent RNA processing genes in our sgRNA library. The purple section indicated: (1) significantly upregulated genes in ES tumor tissues than in normal soft tissues in all four tested GEO datasets (log2FC > 0.6, FDR < 0.05 for ES tissue datasets GSE17679, GSE34620, GSE12102 or GSE142162 versus normal soft tissue dataset GSE38718); (2) significantly upregulated genes in 18 ES cell lines than in normal human mesenchymal stem cells (hMSCs) (log2FC > 0.6, FDR < 0.05 for ES cell datasets GSE17679 or GSE36133 versus hMSC dataset GSE7888). The green section indicated 1.st ranked genes in the pan-cancer analysis from CCLE datasets. B-D Transcriptional profile of ES in tumors (B), cells (C) and among all cancer cell lines in the DepMap database (D). E Representative “Hallmark” molecular signatures revealed by GSEA in 2 ES cohorts. hMSCs: human mesenchymal stem cells. *P < 0.05, **P < 0.01 and *** P < 0.001
In addition, we detected higher KIAA1429 expression in ES tissues than in normal muscle tissues (Fig. 2B), and KIAA1429 expression was higher in ES cells than in human MSCs (hMSCs) (Fig. 2C). In the pan-cancer analysis, KIAA1429 expression was the highest in ES (Fig. 2D). Notably, ES tissue samples showing high expression of KIAA1429 were enriched in gene sets associated with oncogenic malignancies in the GSEA (Fig. 2E).
Inhibition of KIAA1429 suppresses ES cell growth in vitro and in vivo
An analysis of public functional genomics showed that ES tumor growth was highly dependent on KIAA1429 (Fig. 3A). Furthermore, our CRISPR-Cas9 screening showed the ES tumor viability depended on KIAA1429 in ES cells (A673 and SKNMC cells) in vitro (Fig. 3B, C). We then used shRNA approaches to knock down KIAA1429 expression in ES cells (Fig. 3D, E), and KIAA1429 knockdown significantly inhibited ES proliferation in vitro (Fig. 3F). Furthermore, colony formation assays showed that inhibition of KIAA1429 suppressed ES growth (Fig. 3G). We then selected a moderately effective shRNA (shKIAA1429-1) to investigate the effect of KIAA1429 on cell proliferation and cell apoptosis by FACS staining assays, and the results showed that KIAA1429 inhibition reduced cell proliferation and induced cell apoptosis in the two ES lines (Fig. 3H).
Targeting KIAA1429 suppresses ES growth in vitro. A Gene effect data from DepMap showing a strong tumor dependency on KIAA1429 in 13 ES cells with EWS-FLI1 fusion. B-C Dot plots showing the rank of KIAA1429 (left panel) and parallel coordinate plots showing the change of sgRNA (right panel) in CRISPR-Cas9 screening in vitro in A673 (B) and SKNMC (C) cells. D, E KIAA1429 knockdown was measured using Q-RT-PCR and immunoblotting in A673 (D) and SKNMC (E) cells. F The viability of ES cells was measured using CTG after KIAA1429 knockdown. G Colony formation of ES cells after KIAA1429 knockdown. H Cell proliferation and apoptosis analyses of A673 and SKNMC cells after KIAA1429 knockdown by FACS assay. CTG: Cell-Titer-Glo; ES: Ewing sarcoma; shRNA: short hairpin RNA; shSCR: shRNA scrambled control; sgRNA: small guide RNA. *P < 0.05, **P < 0.01 and *** P < 0.001
Our CRISPR-Cas9 screening showed that ES tumor viability depended on KIAA1429 in vivo as well (Fig. 4A). We further validated the inhibitory effect of KIAA1429 suppression on ES growth in vivo by selecting the most powerful shRNA (shKIAA1429-2) to construct a Tet-on system for the induced disruption of KIAA1429 expression. The results showed that dox-induced KIAA1429 knockdown suppressed ES proliferation both in vitro and in vivo (Fig. 4B, C). Histological analysis revealed that KIAA1429 knockdown disrupted tumor microarchitecture (Fig. 4D) and reduced cell proliferation (Fig. 4F). KIAA1429 knockdown and inhibited proliferation were validated using Q-RT-PCR assays (Fig. 4G). Overall, inhibition of KIAA1429 attenuates ES proliferation both in vitro and in vivo.
