Abstract
Background
Root phenotyping aims to characterize root system architecture because of its functional role in resource acquisition. RGB imaging and analysis procedures measure root system traits via colour contrasts between roots and growth media or artificial backgrounds. In the case of plants grown in soil-filled rhizoboxes, where the colour contrast can be poor, it is hypothesized that root imaging based on spectral signatures improves segmentation and provides additional knowledge on physico-chemical root properties.
Results
Root systems of Triticum durum grown in soil-filled rhizoboxes were scanned in a spectral range of 1000–1700 nm with 222 narrow bands and a spatial resolution of 0.1 mm. A data processing pipeline was developed for automatic root segmentation and analysis of spectral root signatures. Spectral- and RGB-based root segmentation did not significantly differ in accuracy even for a bright soil background. Best spectral segmentation was obtained from log-linearized and asymptotic least squares corrected images via fuzzy clustering and multilevel thresholding. Root axes revealed major spectral distinction between center and border regions. Root decay was captured by an exponential function of the difference spectra between water and structural carbon absorption regions.
Conclusions
Fundamentals for root phenotyping using hyperspectral imaging have been established by means of an image processing pipeline for automated segmentation of soil-grown plant roots at a high spatial resolution and for the exploration of spectral signatures encoding physico-chemical root zone properties.
Similar content being viewed by others
Background
The root system is fundamental for plant physiological and ecosystem functioning [39, 50, 61]. Better understanding of root and rhizosphere processes can therefore essentially contribute to enhance resource efficiency in crop production and sustainable soil management [26, 38, 63, 68].
Advances in root system and rhizosphere management critically depend on appropriate measurement methods, making the plants’ “hidden half” [5] accessible to visualization and quantification. Traditionally root research relied on destructive and highly laborious methods (e.g. coring, profiling; [67]). Since the 1990 tie non-destructive imaging methods have become increasingly popular in plant sciences [18, 36]. Imaging of plant root systems has evolved into two directions: (1) deep phenotyping using high resolution 3D methods for small scale processes (e.g. X-ray computer tomography, [45]; magnetic resonance imaging, [69]), and (2) high-throughput phenotyping using optical methods [17]. In spite of their different focus, there is a trend towards approximation owing to technological advance. E.g. current μCT devices yet allow imaging of larger soil volumes up to a range of 1000 cm3 at high resolutions in reasonable measurement time for phenotyping purposes (range: hour; [40]). On the other hand high-throughput applications, traditionally restricted to RGB imaging, are moving towards wider spectral ranges where chemical imaging of rhizosphere components is possible [12, 54]. Nakaji et al. [49] were the first to demonstrate application of hyperspectral imaging (480–972 nm) in order to discriminate between living, senescent and dead roots, leaf debris and soil. For leave tissues, Pandey et al. [51] showed that using an extended hyperspectral range from 550 to 1700 nm enables an accurate prediction of leaf water content and nutritional status.
Most root phenotyping approaches rely on seedling/juvenile plants grown in artificial media such as agar [27], germination paper [33] or hydroponics [10] to facilitate imaging. Extrapolation from such systems towards natural growing conditions has been questioned [75, 80]. Therefore larger soil-filled rhizobox systems have been established for field-near root imaging [37, 47]. RGB root imaging of rhizobox grown plants has been successfully used to characterize distinctive root architectures in breeding germplasm as well as the modification of root architecture in response to variable soil conditions (temperature, water and nutrient supply, compaction; e.g. Price et al. [57], Nagel et al. [46, 48]).
RGB root phenotyping however requires sufficient root-soil colour contrast for (automated) segmentation which is challenging in case of bright coloured soils and/or dark coloured roots. Pierret [54] was the first to suggest spectral imaging of rhizoboxes as a promising advance for root research. Hyperspectral data might overcome problems of root distinction from soil background in the RGB colour space. Furthermore spectral reflectance encodes material properties that can be captured via advanced statistical models (chemometrics; [76]), potentially providing a tool to simultaneously measure root architecture with relevant physico-chemical root zone properties (e.g. water content). Close-range spectral imaging has already been successfully applied in phenotyping aboveground plant parts [43]. However, besides the system described by Nakaji et al. [49], there are still no established spectral imaging systems and data processing pipelines for root phenotyping.
Here a first comprehensive methodological description and evaluation of hyperspectral root phenotyping is presented. Acquisition of high resolution hyperspectral root images and subsequent image processing is described in detail. Root segmentation based on spectral information is compared with standard colour based segmentation. Application of the novel method is exemplified for identification of spectrally distinctive regions within the root and spectral changes during root decay. Classification of such chemometrically distinctive root regions is an important information when aiming to understand possible functional differences between root axes in space and time. The aim here is to establish the fundamentals of hyperspectral imaging for plant root phenotyping and reveal its potential as a novel tool for improved root system and rhizosphere characterization.
Methods
Rhizobox setup
Plants were grown in rhizoboxes (30 × 100 × 1 cm) consisting of a grey PVC back-plate and side frames (thickness 3 cm), and a mineral glass front (thickness 8 mm) fixed by metal angle bars to the sides. At the bottom, drainage holes were drilled into the frame.
Boxes were filled with two substrates to compare different background effects on imaging, i.e. dark topsoil and bright subsoil from a calcareous chernozem [23] sieved to an aggregate size < 2 mm. Table 1 gives an overview of basic soil properties. Soil moisture was adjusted to a matrix potential of h = − 100 cm (drained upper limit for a rhizobox of height/gravitational potential of 100 cm) for a well-watered treatment and h = − 1000 cm for a water limited treatment (retention curve of the two substrates, Additional file 1).
10 g of slow release NPK fertilizer (16% N, 2.6% P, 11.6% K) were mixed into the soil. Rhizoboxes were planted directly with one presoaked seed from Triticum turgidum L. subsp. durum cv. Floradur at a depth of 2 cm below soil surface. The glass surface/root zone was darkened with a black plate and the slit at the top of the boxes was closed with PVC foam to minimize evaporation. Boxes were positioned at 45° angle into a metal frame and transferred into a growth chamber (Light intensity 250 mmol m2 s−1 by six Atum Photon 270 LED; day/night 14 h/10 h; temperature 23 °C/16 °C; Relative humidity 60%).
The experimental design was a CRD with four replicates and substrate (bright vs. dark) and moisture (dry vs. moist) as main factors (i.e. 16 rhizoboxes). Plants were watered every third day upon weighing to keep the initial moisture level. Imaging was done for fully developed root systems (BBCH 44), i.e. when reaching the bottom part of the boxes and showing all types of roots (primary/basal roots, shoot borne roots, laterals; [81]). At this stage, 2.2% of total root length was < 0.2 mm and 48.7% < 0.5 mm diameter.
