Abstract
Background
Porcine circovirus type 3 (PCV3), as an emerging circovirus species, was reported to be widely circulating in the United States, China, South Korea and Poland. Previous studies revealed that PCV3 was mainly concentrated in sick animals with respiratory disease, skin disease, reproductive disorders and so on. However, the circulating status of PCV3 in pigs with other clinical presentations (especilly asymptomatic or diarrhea) was not well established.
Findings
In this study, to conduct a comparative epidemiological survey of PCV3, 80 weaned pig serum samples with severe respiratory disease (SRD), 175 weaned pig serum samples with mild respiratory disease (MRD), 216 asymptomatic weaned pig serum samples, 35 diarrheal weaned pig samples and 35 non-diarrheal weaned pig samples were collected from eight provinces of China. Via qPCR testing, PCV3 was circulating in all sampling provinces, with total positive rates varying from 1.04% to 100%. Interestingly, the PCV3-positive rate was significantly higher in weaned pigs with SRD (63.75%, 51/80) than in those weaned pigs with MRD (13.14%, 23/175) and asymptomatic pigs (1.85%, 4/216) (P < 0.01). Similarly, the PCV3-positive rate was significantly higher in diarrheal weaned pigs (17.14%, 6/35) than in non-diarrheal weaned pigs (2.86%, 1/35) (P < 0.05). Moreover, the lower Ct values of qPCR were frequently found in those weaned pigs or fattening pigs with respiratory disease and diarrhea rather than that in asymptomatic pigs. Sequence analysis showed that low genetic diversity existed among those PCV3 sequences collected from pigs with different clinical presentations.
Conclusions
The present study further extends evidence that newly described PCV3 widely circulates in six additional provinces of Southern and Northern China and has high similarity to previously reported isolates. As an emerging virus of swine, although the present case-control study reveals that PCV3 has a potential association with swine respiratory disease and diarrhea, further investigations into the pathogenesis are needed to ascertain the role of PCV3 in swine health.
Similar content being viewed by others
Background
At present, small non-enveloped DNA circoviruses of concern to animal and human health are classified in two genera (Circovirus and Cyclovirus) in the Circoviridae family. The genus Circovirus consists of 27 species, while the genus Cyclovirus has 43 species [1]. Porcine circoviruses are members of the genus Circovirus with two recognized species, porcine circovirus type 1 (PCV1) and porcine circovirus type 2 (PCV2). PCV1 can infect and replicate in swine, but is not known to cause any obvious disease [2]. While PCV2 infection can result in significant production and economic losses to the worldwide swine industry [3, 4]. Clinically, PCV2 can cause porcine circovirus disease (PCVD) or porcine circovirus-associated disease (PCVAD), which is characterized by postweaning multisystemic wasting syndrome (PMWS), porcine dermatitis and nephrotic syndrome (PDNS), interstitial pneumonia, and reproductive disorders [5, 6].
Recently, a novel circovirus significantly divergent from PCV1 and PCV2, provisionally designated porcine circovirus type 3 (PCV3), was identified in U.S. swine herds [7, 8]. The full-length genome sequence of newly found PCV3 was 2000 nucleotides, which was considerably larger than those of PCV1 and PCV2 (~1760 and 1780 nucleotides, respectively) [9]. Actually, prior to identification of PCV3, PCV3-like sequences were also reported in different farmed animals several years ago, which revealed possible early distribution of PCV3 in animal species [10, 11]. It is interesting that PCV3, similar to PCV2, has a potential association with multi-systemic disease, PDNS and reproductive failure in pigs [7, 8]. At present, PCV3 also has been reported in China, South Korea and Poland, accompanying with widely geographical distribution [12,13,14,15,16,17]. Despite good progress in PCV3 epidemiology, the circulating status of PCV3 in pigs with different clinical presentations (especilly asymptomatic or diarrhea) was not well established. Therefore, the present study aims to perform comparative epidemiology of PCV3 in pigs with different clinical presentations.