Targeting KIAA1429 suppresses ES growth in vivo. A Dot plots showing the rank of KIAA1429 (left panel) and parallel coordinate plots showing the change of sgRNA (right panel) in CRISPR-Cas9 screening in vivo in A673 (upper panel) and SKNMC cells (lower panel). B KIAA1429 transcripts and cell viability were measured using Q-RT-PCR and CTG respectively in Tet-on-shKIAA1429 ES cells without or with Dox treatment. C Photographs and tumor growth curve of nude mice subcutaneously xenografted with Tet-on-shKIAA1429 ES cells without or with Dox in feed. D-E Representative images of HE (D) and IHC (E; Ki-67 staining) in tumor tissue from Tet-on-shKIAA1429 Dox-off and Dox-on mice. F Quantification of the percentage of Ki-67 positive cells. G KIAA1429 and Ki67 transcripts were measured using Q-RT-PCR in tumors from Dox-off and Dox-on mice. Dox: doxycycline; HE: hematoxylin and eosin; IHC: immunohistochemistry; Tet: tetracycline. * P < 0.05, **P < 0.01, and *** P < 0.001
KIAA1429 promotes ES tumorigenesis through multiple cancer-associated pathways
Next, we performed unbiased transcriptome analyses of ES cells (between shSCR and shKIAA1429 samples). GSEA showed that high expression of KIAA1429 was mainly enriched in terms related to ES, malignant cell cycle, ribosome, inflammation, undifferentiation, metabolism and vascular formation (Fig. 5A, B). The signatures of ES, cell cycle and inflammation were the 3 main downstream pathways in ES cells with high KIAA1429 expression (Fig. 5A). Therefore, we combined genes extracted from GSEA and KIAA1429-downregulated genes, and subjected them to PPI analysis to further identify the top 10 hub genes in gene networks of ES, cell cycle and cancer-related inflammation (Fig. 5C). Furthermore, we ranked the 30 hub genes of the three populations by P value and found the great significance of HSP90AA1, PSMD14, NFKBIA and STAT3 (Fig. 5D). We validated the potential downstream genes and found that only STAT3 expression was suppressed in two KIAA1429-knockdown ES cells by Q-RT-PCR (Fig. 5E), suggesting that STAT3 might play an important role in KIAA1429-mediated malignancies.