For the analysis of root decay we followed Nakaji et al. [49], cutting the shoot and subsequently imaging the upper third of the rhizobox (with primary, shoot-borne and lateral roots) at 14, 28, 47, 94, 101 and 201 days after cutting.
Image acquisition setup
The hyperspectral root imaging system (Fig. 1) consists of four main components: (1) A halogen line illumination (45°/− 45°) that provides a homogeneous light sources for imaging the rhizobox; (2) an image spectrograph (ImSpector N17E, Specim, FI) that splits the incoming light into 256 spectral bands between 900 and 1700 nm; (3) a thermo-electrically cooled 14-bit monochrome NIR camera (Xeva, Xenics, BE) that records the spectral NIR bands (900–1700 nm) for each pixel. The resolutions of the camera sensor is 320 by 256 pixels and the frame rate is 100 Hz. The spectral resolution of the imaging system is 3.1 nm (256 bands between 900 and 1700 nm). Due to low sensitivity of the detector chip at the lower edge, resulting in noisy spectra, the spectral bands were limited to the wavelength range of 1000–1700 nm.
The distance between the front lens and the rhizobox is 18 cm with a perpendicular position of the rhizobox with respect to the camera. A white standard (Spectralon tile) is positioned at the top of the rhizobox at the same angle and distance and imaged before each scan. The white standard is covered by the same type of glass as used for the rhizoboxes to compensate for the light transmission path through the glass upon normalization with the white and dark standards (see “Image preparation” section).
A hyperspectral rhizobox image (with x, y as spatial dimensions and z as spectral dimension) is obtained using a push-broom approach, i.e. the imaging system (illumination + spectrograph + camera) is moving in the y-direction while continuously capturing line scans in the x-direction of the rhizobox which are projected into the spectrograph and recorded by the camera. With a field of view (FOV) of 3 cm, maximum spatial resolution is 0.1 mm. A 50 mm lens is used to achieve this image resolution at 10 mm s−1 scanning speed. Via a two-axis positioning system, moving the imaging system up and down, spectral line scans (strides) of 3 cm width with 10% overlap between adjacent strides are acquired. Each stride is saved separately in a compressed file format (SIF) to reduce the size of the files. The single strides are composed to a full rhizobox image during image processing (see “Image processing” section).
Prior to scanning, camera integration time has to be adjusted to the background colour for optimizing image quality. The optimum camera integration time is obtained using the Xenics Xeneth camera software and targeting a bright object (i.e. root) on the rhizobox; it is adjusted in a way that approximately 85% of the full dynamic range of the imaging system are used. Exceeding this maximum will result in data losses during image acquisition, while on the other hand low integration time does not make use of the full capacity of the camera. Here an integration time of 4500 µs for the darker topsoil filled rhizoboxes and 3500 µs for the brighter subsoil filled rhizoboxes were used respectively.
The setup was developed as a prototype for root phenotyping by Carinithian Tech Research (CTR). It is located in a closed room to avoid influence of stray light during image acquisition and keeping the measurement device at stable temperature (20 °C) and air humidity (40%).
Image processing
Figure 2 schematically presents the image processing pipeline. All steps are performed via Matlab scripts (Matlab version R2018a) on an Intel Core i7-6700 PC with 40 GB RAM. Scripts are available from the authors upon request.
Scheme of the image processing pipeline developed for root segmentation and chemometric analysis of root and soil properties from hyperspectral images. Implemented approaches for a pre-processing and b post-processing and image analysis. Abbreviations: ALS asymmetric least squares correction, SNV standard normal variate, MSC multiplicative scatter correction, EMSC extended multiplicative signal correction, SCR simple contrast ratio, WC Weber contrast, MC Michelson contrast, PCA principal component analysis, fft fast Fourier transform, ECOC error correcting output codes, PCR principal component regression, PLSR partial least squares regression, SVMR support vector machine regression. (The Matlab scripts can be obtained from the corresponding author upon request.)
Image preparation
Absolute reflectance of the raw image (Ri,λ) is normalized at each wavelength band λ for each pixel i following Eq. (1) and using the dark (Di,λ) and white (Wi,λ) standards acquired before each scan. The dark standard corrects for the wavelength dependent dark current of the NIR camera, while the white standard represents maximum (100%) reflectance. The normalized image (Ni,λ) is thus obtained by:
Pre-treatment
Following Esquerre et al. [16] three types of data pre-treatments are implemented, i.e. response linearization enhancing low reflectance regions, baseline correction to correct for offset and change in the baseline (polynomial detrending, derivatives, asymmetric least squares, ALS) and multiplicative correction (standard normal variate, SNV, multiplicative scatter correction, MSC, extended multiplicative signal correction, EMSC). For most pre-treatments the mda toolbox for Matlab [31] is used. EMSC is performed via the toolbox from Afseth and Kohler [1]. However, due to excessive computational time of MSC and EMSC for the large-size image data evaluated here, they were not included in further evaluations.
Dead pixels and spikes, containing no information and potentially disturbing segmentation and analysis, are identified via the standard deviation matrix of difference spectra Ni,λ − Ni,λ+1. A threshold is visually defined from the standard deviation histogram and the respective pixels are black masked (i.e. assigned a value of zero over all wavelengths; Vidal and Amigo [72] and Dorrepaal et al. [13]).
Pre-treatment method selection and dimensionality reduction
Identification of adequate pre-treatment methods and spectral bands maximizing feature distinction require selection criteria. A first selection approach targets the entire image: it combines a set of Non-Gaussian indicators (skewness, kurtosis, entropy, negentropy, signal-to-noise ratio, Kullback–Leibler divergence) that measure deviation from random white noise [8, 21]. As single indicators can lead to different results [19] an average rank of all information criteria is used to select the top-ranking pre-treatment method.
The second approach is based on histogram evaluation [29]. Root (foreground) and soil (background) regions of interest (ROIs) are selected and the separation of their histogram peaks at each wavelength is quantified via Bhattacharyya distance [4]. Spectral transformations (differences spectra, Ni,λn − Ni,λm, λm and λn= 1…222) were evaluated with the ROI based histogram method only: due to sharply increasing dimensionality (222 × 222) transformation of entire images for the non-Gaussian indicator approach is not feasible for computational reasons.
The three best pre-treatment methods selected by each approach, i.e. (1) non-Gaussianity based, (2) Bhattacharyya distance based for (a) single spectra as well as (b) difference spectra, were further used for comprehensively testing the influence of pre-treatment on subsequent segmentation.
