Abstract
Rett syndrome (RTT) is a rare disease but still one of the most abundant causes for intellectual disability in females. Typical symptoms are onset at month 6–18 after normal pre- and postnatal development, loss of acquired skills and severe intellectual disability. The type and severity of symptoms are individually highly different. A single mutation in one gene, coding for methyl-CpG-binding protein 2 (MECP2), is responsible for the disease. The most important action of MECP2 is regulating epigenetic imprinting and chromatin condensation, but MECP2 influences many different biological pathways on multiple levels although the molecular pathways from gene to phenotype are currently not fully understood. In this review the known changes in metabolite levels, gene expression and biological pathways in RTT are summarized, discussed how they are leading to some characteristic RTT phenotypes and therefore the gaps of knowledge are identified. Namely, which phenotypes have currently no mechanistic explanation leading back to MECP2 related pathways? As a result of this review the visualization of the biologic pathways showing MECP2 up- and downstream regulation was developed and published on WikiPathways which will serve as template for future omics data driven research. This pathway driven approach may serve as a use case for other rare diseases, too.
Similar content being viewed by others
Background
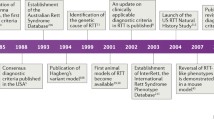
Rett syndrome (RTT; MIM:312750) occurs in 1:10.000 girls at the age of 12 [1]. It is considered a rare disease since it affects fewer than 1 in 2000 individuals [2], but it is still one of the most abundant causes for intellectual disability in females. RTT was first described in 1966 by the Viennese pediatric Andreas Rett, who observed the typical hand movements (“hand washing”) of his patients [3, 4]. Cause of RTT is in most cases a de novo mutation of MECP2 (methyl-CpG-binding protein 2) gene; which was discovered by Amir et al. [5]. However, as stated by Neul et al., “not all mutations in MECP2 cause RTT and not all RTT patients have mutated MECP2”. Some MECP2 mutations cause not RTT but a mild intellectual disability [6] and mutations in two other genes can cause a RTT like phenotype, i.e. FOXG1 and CDKL5. These phenotypes were formerly considered as RTT but are now defined as RTT like syndrome [7].
RTT was considered a neurodevelopmental disorder but since some of the main symptoms were found to be reversible [8] researchers and clinicians tend to categorize it as a neurological disorder now [9]. RTT was also classified as an autism spectrum disorder as patients often develop autistic features like social withdrawal but only during a certain stage of development [10]. Although RTT has some autistic features/phases these usually disappear with time and adult RTT females are quite socially active again [11]. MECP2 mutations are rarely found in autism patients and if, they are termed “Autism with MECP2 mutation” [7].
Recent research was able to find a correlation between certain MECP2 mutations (or MECP2 variants) and some phenotypes, e.g. cardiorespiratory phenotype [12], but most of the biological pathways between gene and phenotype are not yet fully understood. Especially the molecular pathways leading from MECP2 gene to scoliosis, epilepsy or decreased growth are currently not known. In this review, we summarize the knowledge about the molecular interactions of MECP2 gene and protein, their known downstream effects and discuss how pathway and omics data based research can elucidate the pathways towards RTT phenotypes. This review integrates database knowledge from Ensembl [13], OMIM [14], UniProt [15], The Human Protein Atlas [16], and Gene Ontology [17] and a biologic pathway was developed and published on WikiPathways [18] to visualize the mechanistic action of MECP2 and serve as a template for future omics data analysis also in other rare disease models.
Rett syndrome phenotype development within life
Typical development of RTT starts with an “asymptomatic” first stage followed by decreased, arrested and retarded development of motor and communication skills after 6–18 months of normal postnatal development, development of stereotypic movements and loss of purposeful movement. Although the onset of typical disorder symptoms after the age of 6 months is characteristic for RTT, observations of parents that “something is wrong with this child”, are often made before. This matches with newer research which indicates that severe changes in neuronal development are apparent already at this age but due to their mild and uncertain symptoms not able to be diagnosed [19]. After a stagnation stage (2 – 10 years) which can last for years and can include some recovery and secondary gain of abilities the fourth stage typically reduces again mobility (by abnormal muscle tonus and scoliosis) while communication and cognition is preserved. RTT females are typically severely intellectual disabled, have microcephaly and seizures. Additionally, they often develop symptoms like cardiac and breathing abnormalities, gastro-intestinal problems like constipation, low muscle tension, autistic like behavior, scoliosis (and other osteopathies), sleeping problems and hormone disequilibrium. In summary, MECP2 affects epigenetic regulation of gene expression, which changes neurobiological activity, network formation and function, which causes the major phenotype. In summary, MECP2 affects epigenetic regulation of gene expression, which changes neurobiological activity, network formation and function, which causes the major phenotype. Recent longitudinal studies on the lifelong development of RTT stated that survival at the age of 25 years was 77.6% and 59.8% at 37 years [20]. The most abundant causes of death were lower respiratory tract infection, aspiration/asphyxication and respiratory failure. Two-thirds of RTT females had seizures on some point in their lives of which 36.1% were drug resistant [20]. About half of the females completely lost the ability to walk while independent walking was preserved in 17.8%. Scoliosis (85.5%) and abnormal breathing patterns (up to 88.7%) were very abundant [20]. Typical noticeable laboratory results are EEG (and EKG) abnormalities, atypical brain glycolipids, altered neurotransmitter, creatine and growth factor levels, and alkalosis. Most of the symptoms can be related to disturbed neuronal function but some of them are caused or influenced by alterations which are not yet elucidated [4, 21, 22]. It is also unknown whether it is the often in RTT observed changes of carbon dioxide metabolism that cause respiratory problems or that dysfunctional brain stem neurons are responsible for breathing abnormality [4].
MECP2 gene, transcript and protein
The MECP2 gene is highly conserved in Euteleostomi (bony vertebrates). The NCBI HomoloGene/UniGene database gives detailed information about gene homologues in 10 mammalian, 2 amphibian and 1 bony fish species [23]. The human MECP2 is located on chromosome X, position 154,021,573-154,137,103 (reverse strand) (according to Ensembl, version 84, genome build GRCh38.p5 (GCA_000001405.20)) and there are currently 21 transcripts known, two of these are protein coding (Fig. 1). Due to dosage compensation MECP2 is inactivated in one X-chromosome in females and the degree of inactivation is assumed to contribute to the difference in phenotypes for RTT [24]. RTT is more often observed in females due to its location on the X-chromosome. Hemizygous males with a severe mutation are generally not viable, but there are several non-lethal mutations which can lead to severe congenital encephalopathy, RTT-like syndrome, and mild to severe intellectual disability in males [25]. Mosaic expression with only wild-type MECP2 active in females are possible but supposed to be extremely rare [24].