High-expressed KIAA1429 enriches various cancerous pathways in ES. A Functional enrichment map depicting the functional groups of the main GSEA hits for the high-expressed KIAA1429 effects on A673 cells based on shKIAA1429 RNA-seq results. B Representative GSEA of different populations of gene sets in the KIAA1429-high and KIAA1429-low groups in A673 cells. C The “Degree” approach using Cytoscape software (CytoHubba plugin) to show the top 10 hub genes in 3 main populations of gene sets (ES, cell cycle and cancer-associated inflammation). D Volcano plots showing the hub DEGs in A673 cells after KIAA1429 knockdown. The cutoff setting was |log2FC|> 0.3, P < 0.05. E Q-RT-PCR validation of some hub genes in ES cells indicating the common decrease of STAT3. DEGs: differentially expressed genes; ES: Ewing sarcoma; FDR: false discovery rate; GOBP: Gene Ontology, biological process; GSEA: gene set enrichment analysis; Q-RT-PCR: quantitative reverse transcription polymerase chain reaction. *P < 0.05, **P < 0.01, and *** P < 0.001. Nom p value < 0.05 or FDR q value < 0.25 was considered as statistical significance
KIAA1429 and STAT3 might form a positive feedback loop
To further analyze the role of STAT3 in the KIAA1429-associated pathway, we performed GSEA concerning STAT3 signaling and found that high expression of KIAA1429 enriched the STAT3-related signature (Fig. 6A). Knockdown of STAT3 in ES cells significantly suppressed ES growth (Fig. 6B, C), and public functional genomics data also indicated tumor dependency of STAT3 in ES cells (Fig. 6D). Additionally, we observed a positive correlation between STAT3 and KIAA1429 expression in ES tumors (Fig. 6E). Stattic is a type of STAT3 inhibitor, and we generated a killing curve by treating ES cells with this agent (Fig. 6F). Interestingly, inhibition of STAT3 was shown to suppress KIAA1429 transcript expression (Fig. 6G). Moreover, we analyzed several public ChIP-seq datasets of STAT3 in multiple cancers (breast cancer, lymphoma, lung cancer), and the results showed conservative binding of STAT3 at the KIAA1429 promoter (Fig. 6H), and further ChIP-QPCR experiments validated the aforementioned finding in ES cells (A673 and SKNMC) (Fig. 6I), further indicating a potential positive feedback loop between KIAA1429 and STAT3.
KIAA1429 and STAT3 might form a positive feedback loop. A Representative (left panel) and the top 5 (right panel) of GSEA of STAT3-related gene sets in the KIAA1429-high and KIAA1429-low groups in A673 cells. B KIAA1429 knockdown in ES cells (A673 and SKNMC cells) was measured using Q-RT-PCR. C The viability of ES cells was measured using CTG after STAT3 knockdown. D Gene effect data from DepMap showing a relative high level of ES dependency on STAT3. E High correlation between KIAA1429 and STAT3 was visualized in one representative ES tumor dataset. F High drug sensitivity of STAT3 inhibitor (stattic) in ES cells revealed by killing curve of stattic. G KIAA1429 suppression by stattic in ES cells (A673 and SKNMC cells) was measured using Q-RT-PCR. H Gene track showing high binding signals for STAT3 at the KIAA1429 promoter region using public tumor ChIP-seq datasets. Breast cancer cells: MDA-MB-157 and MDA-MB-468. Lymphoma: OCI-Ly10 and OCI-Ly19. Lung cancer: H358. Primer 1–2 indicated the primer locations for ChIP-QPCR. I ChIP-qPCR analyses of STAT3 signals at the promoter of KIAA1429 in A673 and SKNMC cells. ChIP: chromatin immunoprecipitation; ChIP-seq: ChIP sequencing; ES: Ewing sarcoma; FDR: false discovery rate; shSCR: shRNA scrambled control. *P < 0.05, **P < 0.01, and *** P < 0.001
The KIAA1429-related N6-methyladenosine (m6A) network might be induced by NKX2-2
Finally, we aimed to explore the epigenetic/transcriptional upstream signaling of KIAA1429. A pooled analysis was performed using a DepMap dataset of the 1st ranked highly expressed genes, a gene library of epigenetic/transcriptional regulatory genes [15] and more than two ES tumor datasets of genes positively correlated with KIAA1429 to identify 9 genes (Fig. 7A). We ranked them by FC in expression (between ES and other cancer types; Fig. 7B), the results suggested that NKX2-2 might be an upstream regulator in the KIAA1429-mediated pathway with high ES specificity (Fig. 7C). Next, we combined one epigenomic dataset and one transcriptomic dataset, and the analysis showed that the m6A writers KIAA1429 and METTL3 might be downstream genes regulated by NKX2-2 (Figs. 7D, E). Additionally, a positive correlation between NKX2-2 and m6A writers was found in different ES tumor datasets (Fig. 7F).