Simultaneously with pre-treatment evaluation, the most informative wavelengths within the pre-treated images are extracted, while removing all noisy bands. Here the ten best bands with highest frequency of occurrence among the applied evaluation approaches were selected. For comparison PCA was used as reference dimensionality reduction method. Depending on the segmentation algorithm either 2D or 3D data can be used. 3D images contained all ten selected bands, 2D data contained one spectral dimension only: reduction to a single spectral dimension was done via (1) averaging over the ten best bands, (2) extracting the single most informative band and (3) the principal component with highest feature contrast.
Image segmentation
Segmentation is performed with unsupervised (multilevel thresholding, k-means clustering, fuzzy clustering) and supervised (two-class support vector machine) techniques. The implemented fuzzy clustering algorithm was presented by Li et al. [34], while all other algorithms are adapted from standard Matlab scripts. Evaluation of segmentation quality was done based on obtained root length in relation to a manually tracked reference length and image skewness: misclassification can increase false negatives (root pixels wrongly classified as soil background) resulting in a conservative segmentation with low noise (high skewness) at the cost of underestimation of root length; false positives (soil pixels wrongly classified as root) on the contrary result in a noisy image (low skewness) and corresponding overestimation of root length.
Post-processing
Post-processing comprises stitching of all strides to an entire binary root system image and removal of remaining noise in the image. Automatic stitching is done with an image registration algorithm based on cross-correlation of image pixels in Fourier domain [22]. Noise removal is performed by filtering for small circular objects using region property analysis with a manually set minimum extent.
Chemometric root analysis
Chemometric models delimit physico-chemically distinctive regions within the segmented root and/or soil domains based on their specific spectral patterns. Also here unsupervised (fuzzy and k-means clustering, PCA) and supervised (Multiclass error-correcting output codes, ECOC, with optional classifiers, e.g. support vector machine, k-nearest neighbour, discriminant analysis, decision tree) approaches are implemented.
With directly measured reference data spectral regression models can be applied. Scripts for linear and non-linear univariate regression, partial least squares regression (PLSR), principal component regression (PCR) and support vector machine regression (SVMR) were included.
The application of chemometric models is exemplified for the analysis of spectrally distinctive radial (centre to border; supervised ECOC model) pattern on the roots, as well as for root decay following clipping of the shoot (non-linear regression model). For radial differentiation the image was shape-corrected via ALS baseline correction to exclude confounding effects from surface geometry of spherical objects [58] and dimensions were reduced to the first five principal components. Eight radial classes from root centre (0–0.1 mm) to border pixels at 2 pixel increments were defined and the respective pixels were labelled accordingly. An ECOC model with a decision tree classifier [6] was trained with 50% of pixels from the eight radial classes and validated with the remaining 50% of pixels.
For root decay, the most distinctive wavelength separating spectra (untransformed, first derivative, single wavelength and wavelength differences) at different times after clipping was identified via Bhattacharyya distance. Spectral reflectance was then related to decay time via an exponential model and subsequently mapped on each root pixel.
Imaging data
Determination of an adequate image processing strategy (pre-treatment, dimensionality reduction, segmentation) was done on topsoil and subsoil image sections (320 × 3000 pixel, 222 wavebands from the densely rooted upper part of rhizoboxes, i.e. one central stride with one third of the vertical pixels). The reduced file size of 9.4 GB ensured comparatively short calculation time for the single steps, required for evaluating all processing options. The resulting image processing strategy is then applied to all rhizoboxes of the same treatment resulting in binary root system images for structural quantification. With the 40 GB RAM PC used here the Matlab based processing pipeline takes about 40 min to obtain a binary rhizobox image from the spectral raw data.
In this study structural quantification was done with WinRhizo (Version 2013; Regent Inc.) for root length only. Visible root length from spectral segmentation was compared to colour based segmentation from RGB images. RGB images were taken with a Canon EOS 6D camera (resolution 5472 × 3648 pixel) after the spectral scans. Images were converted to TIFF-files and segmented using WinRhizoPro based on five distinctive colour classes for root and soil respectively. Manually tracked roots on RGB images were used as reference. Manual tracking was done in CorelDraw (Version X7) using a graphic tablet (Wacom Intuos®pro) and zooming into the image (500% magnification) to capture all root axes visible on the glass surface.
Visible length was also compared to total length, including the non-visible roots, obtained at the end of the experiment after separating (washing) roots from soil following Himmelbauer et al. [25].
Chemometric analysis of distinctive root regions and root decay is exemplified with a segmented root image from the well-watered topsoil treatment.
Statistical evaluation
The influence of pre-treatment, dimensionality reduction and segmentation methods was evaluated by analysis of variance. The dependent variable was root length, while skewness of the segmented binary image was used as covariate capturing the influence of misclassified pixels. Cases where segmentation failed (noise threshold: skewness < 2.5; no visual identification of the root system) were excluded. Evaluation of spectral versus colour based segmentation in relation to the manually tracked reference was done by regression analysis with zero intercept. Slopes were compared statistically following Sawand [59] to reveal whether the two segmentation approaches differed significantly in capturing visible root length. All statistical analyses were performed in SAS Version 9.4 using PROC MIXED for covariance analysis, PROC REG for regression analysis and PROC GLM for slope comparison.
Results
Image processing strategy
Hyperspectral rhizobox images with 222 bands in the spectral range of 1000–1700 nm and a spatial resolution of 0.1 mm result in an image size of 42.3 GB. Using a representative image section of 320 × 3000 pixels near the plant base, file size was decreased to 9.4 GB for deriving an adequate image processing strategy, while reducing computational time sufficiently to test several possible combinations of processing steps (pre-treatment, dimensionality reduction, segmentation) on a standard PC.
Spectral pre-treatment
Image pre-treatment corrects uneven surface morphology due to soil aggregates, resulting in inhomogeneous illumination with increased reflectance scattering [16]. In total 16 pre-treatment approaches were evaluated via non-Gaussian indicators (whole image section) and Bhattacharyya distance (root and soil ROIs; 0.6% of image pixels). Bhattacharyya distance was also calculated for difference spectra (graphical examples, Additional file 2). Evaluation results are given in Table 2.
The indicators did not point to a unique pre-treatment method to maximize distinction between root foreground and soil background. Particularly in the topsoil there was no significant correlation between ranks from non-Gaussian measures and Bhattacharyya distance. In the subsoil on the contrary correlation coefficients between the methods were significant with an r > 0.61.
Among the three best pre-treatment methods indicated by each of the information criteria, de-trending via ALS baseline correction occurred in 44% of all cases. For the subsoil log-linearization resulted in an average improved foreground–background distinction. Difference spectra increased the distinctiveness of image features (Bhattacharyya distance topsoil 3.0–6.5; subsoil 4.2–8.0). The three top-ranking pre-treatment approaches from each information criterion for topsoil and subsoil were further used in the evaluation of subsequent image processing steps.