MECP2 gene and protein. MECP2 is located on the X chromosome (X:154021573–154137103) on the reverse strand (ensembl, human genome built 8.2). MECP2 gene is about 10505 bp long and has 4 exons which can be spliced to two protein-coding variants e1 and e2. The protein has 498 (486) amino acids and consists of 6 distinct domains whereas the methyl-DNA binding (MDB) and the transcriptional repression domain (TRD) are the most important for function. Mutation positions are marked with red arrows according to Lyst et al. 2013 [57]. Solid red arrows indicate position of mutations of MECP2, which are present in Rett females but not in their parents. Those are found mostly in MDB and the C-terminal end of TRD. Empty red arrows indicate MECP2 mutations, which do not lead to Rett syndrome. The major phorphorylation sites (S80 and S241) are marked in black [118]
The transcription and translation of MECP2 is highly regulated [26] (Table 1 and Fig. 2). There are several cis- and trans-regulatory elements for MECP2 gene expression regulation known. Cis regulatory elements, including promotor elements, are loci on the DNA which act as binding sites for transcription factors and activate or repress gene expression. Trans-elements affect the regulation in an indirect way and can be located close or far away. They can for instance include genes that encode transcription factors for this specific gene. Translation of MECP2 can be regulated by a set of microRNAs [27–35]. MicroRNAs are small non-coding RNAs, that repress translation mRNA into protein by binding to the 3’ untranslated region of the mRNA. The regulation of MECP2 expression, stimulation and repression, is visualized in Fig. 2 (transcriptional and translational regulation of MECP2) which is derived from WikiPathways [18] pathway ID 3584.
The two coding transcripts are isoforms of MECP2, long e1 and short e2, while e1 seems to be the more important one [36, 37] (Fig. 1). Itoh et al. observed that specific deactivation of e2 did not influence normal neurodevelopment while loss of e1 led to RTT [38]. The MECP2 protein has at least six biochemically distinct domains [39]. Two of them are most important for the protein function: the (84 amino acids) methyl-CpG binding domain (MDB) which is the one which selectively binds 5MeCyt and the transcriptional repression domain (TRD) (102 amino acids) which binds cofactors attracting histone deacetylase and finally leading to transcription repression as explained in the chapter 4 (Fig. 1) [39, 40]. Interestingly, MBD is the only structured domain (α-helix) while 60% of MECP2 is unstructured [39]. There are several post translational modifications of MECP2 known which contribute to its multi-functional properties, phosphorylation, acetylation, SUMOylation, and ubiquitination [41].
MECP2 protein is most abundant in brain but also enriched in lung and spleen tissue [42]. However, according to The Human Protein Atlas database MECP2 protein (and its transcript) is found in quite high amounts in almost every tissue, too. This may actually be the most underestimated part in RTT research which typically focus on neuronal development and function [16]. Many phenotypes and symptoms may be as well deriving from dysfunctional cellular regulation in other organs than central nervous system. In neurons an expression level of about 1.6x107 protein copies per nucleus was estimated by Skene et al. It is about the same number as nucleosomes or 5-methyl-cytosine (5MeCyt) spots on the DNA, leading to the suggestion that every spot might be covered by one MECP2 [43].
MECP2 function
MECP2 is a multifunctional protein which influences gene expression and metabolism on many levels [9] (Fig. 3). The main function of MECP2 is to recognize and bind specifically methylated cytosine residues in the DNA (namely 5MeCyt) that are enriched with A/T bases adjacent [44]. MECP2 binds also but with lesser affinity to hydroxymethylated DNA (namely 5-hydroxy methylated cytosine, 5OHMeCyt). Mutations in MECP2, especially in the MDB, which lead to loss of specific 5MeCyt binding functions are known to cause RTT [45] (Fig. 1).
This is in line with the Gene Ontology classification for the main molecular functions of MECP2: DNA binding, namely double-stranded methylated DNA and protein binding, namely histone deacetylase. The actual full Gene Ontology annotation of MECP2 can be found online e.g. Ensembl database MECP2 entry [13].
The molecular functions of MECP2 are known to influence various biological mechanisms, which are summarized and visualized in the pathway Fig. 2, namely 1) MECP2 influences global translation by enhancing the AKT/mTOR signaling pathway [46], 2) Alternative splicing of downstream gene products is affected because MECP2 forms a complex with YB1, an important splicing factor [29, 47–51], 3) Expression of various microRNAs and long non-coding RNAs is regulated by MECP2 (20, 45, 47–49), and 4) MECP2 triggers the chromatin compaction at methylated DNA sites which regulates the transcription of adjacent genes (34, 37–41). The last one is an important (and best investigated) pathway and will be explained in detail below.
MECP2 as epigenetic regulator
Methylation of DNA is part of epigenetic gene expression regulation, where DNA is modified without changing the genetic code. Most transcription factors are unable to bind to methylated DNA so methylation usually silences a gene. Furthermore, methylated DNA is – via MECP2 mediated cofactor binding - a binding site for histone deacetylase (HDAC) which increases DNA compaction by removing certain acetyl residues from lysine at the histone tail allowing them to get closer to each other (Fig. 2, MECP2-HDAC complex). So, methylated DNA is tightly wrapped around histone proteins and the access of transcription factors is physically inhibited [40]. About 1% of DNA is methylated in humans and the methylation sites are often in regions with a high occurrence of CG, so-called CpG islands. CpG islands are present in the promotor regions of most human genes (60%). Methylation patterns play a role in cellular differentiation and tissue specific gene expression already during early development [52–54]. The methylation pattern is continuously modified and maintained during mitosis and cell differentiation throughout life to grant cellular function [55, 56]. Figure 4 visualizes this circular pathway of DNA methylation, hydroxylation and de-methylation and shows the mode of action of involved proteins including MECP2.
MECP2 recognizes and binds specifically to 5MeCyt present in DNA. After binding it attracts co-repressor complexes containing SIN3A, NCOR and SMRT. This co-repressor complex finally recruits histone deacetylase (HDAC) [29, 40, 47, 57] (Fig. 2, see MECP2 – HDAC complex). The complex acts by removing certain acetyl groups from histone proteins leading to chromatin condensation around methylated DNA [58, 59]. Cell culture experiments with an inhibitor of HDAC resulted in the same phenotype as MECP2-KO cells [60]. The NCOR-SMRT interaction domain (NID) of MECP2 is located within the TRD domain and the association with NCOR-SMRT is responsible for transcription repression activity of MECP2.
MECP2 was generally considered as a transcription inhibitor but recent research found also a conditional transcription activation function. Skene et al. identified MECP2, because of its mere amount, being equal to number of histone octamers or methylated DNA sites, as a global damper of gene transcription in mature mouse brain cells [43]. During MECP2 absence H3 (histone family 3) acetylation levels were globally elevated and H1 (histone family 1) levels doubled suggesting MECP2 alters global chromatin state towards condensation and represses transcription. Li et al. on the other hand found global transcription activation by MECP2 in human embryonic stem cells which underwent differentiation to neuronal cells but repression by MECP2 in mature neuronal cells indicating that MECP2 can be both, an activator and repressor [61]. The activation mechanism is explained as follows: MECP2 recruits CREB1 as a cofactor to target gene promotors [62] (Fig. 2, Activation of transcription). MECP2 binding to 5OHMeCyt was even interpreted as a marker of active genes in neurons [62]. MECP2 was also found to form a TET1 containing complex which leads to 5MeCyt hydroxylation and further to demethylation of DNA, enabling transcription [63]. This mechanism was found to activate expression of downstream genes, namely BDNF, SST, OPRK1, GAMT, GPRIN1, and CREB1 [28] and is contradictory to other findings which describe MECP2 to block DNA demethylation by TET complex (Fig. 4) [64].