Methods
In 2016, two small-scale fattening pig farms (about 200 or 300 American Landrace pigs in total), locating in Guangdong province (southern China) and Jilin province (northern China) separated by more than 3, 000 km in distance (Fig. 1), suffered from severe respiratory disease (SRD). Pigs (2-3 months old) on both farms have received immunizations against PCV2, pseudorabies virus (PRV), classical swine fever virus (CSFV) and type 2 porcine reproductive and respiratory syndrome virus (PRRSV). However, the morbidity rate of pigs on the two farms was approximately 60%, the mortality in affected pigs was about 80%. From autopsy results of 18 individual dead pigs (n = 8 for farm A, n = 10 for farm B) (Additional file 1: Table S1), the similar macroscopic lesions were concentrated in the lungs and lymph nodes, which was characterized by PRRSV- or PCV2-like lesions including pulmonary interstitial widening, congestion and lymphadenopathy bleeding. These 18 pig lung samples were defined as the samples with SRD. To compare the detection results of PCV3 based on the same organ and same age of pigs, additional 80 swine (with SRD, 5-8 weeks old) serum samples, 216 asymptomatic weaned swine (5-8 weeks old) serum samples and 175 diseased weaned swine (5-8 weeks old) (with mild respiratory disease [MRD] including cough, softly panting, abdominal breathing) serum samples (Additional file 1: Table S1) were collected from Shandong province (n = 26), Gansu province (n = 120), Guangxi zhuang autonomous region (n = 34), Guangdong province (n = 130), Sichuan province (n = 65) and Yunnan province (n = 96) (Fig. 1). Moreover, 35 diarrheal weaned pig (6-7 weeks old) samples and 35 non-diarrheal weaned pig (6-7 weeks old) samples (Additional file 1: Table S1) were collected from Gansu province (n = 40) and Neimenggu autonomous region (n = 30) (Fig. 1). The above-mentioned samples were stored −80 °C until further use.
Prior to quantitative PCR (qPCR), 200 μL tissue supernatants or serum samples were used to perform viral DNA extraction according to the manufacturer’s instructions (TIANGEN Biotech Co., Ltd. Beijing, China). Viral DNA samples were stored at −80 °C until use. A previously established qPCR protocol was used, consisting of two specific primers (Forward, 5′–AGTGCTCCCCATTGAACG–3′; Reverse, 5′–ACACAGCCGTTACTTCAC–3′) and one modified probe (5′–FAM–ACCCCATGGCTCAACACATATGACC–BHQ1–3′) [8]. Moreover, the detection of PRRSV was performed according to a previous method [18]. The full-length genome sequence of PCV3 was amplified using the two pairs of overlapping PCR primers according to a previous description [13]. These two PCR fragments were cloned and sequenced. The corresponding sequencing results were spliced using SeqMan program (DNAStar software version 7). Sequence alignment and phylogenetic analysis based on three species of PCV sequences were performed using Clustal W program implemented in DNAStar software and MEGA 5.1 software, respectively. In addtion, qPCR detection results in pigs with different clinical presentations were analyzed by chisquare (χ2) test (Ziyue software), the value of P < 0.05 and P < 0.01 was considered significant and very significant, respectively.
Results
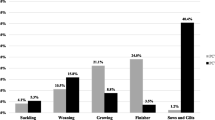
After testing by qPCR, PCV3 was circulating in all sampling provinces, with total positive rates of 100% (8/8) in Jilin, 44.29% (62/140) in Guangdong, 7.69% (2/26) in Shandong, 3.75% (6/160) in Gansu, 8.82% (3/34) in Guangxi, 13.85% (9/65) in Sichuan, 1.04% (1/96) in Yunnan and 23.33% (7/30) in Neimenggu, respectively. Interestingly, the PCV3-positive rate was significantly higher in weaned pigs with SRD (63.75%, 51/80) than in those weaned pigs with MRD (13.14%, 23/175) and asymptomatic pigs (1.85%, 4/216) (P < 0.01) (Fig. 2a). Similarly, the PCV3-positive rate was significantly higher in diarrheal weaned pigs (17.14%, 6/35) than in non-diarrheal weaned pigs (2.86%, 1/35) (P < 0.05) (Fig. 2a). Moreover, the lower Ct values were frequently found in those samples collected from fattening or weaned pigs with respiratory disease and diarrhea rather than those from asymptomatic pigs (Fig. 2b). In addition, co-infection of PCV3 and PRRSV was found in 29 of 140 SRD pigs, not in MRD pigs and clinically asymptomatic pigs.