Dimensionality reduction
Figure 3 gives an overview of the frequency of occurrence of wavelength regions with highest foreground–background contrast identified via band selection indicators (see movie Additional file 3). The highest spectral differentiation between root and soil occurred in the region from 1440 to 1480 nm (topsoil) and 1400 to 1440 nm (subsoil). A second distinctive region was found between 1050 and 1090 nm. Spearman rank correlation indicated that location of the most informative spectral bands in topsoil and subsoil was similar (r = 0.80, P < 0.001).
Segmentation algorithm
Four segmentation methods (fuzzy clustering, k-means clustering, multi-level thresholding and a two-class SVM) were evaluated with dimensionality reduced images. As expected there was a trade-off between segmented length and noise: the higher the number of pixels misclassified as roots the higher the noise from these sparsely distributed pixels (cf. Additional file 4). In the subsoil segmentation failed in 51.0% of the cases with high noise (skewness < 2.5) and no clearly identifiable root axes. This was mainly the case for thresholding (78.6%), while fuzzy clustering only failed in 17.9% of cases. In the topsoil segmentation only failed in 15.9% of all cases (thresholding 28.6%, fuzzy clustering 14.3%, k-means clustering 10.8%, SVM 10.0%).
Significant influences from previous processing steps on the final segmentation result are highlighted in Table 3. The highest significant interaction between processing steps was a soil type specific effect of pre-treatment and segmentation algorithm. Thus adequate pre-treatment methods strongly depended on the soil background as well as the subsequent segmentation method.
Figure 4 shows the resulting mean comparison (Tukey test) for the highest significant interaction (Soil × Pre × Seg). For clarity only the twenty segmentation results next to the manually tracked reference are plotted.
Root length for the twenty best combinations of pre-treatment, band selection and segmentation algorithm in the topsoil (a) and subsoil (b). The manually tracked reference length is shown with ± 10% margins (transparent grey area); grey dotted areas show the range of results without significant difference (Tukey, p < 0.005). NG refers to band selection via non Gaussian measures, BAT to Bhattacharyya distance based selection for single wavelength (BATs) and wavelength differences (BATd); for pre-treatments cf. Table 1; FU (grey) fuzzy clustering, TH (orange) thresholding, SVM (brown) support vector machine classification, KM k-means clustering (turquoise)
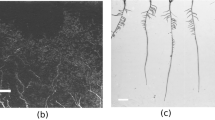
In the topsoil overlap between different strategies was comparatively large, i.e. similar length estimates and thereby accuracy in relation to the manually tracked reference length could be obtained via different image processing strategies. In the brighter subsoil differences were substantially higher and only some processing strategies resulted in segmented root length near to the reference length. Overall fuzzy clustering tended to be more conservative, while thresholding resulted in high number of pixels classified as roots with a tendency to increase noise from misclassified pixels. This is exemplified in Fig. 5: compared to the manually tracked reference (Fig. 5a) it can be seen that fuzzy clustering (Fig. 5b) did not capture some lateral root axes, while thresholding (Fig. 5c) resulted in a noisier image still conserving these lateral axes.
The best segmentation results with root length next to the manually tracked reference and low image noise were obtained for the topsoil with log-linearization and ALS correction as pre-treatment and using thresholding on the single most informative band (difference to manual tracking 3.5%). For the subsoil log-linearization and ALS correction in combination with fuzzy clustering on the single most informative band resulted in the lowest difference to manual tracking (4.6%).
Spectral versus colour based root segmentation
The topsoil and subsoil processing strategies with best segmentation result were then applied to the entire rhizobox images and compared to colour threshold segmented RGB images as well as manually tracked references. Figure 6 shows that both, spectral and colour based image segmentation, reliably predicted visible root length. The spectral approach captured slightly less root length (77.0%) compared to colour based segmentation (83.4%). However, slopes of both segmentation methods were not significantly different (P = 0.225) indicating that both methods had similar performance to predict visible root length for substrates used here.
Overall root length was significantly influenced by the soil material (807.3 cm for topsoil vs. 461.7 cm for subsoil; P = 0.012) and soil moisture (884.2 cm for moist soil vs. 423.1 cm for dry soil; P < 0.001), but not by segmentation method (770.6 cm for manual tracking, 593.5 cm for automated spectral segmentation, 643.0 for automated RGB segmentation; P = 0.377) nor interaction between segmentation method and treatment.
On average visible root axes at the observation window, accessible to optical imaging, were 27.3% of total root length in the rhizoboxes. The relation between visible and total root length was significant with an r2 of 0.74 (P < 0.001; cf. Additional file 5). For the topsoil, surface visibility was significantly higher in the control compared to the dry treatment (37.6% vs. 15.6% of all root length visible at the glass observation window; P = 0.030), while it did not differ in the subsoil.
Spatial differentiation of root spectral pattern
A key advantage of hyperspectral imaging is the chemometric information contained in the spectral images. Radial differentiation in spectral signatures from center to border (cf. Additional file 6) were predicted by a decision tree model with an r = 0.86 for the training data and r = 0.48 for the validation data. Table 4 gives the percentage of pixels allocated to the respective labelled classes by the trained model.
The chemometric model identified two main spectrally distinctive regions: an inner region (0–0.3 mm) where in average 82% of the pixels are allocated to the first and second class, and an outer region where in average 62% of the pixels are allocated to spectral classes > 0.3 mm. Within these two regions, the spectral pattern of pixels largely overlapped. It should be noticed that coarser root axes (> 0.7 mm) made up only 5.3% of all root pixels in the image. Figure 7 shows the classification result with close-ups for the basal and apical region. At the basal parts of root axes pixels with the spectral features of the central classes (dark blue) are more abundant compared to the apical parts with predominance of pixels with spectral characteristics of the outer region (light blue).
Temporal changes of spectral signature of decaying roots
Differentiation in the spectral pattern between the initial and final time was highest for the first derivative difference spectra between wavelengths of 1649–1447 nm with a maximum Bhattacharyya distance of 2.21 (Additional files 7 and 8). Figure 8 shows the exponential model predicting root decay duration as a function of spectral reflectance with two validation time points (28 and 101 days after clipping) not included in curve fitting. Changes in spectral reflectance were closely related to decay duration (r2 = 0.96).
Figure 9 provides a close-up image of root axes at 28 and 101 days after clipping with mapping of the decay model on the root pixels. At 101 of root decay (Fig. 9b) only one major root axes was recognized (segmented) as spectrally different from the soil background, while the coarser shoot-borne root axis and the laterals were apparently strongly decomposed. On the contrary a small part of the main axis was not recognized in the image at 28 days after clipping (Fig. 9a). Fine mapping also revealed a centre-to-border gradient of reflectance values, suggesting chemometric similarity between spatial (radial) and temporal spectral patterns.