MECP2 is additionally responsible for neuronal activity triggered transcription. Neuronal membrane depolarization and Ca2+ influx leads to phosphorylation of MECP2 which makes it detach from DNA, allowing decondensation and transcription (Fig. 2) [65–69]. Specific blocking of MECP2 phosphorylation sites led to RTT like symptoms [70]. For a conclusion, MECP2 is responsible for the epigenetic regulation of gene expression, which changes neurobiological activity, network formation and function which is likely to cause a severe disorder phenotype if the protein is affected by mutation.
Mutations of MECP2 leading to RTT
At the moment, several hundred different mutations have been reported leading to RTT by loss or impaired function of MECP2 protein due to truncation, abnormal folding, or binding instability [25] (see also MECP2 varieties on LOVD database [71]). This contributes to the variety of RTT phenotype and symptom severity.
The effects of total absence of MECP2 protein were investigated in several model systems. Deletion of MECP2 from glial cells had only mild phenotypic consequences [72]. In a mouse model specific deletion of MECP2 in the forebrain caused behavioral abnormalities, limb clasping, impaired motor coordination, anxiety, and abnormal social behavior but not locomotor activity or changes in fear conditioning [73]. In another mouse model, silencing MECP2 in GABAergic neurons led to severe RTT like phenotype [74].
Sixty-seven percent of all MECP2 mutations found in humans are in eight hot spots: R106 (corresponding RS number from dbSNP: rs28934907), R133 (rs28934904), T158 (rs28934906), R168 (rs61748427), R255 (rs61749721), R270 (rs61750240), R294 (rs61751362) and R306 (rs28935468). Most of the mutations which cause RTT occur in the MDB region of MECP2 [70] (Fig. 1). A 100-fold reduction of binding affinity of MECP2 to methylated DNA is documented for the mutations R106W (rs28934907), R133C (rs28934904), and F155S (rs28934905) and binding affinity reduction of about 2-fold was found in mutation T158M (rs28934906) [45].
The most important metabolites, genes and pathways affected by MECP2 in RTT
The examination and investigation of RTT females (and model systems) revealed that an impaired MECP2 influences biological pathways on many levels. Several genes have been found to be increased or decreased in expression, levels of various metabolites are changed and several biological pathways were found to be typically affected although the molecular mechanisms are not yet clear. In this chapter the main metabolites, genes and pathways which are influenced or changed by RTT are summarized and as far as known integrated in the MECP2 mechanistic pathway (Fig. 2). For getting these results, samples from human RTT females were often used but many results come from studies with Mecp2-/y mice (e.g. the Bird model [75]). These mice do not express Mecp2 at all and they display the same symptoms as humans, such as normal development until about 6 weeks, regression, reduced movement, clumsy gait, irregular breathing, and the mice have a reduced life span of about 3 months. Postmortem analysis revealed reduced brain and neuronal cell size which is similar to observations in humans. Other mouse strains or in vitro models with mutated MECP2, reduced or overexpressed MECP2 levels are also commonly used to study RTT. Recently, researchers started to use iPSCs (induced pluripotent stem cells) from human or murine origin [76].
Metabolites
Early autopsies revealed reduced levels of catecholamines, namely dopamine, serotonin and norepinephrine while markers for bioaminergic metabolism in general were higher [77, 78] (summary of metabolites in Table 2 and see also the list of metabolites on WikiPathways [18] (pathway ID 3584). This was confirmed by a study of Panayotiset al. [79] who revealed time-dependent levels of dopamine, norepinephrine, serotonin, and their catabolites in the brain tissues of Mecp2-/y mice. Viemari [80] et al. showed that Mecp2-/y mice have a deficiency in norepinephrine and serotonin content in the medulla and a drastic reduction of medullary TH (catecholamine producing) neurons indicating that dysfunctional neuronal development may be the reason for decreased catecholamine levels. The phosphorylation of MECP2 is triggered by neuronal activity which is caused by release of neurotransmitters (Fig. 2). Hutchinson et al. [81] demonstrated that phosphorylation of MECP2 is dependent on dopamine, serotonin and norepinephrine activated pathways (Fig. 2, MECP2 phosphorylation).
Another metabolite shown to be present in high levels in RTT females is glutamate. Increased glutamate production (or decreased glutamate consumption) may lead to over excitation of glutamatergic neurons and can trigger increased uptake and conversion of glutamate to glutamine. Over excitation feedback may cause the downregulation of the glutamate receptor which was observed before [82]. Moreover, severe downregulation in gene expression for the glutamate D1 receptor GRID1 (GluD1) was found (Fig. 2). This receptor links post and presynaptic compartments and influences not only synapsis function but also neuronal differentiation [83]. Glutamate levels during sleep-wake cycle were also affected in Mecp2 deficient mice [84]. Additionally, BDNF, which is directly regulated by MECP2, is also known to regulate GRID1 expression [85]. Glutamate disposal is energy consumptive which may help explain the increased level of energy metabolism found in brains of Mecp2-/y mice and RTT females in neuroimaging studies [86].
A metabolomics investigation found changed phospholipid profiles in Mecp2-/y mice [82]. Phospholipid metabolism is directly associated with cell growth since it provides membrane material for inner and outer hull structure. Still, it remains unclear whether this is the cause or just one of the consequences of reduced neuronal cell size and network connectivity of RTT. But as MECP2 basic function is global transcription dampener in combination with activity dependent activation of transcription this suggests that reduced membrane material production is a consequence of lack of activity specific transcription activation.
Genes/gene products
MECP2 is a global transcription and translation influencing factor but there are also single specific genes which are found to be up- or downregulated in the absence of functional MECP2. The expression of the MECP2 target genes is affected in human, mouse and in vitro models. In the absence of functional MECP2 the expression of BDNF, GAMT, DLX5, DLX6, FKBP5, SGK1, FXYD1 and MPP1 is upregulated whereas the expression of UBE3A [25] and GRID1 [83] are downregulated (Table 3 and see also the list on pathway 3584 [18]). BDNF, which shows in all investigated models the most consistent effect, is a MECP2 regulated protein and it is necessary for neuronal development and function, too (Table 3). The BDNF molecular pathway and its influence on neuronal development and function is currently the best investigated. For GAMT it is known that this gene is involved in creatine metabolic and biosynthetic process which was found to be dysregulated in some (not all) Rett females and may cause or consequence to breathing problems [4]. DLX5 and DLX6 are homeobox genes and involved in regulation of gene expression in general. DLX5 is especially responsible for ectodermal differentiation processes. The activity of both is highly regulated by methylation and for the maintenance of this methylation pattern MECP2 is required: binding sites for MECP2 have been found in both promoters. There is an ongoing discussion about DLX5 and DLX6 expression being influenced by MECP2 deficiency [87], the final argument seems that it is at least true for DLX5 [88]. FKBP5 and SGK1 were identified as potential MECP2 targets because they are involved in regulation of glucocorticoid responding gene regulation (stress response). Mecp2-0 mice showed anxiety behavior, had elevated levels of those transcripts but normal levels of glucocorticoids [89]. FXYD1 is a transmembrane modulator of Na+/K+ -ATPase activity involved in neurite outgrowth. It was found to be overexpressed in a Mecp2-0 mouse model inhibiting neuronal growth [90] and reduced arborization is generally found in RTT. MPP1 (signal transduction, neuronal homeostasis) and IGF2 (cell proliferation) were identified in a transcriptomics expression study to be strongly overexpressed in a study with human lymphoblastoid cells of Rett patients and controls [91]. MECP2 interacts with E6AP, the protein of the UBE3A gene and they are regulating the expression of several target genes [92]. GRID1 encodes for glutamate D1 receptor (GluD1) and is downregulated in the absence of functional MECP2 [83]. Glutamate receptors play a major role in neuronal signal transduction.