KIAA1429-related m6A network might be induced by NKX2-2. A Venn diagram showing the 9 overlapping highly expressed and KIAA1429-correlated transcripts in ES. B Ranks of 9 ES-specific transcripts based on the FC in expression. C Transcriptional NKX2-2 profile of ES cells among all cancer cell lines in the DepMap database. D Gene track showing high binding signals for NKX2-2 at the KIAA1429 promoter region (upper panel) and the METTL3 promoter region (lower panel) using public ES ChIP-seq datasets. H3K27ac and H3K4me3 as the markers of promoter. E RNA-seq showing decrease in a set of m6A writers after NKX2-2 knockdown. F Matrix showing the gene–gene correlation value among KIAA1429, METTL3, WTAP and YTHDF3 expression in different ES tumor datasets. H Summary diagram describing the KIAA1429 pathway and potential critical phenotypes in ES cells. ChIP: chromatin immunoprecipitation; ChIP-seq: ChIP sequencing; ES: Ewing sarcoma; FC: fold change; *P < 0.05, **P < 0.01, and *** P < 0.001
Collectively, the results indicate that NKX2-2 regulates KIAA1429-associated m6A writers, facilitating the positive feedback loop between KIAA1429 and STAT3 and reinforcing ES-dependent epigenetic changes, the malignant cell cycle and tumor-triggered inflammation in ES (Fig. 7G).
Discussion
ES is sensitive to chemotherapy, but the treatment of metastatic and recurrent ES is still a challenge for researchers and clinicians [1,2,3]. ES is driven by the interplay of epigenetic regulators [6]; therefore, we performed an integrative analysis to identify KIAA1429 as a potential therapeutic target in ES patients. In addition, we revealed that KIAA1429 positively regulated ES cell viability and tumorigenicity. In particular, KIAA1429 regulated various cancer-associated processes and formed a positive feedback loop with STAT3. NKX2-2 was proved to be an upstream regulator. Taken together, these results indicate that NKX2-2 triggers KIAA1429-associated m6A writers to promote ES progression via the KIAA1429-STAT3 pathway.
KIAA1429 is believed to be a “writer” of the m6A modification complex that influences RNA modification. Previous studies have shown an oncogenic role for KIAA1429 in various types of cancers, such as glioma [22], colorectal cancer [23], germ cell tumors [24] and osteosarcoma [25]. To illustrate, Chai et al. [22] concluded that KIAA1429 was a high-risk gene predicting the prognosis of glioma based on the Chinese Glioma Database. Ma et al. showed that KIAA1429 enhanced the proliferation of colorectal cancer cells by downregulating WEE1, which is that regulated by NK-κB [23]. Miranda et al. demonstrated that KIAA1429 contributed to germ tumor cell progression and cisplatin resistance [24]. A recent study indicated that KIAA1429 increased the proliferation, migration and invasion of osteosarcoma cells [25]. In the present study, we also confirmed the tumor-dependent role of KIAA1429 in ES, further broadening the role of KIAA1429 in tumorigenesis.
Previous studies have shown that aberrant STAT3 signaling sustains cell proliferation, facilitates chemotherapeutic resistance, induces metastasis and plays an anti-apoptotic role in ES tumors [26,27,28,29,30]. A completed phase I clinical trial investigated the safety of simvastatin, which was supposed to inhibit STAT3 signaling (NCT02390843). In addition to ES, different types of STAT3 inhibitors have been applied in glioma (NCT01904123), breast cancer (NCT03195699), lymphoma (NCT01563302), etc., in clinical trials. In our study, STAT3 was identified as an oncogenic target gene of KIAA1429 in ES. Moreover, a transcript-level positive feedback loop between KIAA1429 and STAT3 was revealed in the mechanistic study. These results not only reveal the regulatory mechanism of KIAA1429, but also provide additional information supporting the theoretical basis for the approval of STAT3 targeted drugs in clinical trials of ES.