Close-up of the same image region with fine-mapping of regression model for root decay duration (cf. Fig. 8) on root pixels; a root image at 28 days after clipping, b root image at 101 days after clipping. Colour scale: days after clipping
Discussion
Rhizobox experimental system
Inference from root phenotyping towards field environments critically depends on suitable substrates [24] and plant development stage, particularly for monocots with shoot-borne roots emerging upon tillering [74, 80]. The rhizobox setup used in this study represents a field-near system with soil as substrate and sufficiently large containers for unconstraint (cereal) root growth until end of vegetative development. Size of phenotyping systems is particularly important when focussing on root contribution to drought resistance [52, 56]. The rhizoboxes of 1 m height used here closely match the hydraulic behaviour of similarly textured field soils (e.g. Saxon and Rawls [60]: field capacity for silt loam 0.31 cm3 cm−3; cf. Table 3: rhizobox drained upper limit 0.27–0.33 cm3 cm−3).
Imaging setup
Camera model and spectrograph determine the spectral range as well as spectral and spatial resolution of imaging systems. For root imaging high spatial resolution is required as most root axes of annual plants are allocated in the lowest diameter classes (very fine < 0.5 mm; fine 0.5–2 mm) according to Böhm classification [7]; e.g. here 48.7% < 0.5 mm and 99.8% < 2 mm. The spatial resolution limit of the scanning device used here is 0.1 mm. This is only slightly lower compared to common settings of RGB root scanning (400 dpi equivalent 0.063 mm pixel size; Himmelbauer et al. [25]) and μCT (0.056–0.099 mm according to pot size; Metzner et al. [42]). Image acquisition time for a rhizobox of 100 cm height an 30 cm width at this spatial resolution is 16 min. This is substantially shorter to high-resolution 3D methods (e.g. 20 min for a 30 cm high × 8.1 cm inner diameter tube; Metzner et al. [42]) and therefore suitable to mature root phenotyping.
Compared to RGB imaging, a hyperspectral approach substantially increase data size as well as the complexity of evaluation. The automated segmentation pipeline presented here however allows a comparatively rapid data processing to obtain a binary root image (about 40 min). Still comparison of RGB versus spectral segmentation suggested an advantage of NIR bands just in case of very bright soil background. Otherwise VIS bands are suitable for detection of living root axes from soil background [49] with high resolution RGB cameras leading to a potentially superior root-soil segmentation compared to a NIR setup (see discussion on spectral vs. colour based segmentation results below).
Thus the most relevant added value of hyperspectral NIR imaging is related to the physicochemical information contained in this wavelength range. These chemical imaging capacities of a hyperspectral setup are determined by spectral range and resolution. The camera sensor of the device used here allows imaging between 900 to 1700 nm with 256 narrow bands (i.e. spectral resolution of 3.1 nm) covering wavelength bands such as water, cellulose, lignin, starch and protein [15, 30, 64, 70]. Hyperspectral imaging thus has the potential to extend root phenotyping towards such physicochemical root zone properties with relevance for root functionality.
Image processing strategies for feature detection
Exploitation of the information hidden in hyperspectral data critically depends on algorithms capable to detect the features of interest. Root phenotyping, similarly to food quality control, targets biological objects of variable biochemical composition and high tissue water content. Additional challenges in root–soil image processing arises from the complex background with uneven surface morphology from soil aggregates, and non-uniform and time-variable water content. Esquerre et al. [16] suggested chemometric image pre-treatment to suppress surface morphology and improve contrast between image features. Also our results demonstrated that root versus soil pixels could be better separated after spectral pre-treatment. Baseline correction with asymmetric least squares and the use of difference spectra were particularly efficient for enhancing feature contrasts. A major disadvantage of ALS baseline correction is the high computational time required for large datasets (1545.5 s for a 9.4 GB image) compared to other pre-treatment methods with similar performance such as first derivatives (33.4 s) or standard normal variate (6.8 s) of log-linearized spectra.
Beyond visual inspection, different selection methods can be applied for an objective pre-treatment selection and extraction of the most informative wavelengths. Although the different measures (non-Gaussian indicators, Bhattacharyya distance) did not point to a unique wavelength to maximise feature contrast, for both topsoil and subsoil foreground (root)–background (soil) distinction was highest between 1400 and 1480 nm. Secondary regions of distinctive wavebands occurred between 1050–1090 nm and 1620–1700 nm. The region around 1450 nm corresponds to a major water absorption band (e.g. [20]). For leaf tissues Mobasheri and Fatemi [44] also found high correlation to equivalent water thickness around 1050 nm. In the third region of high root-soil contrast several spectral peaks for cellulose (1632 nm), hemicellulose (1668, 1681 nm) and lignin (1672–1674, 1677, 1685 nm) are located [62].
Segmentation accuracy was dependent on soil substrate and pre-treatment. The most efficient pre-treatment for both substrates was log-linearization and ALS correction, while thresholding was most accurate in the topsoil and fuzzy clustering in the subsoil.
Unsupervised approaches such as clustering can be readily applied in automated segmentation of phenotyping data as they do not require a user-labelled training dataset. Still clustering results depend on the user-defined cluster number which implies a certain degree of subjectivity and empiricism [28]: e.g. using two clusters only to represent the data structure, one for root and one for soil, tended to increase noise in segmentation results, while higher cluster number underestimated root length by allocating root pixels to a mixed class. Stability measures can provide some decision-support to assess the clustering result [32]. However, also clustering algorithm and distance metrics influence the segmentation result. Thus optimizing the settings of unsupervised approaches before evaluation of an image series still requires some user interaction.
Supervised machine learning approaches are often considered as particularly adapted for classification of multidimensional data [66, 79]. However, our results did not suggest an advantage of using support vector machine segmentation. Belgiu and Drǎguţ [3] and Anzanello et al. [2] concluded that transformation of spectral data could change the superiority of supervised versus unsupervised approaches. Other studies (e.g. [11]) reported a slightly better performance of supervised classification. Chen and Stow [9] underlined that size and homogeneity of training data are critical for the accuracy of supervised approaches. In our case the labelled data were only 0.6% of image pixels. Also inaccuracy of freehand-labelling could have reduced performance: considering the tiny size of root axes and constraints of camera resolution falsely labelling of soil pixels as root and vice versa, or influence of mixed border pixels can hardly be excluded [55]. Therefore unsupervised approaches were more efficient for automatic segmentation in a phenotyping context, while supervised machine learning approaches were most adapted to subsequent chemometric analyses.
Overall our results demonstrate that an adequate image processing strategy has to be found for an experimental dataset with a specific foreground (species dependent roots)–background (soil) combination. There is no unique combination of pre-treatment, band selection and segmentation algorithms that fits all hyperspectral root images. However, following the image processing pipeline presented here, the appropriate pre-treatment and segmentation methods and best bands for an optimum segmentation result can be found efficiently.