Pathways
Bedogni et al. mentioned the difficulty to identify unique target pathways of MECP2 because MECP2 is both a repressor and an activator of transcription and the balancing and timing of transcription levels seems to contribute more to disorder development than activation of single pathways [70]. Several transcriptomics studies using samples from RTT females, mouse and in vitro model systems showed about 60 significantly enriched pathways, for example inflammation, MAPK signaling, ERBB signaling, neurotropin intracellular cascade, sterol biosynthesis [93], cholesterol metabolism [93], cytoskeleton formation, and apoptosis [70].
While animal and in vitro models often use MECP2 – KO models (e.g. Mecp2-/y mouse) human RTT patient derived samples often have a residual MECP2 activity due to the various mutations which impair the function rather than inhibiting the expression completely. Tanaka et al. [76] investigated gene expression profiles of iPSC lines derived from six different RTT females with different MECP2 mutations. They created and compared expression profiles of two iPSCs cell lines derived from each of the patients, one with the X chromosome active that has the wild type MECP2 and the other one with the chromosome with the mutated gene. The differently expressed genes and altered pathways are very different for each patient. This is an interesting result, as there are so many different MECP2 mutations which lead to RTT. A specific mutation affects protein function differently and may trigger different pathways leading to different phenotypes. Linking genetic data to molecular analysis (transcriptome, metabolome) and phenotype will be a future challenge to elucidate the pathways of RTT.
MECP2-related pathways involved in RTT phenotypes
In this chapter we discuss how mutated MECP2 leads to failure in neuronal synapsis formation and function which are one of the major causes of RTT phenotype. MECP2 acts in a biological (molecular) network of constant interaction by regulating and being regulated. This complex molecular network interaction leads to effects in cellular morphology (e.g. arborization), synapsis function and neuronal network growth, development, and maintenance.
According to Lyst and Bird [9] MECP2 mutations cause RTT by disrupting two major functions: 1. co-repressor recruitment, and 2. chromatin compaction, which are both basic molecular functions. Skene and Bedogni specified this assumption of MECP2 function as a global dampener of transcription in neurons plus the activity dependent transcription activation which leads to proper synapsis function and development [43, 70].
The differences in global gene expression of RTT and wild-type control groups are not substantial – neither in fold change nor number of genes differently expressed - indicating that more subtle dysregulation events in several pathways are responsible for RTT [62, 94–97]. MECP2 is a global repressor of transcription [43], a global activator of gene translation [61] and it reacts on neuronal pathway signals which lead to phosphorylation of MECP2 and detachment from DNA. Therefore, MECP2 dampens neuronal transcription globally and allows activity related responses which seem to be necessary for learning activity and specific synapsis formation [98]. Bedogni et al. found a direct connection between MECP2 and molecular pathways leading to cytoskeleton re-formation [70] indicating structural changes leading towards neuronal network formation. BDNF and FXYD1 seem here to be the link between MECP2 and the cellular phenotype [90]. Li et al. found in human embryonic stem cells, which are developing from stem cells to neuronal precursor cells to neurons, that in a premature state, MECP2 acts as an activator of transcription while transcription repressor activity was only found in mature neurons [61]. The levels of MECP2 start to increase postnatally and the protein is quite abundant in mature nervous systems [43, 99]. The expression of MECP2 is not uniform in different neuronal cell populations [100], parts of the brain and changes with age [101]. Mouse Mecp2-KO neuronal precursor cells are not different from wildtype ones in respect to expression patterns, proliferation, and differentiation (morphology), they change only during maturation [99]. Together with the observation that symptoms of RTT do not appear before about month 6, this led to the assumption that MECP2 has less to do with neurogenesis but more with neuronal function and maintenance, and synapsis formation and function. This may explain why the RTT phenotype becomes visible only at the quite late age of 6–18 months. In RTT females brains a decreased number of synapsis was found [102–104]. Synaptogenesis again is mostly observed in the period of RTT symptom development (month 6 – 18) which may explain the development of learning disability. In MECP2 null mouse model reduced neuronal differentiation [105] and synaptic deficits [98] were observed. Mice studies and postmortem brains of RTT females reveal alterations in neuron structures which may be due to decreased dendritic complexity because of an immature synaptic spine morphology leading to malfunction of synaptic development and plasticity [106–108]. Changed neuronal tubulin expression was found directly in the brain tissue of RTT and Angelman syndrome patients [109]. Dysfunctional MECP2 led also to changes in synaptic transmission, short and long-term synaptic plasticity, deficits in short and long term potentiation (LTP and LTD) in mice [110].
The abnormal levels of neurotransmitters and differently expressed neurotransmitter receptors lead to an imbalance between excitatory and inhibitory neuronal activity (namely imbalance of GABAergic, glutamatergic and dopaminergic neuronal pathways). Such abnormal ratio of excitation/inhibition in brain activity was also found in autistic patients before [111–113] and it is a known effect in Parkinson’s disease which shares the motoric disabilities with RTT [114]. The gene in the neurotransmitter pathway which is known to be downregulated in the absence of functional MECP2 is GRID1 which could here be the link between mutation and phenotype [83]. RTT models using murine and human induced pluripotent stem cells showed some RTT features and these symptoms were documented for MECP2 overexpression models, too, leading again to the assumption that MECP2 function is dose dependent [26]. In summary, MECP2 affects epigenetic regulation of gene expression, which changes neurobiological activity, network formation and function which causes the major phenotype.