NKX2-2 has been reported to be a biomarker for ES, and plays critical oncogenic roles in ES progression [31,32,33]. Notably, NKX2-2 is a key transcription factor in forming core regulatory circuitry (CRC) to drive important signaling pathways for tumor development [34]. In our study, we further confirmed the highest expression level of NKX2-2 in ES in the pan-cancer analysis and confirmed nearly undetectable expression in other tumors, as described previously [34]. Epigenome and transcriptome analyses showed that NKX2-2 directly regulates the transcription of KIAA1429. These findings reveal that the restricted expression pattern of NKX2-2 may induce the specific high expression of KIAA1429 in ES. Moreover, several m6A-associated functional components, including MELLT3, WTAP and YTHDF3, were found to be target genes of NKX2-2, suggesting that NKX2-2 probably plays a critical role in the m6A-associated process in ES.
Although epigenetic/transcriptional regulation has been widely reported to be involved in the development, metastasis and drug resistance of various tumors, for most epigenetic/transcription factors, it is difficult to design targeted drugs due to structural disorder and a lack of defined small-molecule binding pockets [35, 36]. However, recent studies have uncovered new targeting strategies for epigenetic/transcription factors; these include direct and indirect approaches, such as the promising direct protein-degradation approach proteolysis targeting chimera (PROTAC) and indirect transcriptional/posttranscriptional modifications [37]. Notably, drug repurposing has risen in popularity, especially in the context of food and drug administration (FDA) approvals and clinical trials, and known pharmacological properties and clinical safety may facilitate the translation of drugs into the clinic [38]. This study provides a novel strategy and direction to identify drugs that specifically inhibit transcription and growth by acting on epigenetic/transcription factors based on a high-throughput drug screening platform. These advanced targeting strategies will provide potential clinical translational significance by revealing new tumor-dependent epigenetic/transcription factors and agents.
Conclusions
The KIAA1429-mediated m6A network facilitates Ewing sarcoma cell proliferation and tumorigenesis via the STAT3 pathway and is regulated by NKX2-2.
Availability of data and materials
The ES tissue transcriptomic datasets: GSE17679 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17679), GSE34620 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE34620), GSE12102 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12102), GSE142162 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142162). ES cell transcriptomic datasets: GSE17679, GSE36133 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE36133). Normal skeletal muscle datasets: GSE17679, GSE38718 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE38718). Human mesenchymal stem cell (hMSC) dataset: GSE7888 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE7888). Chromatin immunoprecipitation sequencing (ChIP-seq) datasets of NKX2-2: GSE141493 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE141493). All above datasets were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Data on expression and dependency in cancer cell lines were downloaded from the Depmap database (https://depmap.org/portal/; version 22Q2). ChIP-seq datasets of STAT3 were downloaded from the Cistrome database (http://cistrome.org/).
Abbreviations
- BP:
-
Biological process
- CC:
-
Cell component
- CCLE:
-
Cancer cell line encyclopedia
- ChIP:
-
Chromatin immunoprecipitation
- COG:
-
Children oncology group
- CRC:
-
Core regulatory circuit
- CTG:
-
Cell titer Glo
- CVS:
-
Crystal violet staining
- Dox:
-
Doxycycline
- ES:
-
Ewing sarcoma
- FC:
-
Fold change
- FDA:
-
Food and drug administration
- FDR:
-
False discovery rate
- GEO:
-
Gene expression omnibus
- GO:
-
Gene ontology
- GSEA:
-
Gene set enrichment analysis
- HE:
-
Hematoxylin and eosin
- hMSCs:
-
Human mesenchymal stem cells
- IHC:
-
Immunohistochemistry
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- MF:
-
Molecular function
- MOI:
-
Multiplicity of infection
- NES:
-
Normalized enrichment score
- NOM:
-
Normalized
- NTC:
-
Nontargeting control
- PPI:
-
Protein to protein interaction
- Q-RT-PCR:
-
Quantitative reversed transcription polymerase chain reaction
- PROTAC:
-
Proteolysis targeting chimera
- shRNA:
-
Short hairpin RNA
- shSCR:
-
ShRNA scrambled control
- STCs:
-
Stable transfected cells
- WT:
-
Wild type
References
Riggi N, Suva ML, Stamenkovic I. Ewing’s Sarcoma. N Engl J Med. 2021;384:154–64.