Spectral versus colour based segmentation
Our results demonstrated that even for bright subsoil segmentation via colour thresholds did not differ in performance compared to spectral segmentation. Overall the colour based segmentation was even slightly, though not significantly, more accurate than the spectral segmentation. We hypothesize that the main reason for this was the smaller pixel size of high resolution RGB cameras (6.6 μm for the camera model used here) better capturing fine lateral axes. Still there could be substrate types such silica sand (Additional file 9) where colour based segmentation fails, while still spectral patterns allow foreground–background distinction. Also more advanced segmentation algorithms (e.g. sub-pixel mapping approaches; e.g. [41, 71, 77] might further improve detection of tiny structures with the given spatial resolution of the camera.
Importantly all reports on rhizobox-like systems found a significant relation between visible and total root length [47]. Also here total root length could be predicted from visible axes (r2 = 0.74) with a similar percentage of visible roots as reported by Pfeifer et al. [53] for barley (between 20 and 30%) in a comparable setup.
Chemometric root analysis
Chemometric classification of the segmented root axes with a decision tree model revealed major spectral distinction between a central region extending to an average radius of 0.3 mm and the region beyond 0.3 mm to the outer border of root axes. It is hypothesized that the model captured distinctive constituents of the root stele via their spectral pattern (particularly related to water absorption; cf. Additional file 6). For example Watt et al. [73] reported stele diameter of cereal primary roots in the range of 0.1–0.2 mm.
Spectral root signatures changed with root decay time after clipping. The temporal change was most pronounced at a wavelength difference of 1649 minus 1447 nm. The region around 1649 nm contains several bands for cellulose, hemicellulose and lignin [35, 62], while the region around 1447 nm has strong water absorption properties [20]. It is thus suggested that the spectral pattern could indicate a change in tissue water content relative to the concentration in structural carbohydrates. The dynamics of changing spectral reflectance were modelled by an exponential curve which is common in plant (root) litter decomposition [14, 65]. Fine-mapping of the model on root pixels revealed that spatial (mainly radial) differences of root constituents were encoded by the same spectral signature as decay duration. Upon calibration with measured root chemical components (e.g. C:N, lignin, cellulose, hemicellulose, calcium; [78]) it can be expected that hyperspectral imaging provides relevant insights into the spatial and temporal biochemical patterns of plant roots and a better understanding of root functioning.
Conclusions
Hyperspectral imaging is a novel approach for root phenotyping of soil grown plants. Although acquisition and processing time limits throughput compared to RGB imaging, spectral signatures provide potential added-value by encoding physico-chemical root/soil constituents. Hyperspectral imaging thus bridges between high-throughput RGB and CT/MRI deep root phenotyping technologies. Phenotyping requires a high degree of automation in image processing. Adequate methods for root segmentation and chemometric analysis are therefore critical. With the hyperspectral image processing pipeline presented here an adapted strategy for specific experimental settings can be found efficiently. Pre-treated images were successfully segmented with unsupervised clustering and thresholding approaches. Due to lower resolution of spectral images, still there was no segmentation advantage over colour thresholding from RGB images. However using chemometric models, spectral signatures allowed to infer on distinctive radial composition of root axes and their decomposition dynamics. These first results demonstrate that the developed data processing pipeline facilitates application of hyperspectral imaging as promising technology for advances in root research. Further investigations will highlight the relevance of distinctive spectral root properties in relation to root functionality. Finally, exploration of large-size hyperspectral root data will contribute to define target wavelengths relevant for the design of multispectral, high-throughput structural–functional root phenotyping systems.
References
Afseth NK, Kohler A. Extended multiplicative signal correction in vibrational spectroscopy, a tutorial. Chemometr Intell Lab. 2012;117:92–9.
Anzanello MJ, Ortiz RS, Limberger R, Mariotti K. Performance of some supervised and unsupervised multivariate techniques for grouping authentic and unauthentic Viagra and Cialis. Egypt J Forensic Sci. 2014;4:83–9.
Belgiu M, Drǎguţ L. Comparing supervised and unsupervised multiresolution segmentation approaches for extracting buildings from very high resolution imagery. ISPRS J Photogramm. 2014;96:67–75.
Bhattacharyya A. On a measure of divergence between two multinomial populations. Sankhyā Ind J Stat. 1946;7:401–6.
Bishopp A, Lynch JP. The hidden half of crop yields. Nat Plants. 2015;1:15117.
Breiman L. Classification and regression trees. New York: Taylor & Francis; 2017.
Böhm W. Methods of studying root systems. Berlin: Springer; 1979.
Chang C-I, Safavi H. Progressive dimensionality reduction by transform for hyperspectral imagery. Pattern Recogn. 2011;44:2760–73.
Chen D, Stow D. The effect of training strategies on supervised classification at different spatial resolutions. Photogramm Eng Remote Sens. 2002;68:1155–62.
Clark RT, Famoso AN, Zhao K, Shaff JE, Craft EJ, Bustamante CD, McCouch SR, Aneshansley DJ, Kochian LV. High-throughput two-dimensional root system phenotyping platform facilitates genetic analysis of root growth and development. Plant Cell Environ. 2013;36:454–66.
Dai Q, Sun DW, Xiong Z, Cheng JH, Zeng XA. Recent advances in data mining techniques and their applications in hyperspectral image processing for the food industry. Compr Rev Food Sci Food Saf. 2014;13:891–905.
Dhondt S, Wuyts N, Inzé D. Cell to whole-plant phenotyping: the best is yet to come. Trends Plant Sci. 2013;18:428–39.
Dorrepaal R, Malegori C, Gowen A. Tutorial: time series hyperspectral image analysis. J Near Infrared Spec. 2016;24:89–107.
Dornbush ME, Isenhart TM, Raich JW. Quantifying fine-root decomposition: an alternative to buried litterbags. Ecology. 2002;83:2985–90.
Elvidge CD. Visible and near infrared reflectance characteristics of dry plant materials. Remote Sens. 1990;11:1775–95.
Esquerre C, Gowen AA, Burger J, Downey G, O’Donnell CP. Suppressing sample morphology effects in near infrared spectral imaging using chemometric data pre-treatments. Chemometr Intell Lab. 2012;117:129–37.
Fahlgren N, Gehan MA, Baxter I. Lights, camera, action: high-throughput plant phenotyping is ready for a close-up. Curr Opin Plant Biol. 2015;24:93–9.
Fiorani F, Schurr U. Future scenarios for plant phenotyping. Ann Rev Plant Biol. 2013;64:267–91.
Fisher K, Chang C-I. Progressive band selection. Proc SPIE. 2009;7457:745709.
Gausman HW, Allen WA. Optical parameters of leaves of 30 plant species. Plant Physiol. 1973;52:57–62.