Current gaps in understanding RTT pathways
Although these processes could indeed explain many neuronal function related symptoms of RTT there is still lack of evidence for other phenotypes, especially those which occur in many but not in all RTT females. 1) Breathing patterns: breathing is regulated by brain stem function which gets its signals from receptors for blood pH, carbon dioxide and (to a lower amount) oxygen levels. Abnormal RTT breathing patterns could be caused by neuronal dysfunction of the brain stem or neuronal pathways but metabolic dysregulation involving abnormal creatine levels due to GAMT over/under expression is also an influencing factor [4]. GAMT is one of the MECP2 downstream activated genes. 2) Cardiac abnormalities: Heart beat is generally regulated by central nervous system but has its own nerve knots for signal production, too. Specialized neurons in the heart ensure proper electric signal transition. These might be affected by RTT directly, by brain stem function, or both. RTT patients are indeed known for higher incidence of sudden death and heart problems like tachy-, brady- and arythmia were observed before (. Furthermore, vascular dysfunctions have been found which are directly related to MECP2 dysfunction although the molecular pathway between MECP2 and effector genes is not yet elucidated [115]. 3) Digestion and nutrient uptake problems: Stomach and intestines are covered with a complex network of nerve cells. Dysfunctional nerve cells may lead to constipation and malabsorption of nutrients, e.g. Vitamin D but there may also be other nutrient processing pathways involved. 4) Tissue specific effects: Tissue specific neuronal cells are differently influenced by MECP2 mutation, e.g. olfactory sensory vs. visual system (Table 4). Recently it was found that MECP2 mutations contribute to hypersensitivity of mechanoreceptors [116] which aligns with the observation of clinicians and caregivers. It is currently unknown in which neuronal cell subpopulations MECP2 is more or less necessary for normal function although there are indications that there are differences [100].
To understand these processes integration of different levels of biological knowledge and research results is necessary, and interactive biological pathways help to organize, analyze, and visualize existing knowledge. To integrate the information the exact mutation of MECP2 gene needs to be combined with molecular data (e.g. gene expression data, metabolomics) and a detailed description of the RTT females’ phenotype (including clinical laboratory measurements). This process will increase the understanding of the underlying pathways of the variety of RTT phenotypes. Gathering this knowledge and bringing it properly together needs collaboration of biomedical and bioinformatics researchers, physicians and patients [117]. Furthermore, knowing the essential pathways and their components which contribute to a certain phenotype or symptom may lead to the discovery of drug targets. These are not likely to be able to cure RTT itself but may help to reduce the severity of specific symptoms.
Conclusions
The present review summarizes the current knowledge about MECP2 structure and function, how it influences levels of metabolites, gene expression and biological pathways, and tries to bridge the different types of data available to explain the development of typical RTT phenotype by visualizing in form of a pathway (Fig. 2). Although the dysregulation events in neuronal cells can be already be explained quite well, the mechanistic explanation of several additional symptoms is still missing. Integrating the knowledge about the individual mutation, molecular data and phenotype information will help to find biological pathways and therefore explanations for these symptoms. Finding the right target genes, proteins, or metabolites can build a bridge between genotype and phenotype, and possibly to drug targets and pathways like this will be a great help in visualization and analyses.
Abbreviations
- Genes:
-
Transcripts and proteins are abbreviated according to the human genome name consortium.
- RTT:
-
Rett syndrome
- 5MeCyt:
-
5-methyl cytosine
- 5OHMeCyt:
-
5-hydroxy methyl cytosine
- iPSCs:
-
Induced pluripotent stem cells
References
Laurvick CL, de Klerk N, Bower C, Christodoulou J, Ravine D, Ellaway C, Williamson S, Leonard H. Rett syndrome in Australia: a review of the epidemiology. J Pediatr. 2006;148(3):347–52.
Comission E. Useful Information on Rare Diseases from an EU Perspective. 2009
Rett A. On a unusual brain atrophy syndrome in hyperammonemia in childhood. Wien Med Wochenschr. 1966;116(37):723–6.
Halbach NS, Smeets EE, Bierau J, Keularts IM, Plasqui G, Julu PO, Engerstrom IW, Bakker JA, Curfs LM. Altered carbon dioxide metabolism and creatine abnormalities in rett syndrome. JIMD Rep. 2012;3:117–24.
Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23(2):185–8.
Lambert S, Maystadt I, Boulanger S, Vrielynck P, Destree A, Lederer D, Moortgat S. Expanding phenotype of p.Ala140Val mutation in MECP2 in a 4 generation family with X-linked intellectual disability and spasticity. Eur J Med Genet. 2016;59:522–5.
Neul JL, Kaufmann WE, Glaze DG, Christodoulou J, Clarke AJ, Bahi-Buisson N, Leonard H, Bailey ME, Schanen NC, Zappella M, et al. Rett syndrome: revised diagnostic criteria and nomenclature. Ann Neurol. 2010;68(6):944–50.
Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315(5815):1143–7.
Lyst MJ, Bird A. Rett syndrome: a complex disorder with simple roots. Nat Rev Genet. 2015;16(5):261–75.
Katz DM, Bird A, Coenraads M, Gray SJ, Menon DU, Philpot BD, Tarquinio DC. Rett Syndrome: Crossing the Threshold to Clinical Translation. Trends Neurosci. 2016;39(2):100–13.
Schwartzman JS, Velloso Rde L, D'Antino ME, Santos S. The eye-tracking of social stimuli in patients with Rett syndrome and autism spectrum disorders: a pilot study. Arq Neuropsiquiatr. 2015;73(5):402–7.
Halbach N, Smeets EE, Julu P, Witt-Engerstrom I, Pini G, Bigoni S, Hansen S, Apartopoulos F, Delamont R, van Roozendaal K, et al. Neurophysiology versus clinical genetics in Rett syndrome: A multicenter study. Am J Med Genet A. 2016;170(9):2301–9.
Ensembl: http://www.ensembl.org
OMIM: www.omim.org.
UniProt: www.uniprot.org
TheHumanProteinAtlas: www.proteinatlas.org.
GeneOntology: http://geneontology.org/
WikiPathways: http://www.wikipathways.org
Feldman D, Banerjee A, Sur M. Developmental Dynamics of Rett Syndrome. Neural Plast. 2016;2016:6154080.
Anderson A, Wong K, Jacoby P, Downs J, Leonard H. Twenty years of surveillance in Rett syndrome: what does this tell us? Orphanet J Rare Dis. 2014;9:87.
Oddy WH, Webb KG, Baikie G, Thompson SM, Reilly S, Fyfe SD, Young D, Anderson AM, Leonard H. Feeding experiences and growth status in a Rett syndrome population. J Pediatr Gastroenterol Nutr. 2007;45(5):582–90.
Reilly S, Cass H. Growth and nutrition in Rett syndrome. Disabil Rehabil. 2001;23(3–4):118–28.
Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annu Rev Cell Dev Biol. 2011;27:631–52.
Villard L, Kpebe A, Cardoso C, Chelly PJ, Tardieu PM, Fontes M. Two affected boys in a Rett syndrome family: clinical and molecular findings. Neurology. 2000;55(8):1188–93.
Bienvenu T, Chelly J. Molecular genetics of Rett syndrome: when DNA methylation goes unrecognized. Nat Rev Genet. 2006;7(6):415–26.
Collins AL, Levenson JM, Vilaythong AP, Richman R, Armstrong DL, Noebels JL, David Sweatt J, Zoghbi HY. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum Mol Genet. 2004;13(21):2679–89.