Grunewald TGP, Cidre-Aranaz F, Surdez D, Tomazou EM, de Alava E, Kovar H, Sorensen PH, Delattre O, Dirksen U. Ewing sarcoma. Nat Rev Dis Primers. 2018;4:5.
Gaspar N, Hawkins DS, Dirksen U, Lewis IJ, Ferrari S, Le Deley MC, Kovar H, Grimer R, Whelan J, Claude L, et al. Ewing Sarcoma: current management and future approaches through collaboration. J Clin Oncol. 2015;33:3036–46.
Shulman DS, Whittle SB, Surdez D, Bailey KM, de Alava E, Yustein JT, Shlien A, Hayashi M, Bishop AJR, Crompton BD, et al. An international working group consensus report for the prioritization of molecular biomarkers for Ewing sarcoma. NPJ Precis Oncol. 2022;6:65.
Tan F, Zhao M, Xiong F, Wang Y, Zhang S, Gong Z, Li X, He Y, Shi L, Wang F, et al. N6-methyladenosine-dependent signalling in cancer progression and insights into cancer therapies. J Exp Clin Cancer Res. 2021;40:146.
Schmidt O, Nehls N, Prexler C, von Heyking K, Groll T, Pardon K, Garcia HD, Hensel T, Gurgen D, Henssen AG, et al. Class I histone deacetylases (HDAC) critically contribute to Ewing sarcoma pathogenesis. J Exp Clin Cancer Res. 2021;40:322.
Ma X, Liu Y, Liu Y, Alexandrov LB, Edmonson MN, Gawad C, Zhou X, Li Y, Rusch MC, Easton J, et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature. 2018;555:371–6.
Grobner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, Johann PD, Balasubramanian GP, Segura-Wang M, Brabetz S, et al. The landscape of genomic alterations across childhood cancers. Nature. 2018;555:321–7.
Zhang Y, Geng X, Li Q, Xu J, Tan Y, Xiao M, Song J, Liu F, Fang C, Wang H. m6A modification in RNA: biogenesis, functions and roles in gliomas. J Exp Clin Cancer Res. 2020;39:192.
Tan K, Wu W, Zhu K, Lu L, Lv Z. Identification and Characterization of a Glucometabolic Prognostic Gene Signature in Neuroblastoma based on N6-methyladenosine Eraser ALKBH5. J Cancer. 2022;13:2105–25.
Jiang J, Fan Q, Qu H, Liu C, Liang T, Chen L, Huang S, Sun X, Chen J, Chen T, et al. Novel prognostic biomarkers, METTL14 and YTHDF2, associated with RNA methylation in Ewing’s sarcoma. Sci Rep. 2022;12:7041.
Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, Sanjana NE, Zhang F. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc. 2017;12:828–63.
Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu XS. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554.
Mo J, Liu F, Sun X, Huang H, Tan K, Zhao X, Li R, Jiang W, Sui Y, Chen X, et al. Inhibition of the FACT Complex Targets Aberrant Hedgehog Signaling and Overcomes Resistance to Smoothened Antagonists. Cancer Res. 2021;81:3105–20.
Tan K, Mo J, Li M, Dong Y, Han Y, Sun X, Ma Y, Zhu K, Wu W, Lu L, et al. SMAD9-MYCN positive feedback loop represents a unique dependency for MYCN-amplified neuroblastoma. J Exp Clin Cancer Res. 2022;41:352.
Mo J, Tan K, Dong Y, Lu W, Liu F, Mei Y, Huang H, Zhao K, Lv Z, Ye Y, Tang Y. Therapeutic targeting the oncogenic driver EWSR1::FLI1 in Ewing sarcoma through inhibition of the FACT complex. Oncogene. 2023 ;42:11-25.