Geng X, Sun K, Ji L, Tang H, Zhao Y. Joint skewness and its application in unsupervised band selection for small target detection. Sci Rep. 2015;5:9915.
Guizar-Sicairos M, Thurman ST, Fienup JR. Efficient subpixel image registration algorithms. Opt Lett. 2008;33:156–8.
IUSS. World reference base for soil resources international soil classification system for naming soils and creating legends for soil maps. Rome: FAO; 2014. p. 2014.
Hargreaves CE, Gregory PJ, Bengough AG. Measuring root traits in barley (Hordeum vulgare ssp vulgare and ssp spontaneum) seedlings using gel chambers, soil sacs and X-ray microtomography. Plant Soil. 2009;316:285–97.
Himmelbauer ML, Loiskandl W, Kastanek F. Estimating length, average diameter and surface area of roots using two different image analyses systems. Plant Soil. 2004;260:111–20.
Hochholdinger F. Untapping root system architecture for crop improvement. J Exp Bot. 2016;67:4431–3.
Iyer-Pascuzzi AS, Symonova O, Mileyko Y, Hao Y, Belcher H, Harer J, Weitz JS, Benfey PN. Imaging and analysis platform for automatic phenotyping and trait ranking of plant root systems. Plant Physiol. 2010;152:1148–57.
Jain AK. Data clustering: 50 years beyond K-means. Pattern Recogn Lett. 2010;31:651–66.
Kim DM, Zhang H, Zhou H, Du T, Wu Q, Mockler TC, Berezin MY. Highly sensitive image-derived indices of water-stressed plants using hyperspectral imaging in SWIR and histogram analysis. Sci Rep. 2015;5:15919.
Kokaly RF, Clark RN, Swayze GA, Livo KE, Hoefen TM, Pearson NC, Wise RA, Benzel WM, Lowers HA, Driscoll RL. USGS Spectral Library Version 7 Data: US Geological Survey data release. 2017.
Kucheryavskiy S. Multivariate data analysis toolbox for MATLAB. 2014. https://github.com/svkucheryavski/mdatoolsm. Accessed 12 July 2018.
Lange T, Roth V, Braun ML, Buhmann JM. Stability-based validation of clustering solutions. Neural Comput. 2004;16:1299–323.
Le Marié C, Kirchgessner N, Marschall D, Walter A, Hund A. Rhizoslides: paper-based growth system for non-destructive, high throughput phenotyping of root development by means of image analysis. Plant Methods. 2014;10:1.
Li BN, Chui CK, Chang S, Ong SH. Integrating spatial fuzzy clustering with level set methods for automated medical image segmentation. Comput Biol Med. 2011;41:1–10.
Li X, Sun C, Zhou B, He Y. Determination of hemicellulose, cellulose and lignin in moso bamboo by near infrared spectroscopy. Sci Rep. 2015;5:17210.
Li L, Zhang Q, Huang D. A review of imaging techniques for plant phenotyping. Sensors. 2014;14:20078–111.
Lobet G, Draye X. Novel scanning procedure enabling the vectorization of entire rhizotron-grown root systems. Plant Methods. 2013;9:1.
Lynch JP, Brown KM. New roots for agriculture: exploiting the root phenome. Philos Trans R Soc B. 2012;367:1598–604.
Ma Z, Guo D, Xu X, Lu M, Bardgett RD, Eissenstat DM, McCormack ML, Hedin LO. Evolutionary history resolves global organization of root functional traits. Nature. 2018;555:94–7.
Mairhofer S, Zappala S, Tracy S, Sturrock C, Bennett MJ, Mooney SJ, Pridmore TP. Recovering complete plant root system architectures from soil via X-ray μ-computed tomography. Plant Methods. 2013;9:1.
Mertens KC, Verbeke LPC, Westra T, De Wulf RR. Sub-pixel mapping and sub-pixel sharpening using neural network predicted wavelet coefficients. Remote Sens Environ. 2004;91:225–36.
Metzner R, Eggert A, van Dusschoten D, Pflugfelder D, Gerth S, Schurr U, Uhlmann N, Jahnke S. Direct comparison of MRI and X-ray CT technologies for 3D imaging of root systems in soil: potential and challenges for root trait quantification. Plant Methods. 2015;11:1.
Mishra P, Asaari MSM, Herrero-Langreo A, Lohumi S, Diezma B, Scheunders P. Close range hyperspectral imaging of plants: a review. Biosyst Eng. 2017;164:49–67.
Mobasheri MR, Fatemi SB. Leaf equivalent water thickness assessment using reflectance at optimum wavelengths. Theor Exp Plant Physiol. 2013;25:196–202.
Mooney SJ, Pridmore TP, Helliwell J, Bennett MJ. Developing X-ray computed tomography to non-invasively image 3-D root systems architecture in soil. Plant Soil. 2012;352:1–22.
Nagel KA, Kastenholz B, Jahnke S, Van Dusschoten D, Aach T, Mühlich M, Truhn D, Scharr H, Terjung S, Walter A, Schurr U. Temperature responses of roots: impact on growth, root system architecture and implications for phenotyping. Funct Plant Biol. 2009;36:947–59.
Nagel KA, Putz A, Gilmer F, Heinz K, Fischbach A, Pfeifer J, Faget M, Blossfeld S, Ernst M, Dimaki C, et al. GROWSCREEN-Rhizo is a novel phenotyping robot enabling simultaneous measurements of root and shoot growth for plants grown in soil-filled rhizotrons. Funct Plant Biol. 2012;39:891–904.
Nagel KA, Bonnett D, Furbank R, Walter A, Schurr U, Watt M. Simultaneous effects of leaf irradiance and soil moisture on growth and root system architecture of novel wheat genotypes: implications for phenotyping. J Exp Bot. 2015;66:5441–52.
Nakaji T, Noguchi K, Oguma H. Classification of rhizosphere components using visible–near infrared spectral images. Plant Soil. 2008;310:245–61.
Osmont KS, Sibout R, Hardtke CS. Hidden branches: developments in root system architecture. Ann Rev Plant Biol. 2007;58:93–113.
Pandey P, Ge Y, Stoerger V, Schnable JC. High throughput in vivo analysis of plant leaf chemical properties using hyperspectral imaging. Front Plant Sci. 2017;8:1348.
Passioura JB. The perils of pot experiments. Funct Plant Biol. 2006;33:1075–9.
Pfeifer J, Faget M, Walter A, Blossfeld S, Fiorani F, Schurr U, Nagel KA. Spring barley shows dynamic compensatory root and shoot growth responses when exposed to localised soil compaction and fertilisation. Funct Plant Biol. 2014;41:581–97.