Liu J, Francke U. Identification of cis-regulatory elements for MECP2 expression. Hum Mol Genet. 2006;15(11):1769–82.
Singh J, Saxena A, Christodoulou J, Ravine D. MECP2 genomic structure and function: insights from ENCODE. Nucleic Acids Res. 2008;36(19):6035–47.
Liyanage VR, Rastegar M. Rett syndrome and MeCP2. Neuromolecular Med. 2014;16(2):231–64.
Newnham CM, Hall-Pogar T, Liang S, Wu J, Tian B, Hu J, Lutz CS. Alternative polyadenylation of MeCP2: Influence of cis-acting elements and trans-acting factors. RNA Biol. 2010;7(3):361–72.
Burmistrova OA, Goltsov AY, Abramova LI, Kaleda VG, Orlova VA, Rogaev EI. MicroRNA in schizophrenia: genetic and expression analysis of miR-130b (22q11). Biochemistry (Mosc). 2007;72(5):578–82.
Han K, Gennarino VA, Lee Y, Pang K, Hashimoto-Torii K, Choufani S, Raju CS, Oldham MC, Weksberg R, Rakic P, et al. Human-specific regulation of MeCP2 levels in fetal brains by microRNA miR-483-5p. Genes Dev. 2013;27(5):485–90.
Guo Y, Chen Y, Carreon S, Qiang M. Chronic intermittent ethanol exposure and its removal induce a different miRNA expression pattern in primary cortical neuronal cultures. Alcohol Clin Exp Res. 2012;36(6):1058–66.
Volkmann I, Kumarswamy R, Pfaff N, Fiedler J, Dangwal S, Holzmann A, Batkai S, Geffers R, Lother A, Hein L, et al. MicroRNA-mediated epigenetic silencing of sirtuin1 contributes to impaired angiogenic responses. Circ Res. 2013;113(8):997–1003.
Lusardi TA, Farr CD, Faulkner CL, Pignataro G, Yang T, Lan J, Simon RP, Saugstad JA. Ischemic preconditioning regulates expression of microRNAs and a predicted target, MeCP2, in mouse cortex. J Cereb Blood Flow Metab. 2010;30(4):744–56.
Kriaucionis S, Bird A. The major form of MeCP2 has a novel N-terminus generated by alternative splicing. Nucleic Acids Res. 2004;32(5):1818–23.
Mnatzakanian GN, Lohi H, Munteanu I, Alfred SE, Yamada T, MacLeod PJ, Jones JR, Scherer SW, Schanen NC, Friez MJ, et al. A previously unidentified MECP2 open reading frame defines a new protein isoform relevant to Rett syndrome. Nat Genet. 2004;36(4):339–41.
Itoh M, Tahimic CG, Ide S, Otsuki A, Sasaoka T, Noguchi S, Oshimura M, Goto Y, Kurimasa A. Methyl CpG-binding protein isoform MeCP2_e2 is dispensable for Rett syndrome phenotypes but essential for embryo viability and placenta development. J Biol Chem. 2012;287(17):13859–67.
Adams VH, McBryant SJ, Wade PA, Woodcock CL, Hansen JC. Intrinsic disorder and autonomous domain function in the multifunctional nuclear protein, MeCP2. J Biol Chem. 2007;282(20):15057–64.
Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88(4):471–81.
Ausio J. MeCP2 and the enigmatic organization of brain chromatin. Implications for depression and cocaine addiction. Clin Epigenetics. 2016;8:58.
Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY. Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum Mol Genet. 2002;11(2):115–24.
Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37(4):457–68.
Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol Cell. 2005;19(5):667–78.
Ballestar E, Yusufzai TM, Wolffe AP. Effects of Rett syndrome mutations of the methyl-CpG binding domain of the transcriptional repressor MeCP2 on selectivity for association with methylated DNA. Biochemistry. 2000;39(24):7100–6.
Ricciardi S, Boggio EM, Grosso S, Lonetti G, Forlani G, Stefanelli G, Calcagno E, Morello N, Landsberger N, Biffo S, et al. Reduced AKT/mTOR signaling and protein synthesis dysregulation in a Rett syndrome animal model. Hum Mol Genet. 2011;20(6):1182–96.
Gonzales ML, Adams S, Dunaway KW, LaSalle JM. Phosphorylation of distinct sites in MeCP2 modifies cofactor associations and the dynamics of transcriptional regulation. Mol Cell Biol. 2012;32(14):2894–903.
Long SW, Ooi JY, Yau PM, Jones PL. A brain-derived MeCP2 complex supports a role for MeCP2 in RNA processing. Biosci Rep. 2011;31(5):333–43.
Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, Kang D, Richman R, Johnson JM, Berget S, et al. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc Natl Acad Sci U S A. 2005;102(49):17551–8.
Nomura T, Kimura M, Horii T, Morita S, Soejima H, Kudo S, Hatada I. MeCP2-dependent repression of an imprinted miR-184 released by depolarization. Hum Mol Genet. 2008;17(8):1192–9.
Smith RM, Sadee W. Synaptic signaling and aberrant RNA splicing in autism spectrum disorders. Front Synaptic Neurosci. 2011;3:1.
Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–54.
Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293(5532):1089–93.
Fan G, Hutnick L. Methyl-CpG binding proteins in the nervous system. Cell Res. 2005;15(4):255–61.
Ehrlich M. Expression of various genes is controlled by DNA methylation during mammalian development. J Cell Biochem. 2003;88(5):899–910.
Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445(7128):671–5.
Lyst MJ, Ekiert R, Ebert DH, Merusi C, Nowak J, Selfridge J, Guy J, Kastan NR, Robinson ND, de Lima AF, et al. Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat Neurosci. 2013;16(7):898–902.
Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14(12):1607–16.
Adkins NL, Georgel PT. MeCP2: structure and function. Biochem Cell Biol. 2011;89(1):1–11.
Nelson ED, Kavalali ET, Monteggia LM. MeCP2-dependent transcriptional repression regulates excitatory neurotransmission. Curr Biol. 2006;16(7):710–6.
Li Y, Wang H, Muffat J, Cheng AW, Orlando DA, Loven J, Kwok SM, Feldman DA, Bateup HS, Gao Q, et al. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell Stem Cell. 2013;13(4):446–58.
Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320(5880):1224–9.
Cartron PF, Nadaradjane A, Lepape F, Lalier L, Gardie B, Vallette FM. Identification of TET1 Partners That Control Its DNA-Demethylating Function. Genes Cancer. 2013;4(5–6):235–41.
Wang J, Tang J, Lai M, Zhang H. 5-Hydroxymethylcytosine and disease. Mutat Res Rev Mutat Res. 2014;762:167–75.
Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302(5646):885–9.
Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52(2):255–69.
Ebert DH, Greenberg ME. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493(7432):327–37.
Tao J, Hu K, Chang Q, Wu H, Sherman NE, Martinowich K, Klose RJ, Schanen C, Jaenisch R, Wang W, et al. Phosphorylation of MeCP2 at Serine 80 regulates its chromatin association and neurological function. Proc Natl Acad Sci U S A. 2009;106(12):4882–7.