Sanchez-Molina S, Figuerola-Bou E, Blanco E, Sanchez-Jimenez M, Taboas P, Gomez S, Ballare C, Garcia-Dominguez DJ, Prada E, Hontecillas-Prieto L, et al. RING1B recruits EWSR1-FLI1 and cooperates in the remodeling of chromatin necessary for Ewing sarcoma tumorigenesis. Sci Adv. 2020;6:eaba3058.
Flores G, Everett JH, Boguslawski EA, Oswald BM, Madaj ZB, Beddows I, Dikalov S, Adams M, Klumpp-Thomas CA, Kitchen-Goosen SM, et al. CDK9 blockade exploits context-dependent transcriptional changes to improve activity and limit toxicity of Mithramycin for Ewing sarcoma. Mol Cancer Ther. 2020;19:1183–96.
Gollavilli PN, Pawar A, Wilder-Romans K, Natesan R, Engelke CG, Dommeti VL, Krishnamurthy PM, Nallasivam A, Apel IJ, Xu T, et al. EWS/ETS-driven Ewing sarcoma requires BET bromodomain proteins. Cancer Res. 2018;78:4760–73.
Li G, Zhang P, Zhang W, Lei Z, He J, Meng J, Di T, Yan W. Identification of key genes and pathways in Ewing’s sarcoma patients associated with metastasis and poor prognosis. Onco Targets Ther. 2019;12:4153–65.
Mo J, Tan K, Dong Y, Lu W, Liu F, Mei Y, Huang H, Zhao K, Lv Z, Ye Y, Tang Y. Therapeutic targeting the oncogenic driver EWSR1::FLI1 in Ewing sarcoma through inhibition of the FACT complex. Oncogene. 2023;42:11–25.
Chai RC, Wu F, Wang QX, Zhang S, Zhang KN, Liu YQ, Zhao Z, Jiang T, Wang YZ, Kang CS. m(6)A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gliomas. Aging (Albany NY). 2019;11:1204–25.
Ma L, Lin Y, Sun SW, Xu J, Yu T, Chen WL, Zhang LH, Guo YC, Wang YW, Chen T, et al. KIAA1429 is a potential prognostic marker in colorectal cancer by promoting the proliferation via downregulating WEE1 expression in an m6A-independent manner. Oncogene. 2022;41:692–703.
Miranda-Goncalves V, Lobo J, Guimaraes-Teixeira C, Barros-Silva D, Guimaraes R, Cantante M, Braga I, Mauricio J, Oing C, Honecker F, et al. The component of the m(6)A writer complex VIRMA is implicated in aggressive tumor phenotype, DNA damage response and cisplatin resistance in germ cell tumors. J Exp Clin Cancer Res. 2021;40:268.
Luo J, Wang X, Chen Z, Zhou H, Xiao Y. The role and mechanism of JAK2/STAT3 signaling pathway regulated by m6A methyltransferase KIAA1429 in osteosarcoma. J Bone Oncol. 2023;39:100471.
Lai R, Navid F, Rodriguez-Galindo C, Liu T, Fuller CE, Ganti R, Dien J, Dalton J, Billups C, Khoury JD. STAT3 is activated in a subset of the Ewing sarcoma family of tumours. J Pathol. 2006;208:624–32.
Lissat A, Joerschke M, Shinde DA, Braunschweig T, Meier A, Makowska A, Bortnick R, Henneke P, Herget G, Gorr TA, Kontny U. IL6 secreted by Ewing sarcoma tumor microenvironment confers anti-apoptotic and cell-disseminating paracrine responses in Ewing sarcoma cells. BMC Cancer. 2015;15:552.