Pierret A. Multi-spectral imaging of rhizobox systems: new perspectives for the observation and discrimination of rhizosphere components. Plant Soil. 2008;310:263–8.
Plaza J, Plaza A, Perez R, Martinez P. On the use of small training sets for neural network-based characterization of mixed pixels in remotely sensed hyperspectral images. Pattern Recogn. 2009;42:3032–45.
Poorter H, Bühler J, Dusschoten D, Climent J, Postma JA. Pot size matters: a meta-analysis of the effects of rooting volume on plant growth. Funct Plant Biol. 2012;39:839–50.
Price AH, Steele KA, Gorham J, Bridges JM, Moore BJ, Evans JL, Richardson P, Jones RGW. Upland rice grown in soil-filled chambers and exposed to contrasting water-deficit regimes: I. Root distribution, water use and plant water status. Field Crops Res. 2002;76:11–24.
Pu YY, Feng YZ, Sun DW. Recent progress of hyperspectral imaging on quality and safety inspection of fruits and vegetables: a review. Compr Rev Food Sci Food Saf. 2015;14:176–88.
Sawand S. Algorithm to compare the slopes (regression coefficients) between the subgroups in simple/multiple regression using PROC REG. 2012. http://www.pharmasug.org/proceedings/2012/PO/PharmaSUG-2012-PO04.pdf. Accessed 12 July 2018.
Saxton KE, Rawls WJ. Soil water characteristic estimates by texture and organic matter for hydrologic solutions. Soil Sci Soc Am J. 2006;70:1569–78.
Schenk HJ, Jackson RB. The global biogeography of roots. Ecol Monogr. 2002;72:311–28.
Schwanninger M, Rodrigues JC, Fackler K. A review of band assignments in near infrared spectra of wood and wood components. J Near Infrared Spec. 2011;19:287–308.
Shen J, Li C, Mi G, Li L, Yuan L, Jiang R, Zhang F. Maximizing root/rhizosphere efficiency to improve crop productivity and nutrient use efficiency in intensive agriculture of China. J Exp Bot. 2012;64:1181–92.
Shenk JS, Workman JJ, Westerhaus MO. Application of NIR spectroscopy to agricultural products. Prac Spectrosc Ser. 2001;27:419–74.
Silver WL, Miya RK. Global patterns in root decomposition: comparisons of climate and litter quality effects. Oecologia. 2001;129:407–19.
Singh A, Ganapathysubramanian B, Singh AK, Sarkar S. Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci. 2016;21:110–24.
Smit AL, Bengough AG, Engels C, van Noordwijk M, Pellerin S, van de Geijn SC. Root methods: a handbook. Berlin: Springer; 2013.
Sposito G. Green water and global food security. Vadose Zone J. 2013. https://doi.org/10.2136/vzj2013.02.0041.
Stingaciu L, Schulz H, Pohlmeier A, Behnke S, Zilken H, Javaux M, Vereecken H. In situ root system architecture extraction from magnetic resonance imaging for water uptake modeling. Vadose Zone J. 2013. https://doi.org/10.2136/vzj2012.0019.
Thenkabail PS, Lyon JG, Huete A. Hyperspectral remote sensing of vegetation. Boca Raton: CRC Press; 2016.
Trujillo-Pino A, Krissian K, Alemán-Flores M, Santana-Cedrés D. Accurate subpixel edge location based on partial area effect. Image Vis Comput. 2013;31:103–16.
Vidal M, Amigo JM. Pre-processing of hyperspectral images. Essential steps before image analysis. Chemometr Intell Lab. 2012;117:138–48.
Watt M, Magee LJ, McCully ME. Types, structure and potential for axial water flow in the deepest roots of field-grown cereals. New Phytol. 2008;178:135–46.
Watt M, Moosavi S, Cunningham SC, Kirkegaard JA, Rebetzke GJ, Richards RA. A rapid, controlled-environment seedling root screen for wheat correlates well with rooting depths at vegetative, but not reproductive, stages at two field sites. Ann Bot. 2013;112:447–55.
Wojciechowski T, Gooding MJ, Ramsay L, Gregory PJ. The effects of dwarfing genes on seedling root growth of wheat. J Exp Bot. 2009;60:2565–73.
Wold S. Chemometrics; what do we mean with it, and what do we want from it? Chemometr Intell Lab. 1995;30:109–15.
Xiao X, Geyer VF, Bowne-Anderson H, Howard J, Sbalzarini IF. Automatic optimal filament segmentation with sub-pixel accuracy using generalized linear models and B-spline level-sets. Med Image Anal. 2016;32:157–72.
Zhang X, Wang W. The decomposition of fine and coarse roots: their global patterns and controlling factors. Sci Rep. 2015;5:9940.
Zhao J, Bodner G, Rewald B. Phenotyping: using machine learning for improved pairwise genotype classification based on root traits. Front Plant Sci. 2016;7:1864.
Zhao J, Bodner G, Rewald B, Leitner D, Nagel KA, Nakhforoosh A. Root architecture simulation improves the inference from seedling root phenotyping towards mature root systems. J Exp Bot. 2017;68:965–82.
Zobel RW, Waisel Y. A plant root system architectural taxonomy: a framework for root nomenclature. Plant Biosys. 2010;144:507–12.
Authors’ contributions
GB evaluated the data and wrote the manuscript; AN performed the experiments; TA constructed the imaging setup and programmed the control software; DL and GB developed the Matlab scripts for image processing and image analysis. All authors read and approved the final manuscript.
Acknowledgements
The authors acknowledge the funding received for this study by the Austrian Science Fund (FWF) in the context of the project “The roots of drought resistance” (P25190-B16) and the financial support from the Government of Lower Austria (K3 F-282/001-2012).
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The Matlab scripts and datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
The study was financed by the Austrian Science Fund (FWF; Project Number P25190-B16) and the Government of Lower Austria (Project Number K3 F-282/001-2012).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1.
Soil water retention curves of top- and subsoil.
Additional file 2.
Example of different pre-treatment evaluation and band selection approaches.
Additional file 3.
Movie over all wavelength for topsoil image with log-linearization and ALS baseline correction.
Additional file 4.
Relation between segmented root length and image noise.
Additional file 5.
Relation between visible and total root length.
Additional file 6.
Raw spectra of root pixels from center to border region.
Additional file 7.
Untransformed and first derivative spectra during root decay.
Additional file 8.
Histograms at wavelengths with maximum separation between initial and final decay time for different pre-treatments.
Additional file 9.
RGB image of sand grown root system with spectral segmentation and manual tracking.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Bodner, G., Nakhforoosh, A., Arnold, T. et al. Hyperspectral imaging: a novel approach for plant root phenotyping. Plant Methods 14, 84 (2018). https://doi.org/10.1186/s13007-018-0352-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13007-018-0352-1