Ebert DH, Gabel HW, Robinson ND, Kastan NR, Hu LS, Cohen S, Navarro AJ, Lyst MJ, Ekiert R, Bird AP, et al. Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature. 2013;499(7458):341–5.
Bedogni F, Rossi RL, Galli F, Cobolli Gigli C, Gandaglia A, Kilstrup-Nielsen C, Landsberger N. Rett syndrome and the urge of novel approaches to study MeCP2 functions and mechanisms of action. Neurosci Biobehav Rev. 2014;46(Pt 2):187–201.
Leiden Open Variation Database (LOVD): http://databases.lovd.nl/shared/genes/MECP2
Lioy DT, Garg SK, Monaghan CE, Raber J, Foust KD, Kaspar BK, Hirrlinger PG, Kirchhoff F, Bissonnette JM, Ballas N, et al. A role for glia in the progression of Rett's syndrome. Nature. 2011;475(7357):497–500.
Gemelli T, Berton O, Nelson ED, Perrotti LI, Jaenisch R, Monteggia LM. Postnatal loss of methyl-CpG binding protein 2 in the forebrain is sufficient to mediate behavioral aspects of Rett syndrome in mice. Biol Psychiatry. 2006;59(5):468–76.
Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, Neul JL, Gong S, Lu HC, Heintz N, et al. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468(7321):263–9.
Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27(3):322–6.
Tanaka Y, Kim KY, Zhong M, Pan X, Weissman SM, Park IH. Transcriptional regulation in pluripotent stem cells by methyl CpG-binding protein 2 (MeCP2). Hum Mol Genet. 2014;23(4):1045–55.
Lekman A, Witt-Engerstrom I, Gottfries J, Hagberg BA, Percy AK, Svennerholm L. Rett syndrome: biogenic amines and metabolites in postmortem brain. Pediatr Neurol. 1989;5(6):357–62.
Brucke T, Sofic E, Killian W, Rett A, Riederer P. Reduced concentrations and increased metabolism of biogenic amines in a single case of Rett-syndrome: a postmortem brain study. J Neural Transm. 1987;68(3–4):315–24.
Panayotis N, Ghata A, Villard L, Roux JC. Biogenic amines and their metabolites are differentially affected in the Mecp2-deficient mouse brain. BMC Neurosci. 2011;12:47.
Viemari JC, Roux JC, Tryba AK, Saywell V, Burnet H, Pena F, Zanella S, Bevengut M, Barthelemy-Requin M, Herzing LB, et al. Mecp2 deficiency disrupts norepinephrine and respiratory systems in mice. J Neurosci. 2005;25(50):11521–30.
Hutchinson AN, Deng JV, Aryal DK, Wetsel WC, West AE. Differential regulation of MeCP2 phosphorylation in the CNS by dopamine and serotonin. Neuropsychopharmacology. 2012;37(2):321–37.
Viola A, Saywell V, Villard L, Cozzone PJ, Lutz NW. Metabolic fingerprints of altered brain growth, osmoregulation and neurotransmission in a Rett syndrome model. PLoS One. 2007;2(1):e157.
Livide G, Patriarchi T, Amenduni M, Amabile S, Yasui D, Calcagno E, Lo Rizzo C, De Falco G, Ulivieri C, Ariani F, et al. GluD1 is a common altered player in neuronal differentiation from both MECP2-mutated and CDKL5-mutated iPS cells. Eur J Hum Genet. 2015;23(2):195–201.
Johnston M, Blue ME, Naidu S: Recent advances in understanding synaptic abnormalities in Rett syndrome. F1000Res 2015, 4.
Ballas N, Lioy DT, Grunseich C, Mandel G. Non-cell autonomous influence of MeCP2-deficient glia on neuronal dendritic morphology. Nat Neurosci. 2009;12(3):311–7.
Naidu S, Kaufmann WE, Abrams MT, Pearlson GD, Lanham DC, Fredericksen KA, Barker PB, Horska A, Golay X, Mori S, et al. Neuroimaging studies in Rett syndrome. Brain Dev. 2001;23 Suppl 1:S62–71.
Schule B, Li HH, Fisch-Kohl C, Purmann C, Francke U. DLX5 and DLX6 expression is biallelic and not modulated by MeCP2 deficiency. Am J Hum Genet. 2007;81(3):492–506.
Miyano M, Horike S, Cai S, Oshimura M, Kohwi-Shigematsu T. DLX5 expression is monoallelic and Dlx5 is up-regulated in the Mecp2-null frontal cortex. J Cell Mol Med. 2008;12(4):1188–91.
Nuber UA, Kriaucionis S, Roloff TC, Guy J, Selfridge J, Steinhoff C, Schulz R, Lipkowitz B, Ropers HH, Holmes MC, et al. Up-regulation of glucocorticoid-regulated genes in a mouse model of Rett syndrome. Hum Mol Genet. 2005;14(15):2247–56.
Deng V, Matagne V, Banine F, Frerking M, Ohliger P, Budden S, Pevsner J, Dissen GA, Sherman LS, Ojeda SR. FXYD1 is an MeCP2 target gene overexpressed in the brains of Rett syndrome patients and Mecp2-null mice. Hum Mol Genet. 2007;16(6):640–50.
Ballestar E, Ropero S, Alaminos M, Armstrong J, Setien F, Agrelo R, Fraga MF, Herranz M, Avila S, Pineda M, et al. The impact of MECP2 mutations in the expression patterns of Rett syndrome patients. Hum Genet. 2005;116(1–2):91–104.
Kim S, Chahrour M, Ben-Shachar S, Lim J. Ube3a/E6AP is involved in a subset of MeCP2 functions. Biochem Biophys Res Commun. 2013;437(1):67–73.
Lekman AY, Hagberg BA, Svennerholm LT. Membrane cerebral lipids in Rett syndrome. Pediatr Neurol. 1991;7(3):186–90.
Colantuoni C, Jeon OH, Hyder K, Chenchik A, Khimani AH, Narayanan V, Hoffman EP, Kaufmann WE, Naidu S, Pevsner J. Gene expression profiling in postmortem Rett Syndrome brain: differential gene expression and patient classification. Neurobiol Dis. 2001;8(5):847–65.
Nagarajan RP, Hogart AR, Gwye Y, Martin MR, LaSalle JM. Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics. 2006;1(4):e1–11.
Tudor M, Akbarian S, Chen RZ, Jaenisch R. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc Natl Acad Sci U S A. 2002;99(24):15536–41.
Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum Mol Genet. 2009;18(13):2431–42.
Monteggia LM, Kavalali ET. Rett syndrome and the impact of MeCP2 associated transcriptional mechanisms on neurotransmission. Biol Psychiatry. 2009;65(3):204–10.
Kishi N, Macklis JD. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol Cell Neurosci. 2004;27(3):306–21.
Sugino K, Hempel CM, Okaty BW, Arnson HA, Kato S, Dani VS, Nelson SB. Cell-type-specific repression by methyl-CpG-binding protein 2 is biased toward long genes. J Neurosci. 2014;34(38):12877–83.