Zhang Z, Li Y, Huang L, Xiao Q, Chen X, Zhong J, Chen Y, Yang D, Han Z, Shu Y, et al. Let-7a suppresses macrophage infiltrations and malignant phenotype of Ewing sarcoma via STAT3/NF-kappaB positive regulatory circuit. Cancer Lett. 2016;374:192–201.
Lamhamedi-Cherradi SE, Menegaz BA, Ramamoorthy V, Vishwamitra D, Wang Y, Maywald RL, Buford AS, Fokt I, Skora S, Wang J, et al. IGF-1R and mTOR Blockade: Novel Resistance Mechanisms and Synergistic Drug Combinations for Ewing Sarcoma. J Natl Cancer Inst. 2016;108:djw182.
Yu PY, Gardner HL, Roberts R, Cam H, Hariharan S, Ren L, LeBlanc AK, Xiao H, Lin J, Guttridge DC, et al. Target specificity, in vivo pharmacokinetics, and efficacy of the putative STAT3 inhibitor LY5 in osteosarcoma, Ewing’s sarcoma, and rhabdomyosarcoma. PLoS ONE. 2017;12:e0181885.
Fadul J, Bell R, Hoffman LM, Beckerle MC, Engel ME, Lessnick SL. EWS/FLI utilizes NKX2-2 to repress mesenchymal features of Ewing sarcoma. Genes Cancer. 2015;6:129–43.
Yoshida A, Sekine S, Tsuta K, Fukayama M, Furuta K, Tsuda H. NKX2.2 is a useful immunohistochemical marker for Ewing sarcoma. Am J Surg Pathol. 2012;36:993–9.
Owen LA, Kowalewski AA, Lessnick SL. EWS/FLI mediates transcriptional repression via NKX2.2 during oncogenic transformation in Ewing’s sarcoma. PLoS One. 2008;3:e1965.
Shi X, Zheng Y, Jiang L, Zhou B, Yang W, Li L, Ding L, Huang M, Gery S, Lin DC, Koeffler HP. EWS-FLI1 regulates and cooperates with core regulatory circuitry in Ewing sarcoma. Nucleic Acids Res. 2020;48:11434–51.
Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–26.
Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–88.
Henley MJ, Koehler AN. Advances in targeting “undruggable” transcription factors with small molecules. Nat Rev Drug Discov. 2021;20:669–88.
Chen P, Bao T, Yu X, Liu Z. A drug repositioning algorithm based on a deep autoencoder and adaptive fusion. BMC Bioinformatics. 2021;22:532.
Acknowledgements
We thank Dr. Yongqing Chang for writing assistance.
Funding
The work was partially supported by Natural Science Fund of Shanghai Children’s Hospital (2022YGZQ03).
Author information
Authors and Affiliations
Contributions
KT performed functional genomic screening, carried out the molecular genetic studies and bioinformatic analysis, and drafted the manuscript. WL performed functional genomic screening, carried out the immunoassays and FACS assays. FC carried out the bioinformatic analysis and performed the statistical analysis. HS carried out the bioinformatic analysis. YM carried out the bioinformatic analysis. ZC performed the statistical analysis. WW performed the statistical analysis. ZL participated in the design of the study and coordination and helped to draft the manuscript. JM conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Institutional Review Board (IRB) of Shanghai Children’s Hospital (SCH), Shanghai Jiao Tong University School of Medicine, in accordance with the principles of Declaration of Helsinki. In addition, the medical experimental animal administrative committee in Shanghai approved all animal experiment protocols.
Consent for publication
All authors have read and approved the final version of the manuscript, and declared that there was no image duplication, image manipulation and visual plagiarism.
Competing interests
The authors have declared that no competing interest exists.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Tan, K., Lu, W., Chen, F. et al. CRISPR-Cas9 knockout screening identifies KIAA1429 as an essential gene in Ewing sarcoma. J Exp Clin Cancer Res 42, 250 (2023). https://doi.org/10.1186/s13046-023-02828-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13046-023-02828-5