The detection results of PCV3 in pigs with different clinical presentations. a PCV3-positive rate in pigs with different clinical presentations; b Ct value of qPCR for the detection of PCV3 in pigs with different clinical presentations. Note: SRD means severe respiratory disease, while MRD means mild respiratory disease
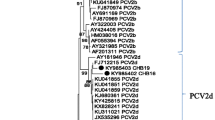
To further genetically characterize PCV3 in those positive samples with low Ct values (< 22), two representative genome sequences (PCV3-CHN/GD2016 and PCV3-CHN/CC2016) were obtained from south China and north China, respectively, and were deposited into GenBank database under accession numbers KY421347-KY421348. Like previously reported PCV3 sequences, their genomes were 2000 nucleotides in length. Multiple sequence alignment results showed that the two current PCV3 genome sequences shared > 97% nucleotide similarity with other known PCV3 sequences, revealing low levels of genetic variation among PCV3 isolates. Online Blastn alignment results showed that they mainly differed in ORF2 gene that encodes viral capsid protein (Data not shown). In addition, phylogenetic analysis showed that PCV3-CHN/GD2016 and PCV3-CHN/CC2016 clustered with those PCV3 isolates from the United States and Korea, and were distantly related to PCV1 and PCV2 (Fig. 3).
Phylogenetic analysis based on complete genome sequences of PCV1, PCV2 and PCV3. The phylogenetic tree was constructed by the Maximum Likelihood method using MEGA 5.1 software. One thousand bootstrap replicates (1000 times) were used to assess tree topology. Only bootstrap scores of at least 50 were retained. Scale bar indicates 10% nucleotide sequence divergence. The PCV3 isolates of PCV3-CHN/GD2016 and PCV3-CHN/CC2016 obtained in this study are indicated by gray shading
Discussion
Previous studies suggested that PCV3 was common in PDNS cases and that it might play a role in reproductive failure and multi-systemic inflammation in U.S swine herds [7, 8]. Although there were other swine pathogens (including torque teno sus virus 1, TTSuV1; porcine hemagglutinating encephalomyelitis virus, PHEV; porcine astrovirus 4, PAstV4) present in PCV3-infected tissues, high numbers of PCV3 sequence reads by metagenomic sequencing suggested that PCV3 might play a potentially pathogenic role in those clinical cases [7, 8]. Similarly, immunohistochemistry localized PCV3 in typical PDNS lesions in the absence of PCV2 [8]. However, in four latest studies from China, single PCV3 infection and/or co-infection with PCV2 were reported in clinical cases [12, 13, 15, 16]. Surprisingly, during one of four studies, PCV3 had an abnormally high positive rate (85.7%, 12/14) in pig samples with productive failure, but PCV2, PRRSV and PRV were negative in these pigs [15]. In this study, we identified co-infection of PCV3 with PRRSV in SRD pigs, not in other pigs. Co-infection of pigs with PCV2 and PRRSV is known to exacerbate disease severity [19, 20]. A possible synergy between PCV3 and PRRSV or PCV2 co-infection with disease severity warrants further investigation. Additionally, low Ct values of PCV3 were frequently found in diseased pigs, not in asymptomatic pigs (Fig. 2b), which revealed that the high titers of PCV3, similar to PCV2, could contribute to the occurrence of clinical diseases in pigs [21]. Another interesting finding was that PCV3 was detected in fecal samples, which may suggest for a fecal-oral transmission by PCV3.