Jain D, Singh K, Chirumamilla S, Bibat GM, Blue ME, Naidu SR, Eberhart CG. Ocular MECP2 protein expression in patients with and without Rett syndrome. Pediatr Neurol. 2010;43(1):35–40.
Belichenko PV, Wright EE, Belichenko NP, Masliah E, Li HH, Mobley WC, Francke U. Widespread changes in dendritic and axonal morphology in Mecp2-mutant mouse models of Rett syndrome: evidence for disruption of neuronal networks. J Comp Neurol. 2009;514(3):240–58.
Matarazzo V, Cohen D, Palmer AM, Simpson PJ, Khokhar B, Pan SJ, Ronnett GV. The transcriptional repressor Mecp2 regulates terminal neuronal differentiation. Mol Cell Neurosci. 2004;27(1):44–58.
Palmer A, Qayumi J, Ronnett G. MeCP2 mutation causes distinguishable phases of acute and chronic defects in synaptogenesis and maintenance, respectively. Mol Cell Neurosci. 2008;37(4):794–807.
Zhao X, Ueba T, Christie BR, Barkho B, McConnell MJ, Nakashima K, Lein ES, Eadie BD, Willhoite AR, Muotri AR, et al. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc Natl Acad Sci U S A. 2003;100(11):6777–82.
Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex. 2000;10(10):981–91.
Jugloff DG, Jung BP, Purushotham D, Logan R, Eubanks JH. Increased dendritic complexity and axonal length in cultured mouse cortical neurons overexpressing methyl-CpG-binding protein MeCP2. Neurobiol Dis. 2005;19(1–2):18–27.
Francke U. Mechanisms of disease: neurogenetics of MeCP2 deficiency. Nat Clin Pract Neurol. 2006;2(4):212–21.
Abuhatzira L, Shemer R, Razin A. MeCP2 involvement in the regulation of neuronal alpha-tubulin production. Hum Mol Genet. 2009;18(8):1415–23.
Asaka Y, Jugloff DG, Zhang L, Eubanks JH, Fitzsimonds RM. Hippocampal synaptic plasticity is impaired in the Mecp2-null mouse model of Rett syndrome. Neurobiol Dis. 2006;21(1):217–27.
Purcell AE, Jeon OH, Zimmerman AW, Blue ME, Pevsner J. Postmortem brain abnormalities of the glutamate neurotransmitter system in autism. Neurology. 2001;57(9):1618–28.
Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003;2(5):255–67.
Serajee FJ, Zhong H, Nabi R, Huq AH. The metabotropic glutamate receptor 8 gene at 7q31: partial duplication and possible association with autism. J Med Genet. 2003;40(4), e42.
di Michele F, Luchetti S, Bernardi G, Romeo E, Longone P. Neurosteroid and neurotransmitter alterations in Parkinson's disease. Front Neuroendocrinol. 2013;34(2):132–42.
Panighini A, Duranti E, Santini F, Maffei M, Pizzorusso T, Funel N, Taddei S, Bernardini N, Ippolito C, Virdis A, et al. Vascular dysfunction in a mouse model of Rett syndrome and effects of curcumin treatment. PLoS One. 2013;8(5):e64863.
Orefice LL, Zimmerman AL, Chirila AM, Sleboda SJ, Head JP, Ginty DD. Peripheral Mechanosensory Neuron Dysfunction Underlies Tactile and Behavioral Deficits in Mouse Models of ASDs. Cell. 2016;166:299–313.
Townend GS, van Kranen HJ, van der Stel R, van den Berg M, Smeets E, van Waardenburg D, Curfs LM. Rett Syndrome as a Rare Disease: A European Perspective. Public Health Genomics. 2015;18(4):233–6.
Chao HT, Zoghbi HY. The yin and yang of MeCP2 phosphorylation. Proc Natl Acad Sci U S A. 2009;106(12):4577–8.
Ronnett GV, Leopold D, Cai X, Hoffbuhr KC, Moses L, Hoffman EP, Naidu S. Olfactory biopsies demonstrate a defect in neuronal development in Rett's syndrome. Ann Neurol. 2003;54(2):206–18.
Cohen DR, Matarazzo V, Palmer AM, Tu Y, Jeon OH, Pevsner J, Ronnett GV. Expression of MeCP2 in olfactory receptor neurons is developmentally regulated and occurs before synaptogenesis. Mol Cell Neurosci. 2003;22(4):417–29.
Degano AL, Park MJ, Penati J, Li Q, Ronnett GV. MeCP2 is required for activity-dependent refinement of olfactory circuits. Mol Cell Neurosci. 2014;59:63–75.
Saunders KJ, McCulloch DL, Kerr AM. Visual function in Rett syndrome. Dev Med Child Neurol. 1995;37(6):496–504.
Lee W, Yun JM, Woods R, Dunaway K, Yasui DH, Lasalle JM, Gong Q. MeCP2 regulates activity-dependent transcriptional responses in olfactory sensory neurons. Hum Mol Genet. 2014;23(23):6366–74.
Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, Ernst C, Hanscom C, Rossin E, Lindgren AM, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149(3):525–37.
Shen K, Fetter RD, Bargmann CI. Synaptic specificity is generated by the synaptic guidepost protein SYG-2 and its receptor, SYG-1. Cell. 2004;116(6):869–81.
Song C, Feodorova Y, Guy J, Peichl L, Jost KL, Kimura H, Cardoso MC, Bird A, Leonhardt H, Joffe B, et al. DNA methylation reader MECP2: cell type- and differentiation stage-specific protein distribution. Epigenetics Chromatin. 2014;7:17.
Durand S, Patrizi A, Quast KB, Hachigian L, Pavlyuk R, Saxena A, Carninci P, Hensch TK, Fagiolini M. NMDA receptor regulation prevents regression of visual cortical function in the absence of Mecp2. Neuron. 2012;76(6):1078–90.
Noutel J, Hong YK, Leu B, Kang E, Chen C. Experience-dependent retinogeniculate synapse remodeling is abnormal in MeCP2-deficient mice. Neuron. 2011;70(1):35–42.
Acknowledgements
The authors would like to thank the WikiPathways curation team namely Kristina Hanspers, Martina Summer-Kutmon, Egon Willighagen, and Ryan Miller for their contribution to the MECP2 pathway.
Funding
This study was funded by Stichting Terre – Rett Syndroom Fonds.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Authors’ contributions
FE did the protocol and project development, drafted the paper and made the MECP2 pathway, SC added database information and critical revision, EC contributed details in MECP2 gene variety information, ES added information about RTT diagnosis, use of diagnostic terms over time and clinical information, CE contributed bioinformatics details and critical revision, LC contributed to protocol and project development, manuscript editing and clinical information. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ehrhart, F., Coort, S.L.M., Cirillo, E. et al. Rett syndrome – biological pathways leading from MECP2 to disorder phenotypes. Orphanet J Rare Dis 11, 158 (2016). https://doi.org/10.1186/s13023-016-0545-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13023-016-0545-5