Conclusions
To conclude, the present study further extends evidence that newly described PCV3 widely circulates in six additional provinces of Southern and Northern China and has high similarity to previously reported isolates. As an emerging virus of swine, although the present case-control study reveals that PCV3 has a potential association with swine respiratory disease and diarrhea, further investigations into the pathogenesis are needed to ascertain the role of PCV3 in swine health. Similar to PCV2 [22], the usage of viral infectious clone accompanying by clinical and histological methods could resolve this scientific issue.
Abbreviations
- CSFV:
-
classical swine fever virus
- MRD:
-
mild respiratory disease
- PAstV4:
-
porcine astrovirus 4
- PCV1:
-
Porcine circovirus type 1
- PCV2:
-
Porcine circovirus type 2
- PCV3:
-
Porcine circovirus type 3
- PCVAD:
-
porcine circovirus-associated disease
- PCVD:
-
porcine circovirus disease
- PDNS:
-
porcine dermatitis and nephrotic syndrome
- PHEV:
-
porcine hemagglutinating encephalomyelitis virus
- PMWS:
-
postweaning multisystemic wasting syndrome
- PRRSV:
-
porcine reproductive and respiratory syndrome virus
- PRV:
-
pseudorabies virus
- qPCR:
-
quantitative PCR
- SRD:
-
severe respiratory disease
- TTSuV1:
-
torque teno sus virus 1
References
Rosario K, Breitbart M, Harrach B, Segalés J, Delwart E, Biagini P, Varsani A. Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch Virol. 2017;162:1447–63.
Tischer I, Mields W, Wolff D, Vagt M, Griem W. Studies on epidemiology and pathogenicity of porcine circovirus. Arch Virol. 1986;91:271–6.
Finsterbusch T, Mankertz A. Porcine circoviruses--small but powerful. Virus Res. 2009;143:177–83.
Ramamoorthy S, Meng XJ. Porcine circoviruses: a minuscule yet mammoth paradox. Anim Health Res Rev. 2009;10:1–20.
Segalés J, Allan GM, Domingo M. Porcine circovirus diseases. Anim Health Res Rev. 2005;6:119–42.
Opriessnig T, Meng XJ, Halbur PG. Porcine circovirus type 2 associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J Vet Diagn Investig. 2007;19:591–615.
Phan TG, Giannitti F, Rossow S, Marthaler D, Knutson TP, Li L, Deng X, Resende T, Vannucci F, Delwart E. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol J. 2016;13:184.
Palinski R, Piñeyro P, Shang P, Yuan F, Guo R, Fang Y, Byers E, Hause BM. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J Virol. 2017;91:e01879–16.
Zhai SL, Chen SN, Xu ZH, Tang MH, Wang FG, Li XJ, Sun BB, Deng SF, Hu J, Lv DH, Wen XH, Yuan J, Luo ML, Wei WK. Porcine circovirus type 2 in China: an update on and insights to its prevalence and control. Virol J. 2014;11:88.
Li L, Shan T, Soji OB, Alam MM, Kunz TH, Zaidi SZ, Delwart E. Possible cross-species transmission of circoviruses and cycloviruses among farm animals. J Gen Virol. 2011;92:768–72.
Zhang W, Li L, Deng X, Kapusinszky B, Delwart E. What is for dinner? Viral metagenomics of US store bought beef, pork, and chicken. Virology. 2014;468-470:303–10.
Shen H, Liu X, Zhang P, Wang L, Liu Y, Zhang L, Liang P, Song C. Genome characterization of a porcine circovirus type 3 in South China. Transbound Emerg Dis. 2017. doi:10.1111/tbed.12639.
Ku X, Chen F, Li P, Wang Y, Yu X, Fan S, Qian P, Wu M, He Q. Identification and genetic characterization of porcine circovirus type 3 in China. Transbound Emerg Dis. 2017;64:703–8.
Kwon T, Yoo SJ, Park CK, Lyoo YS. Prevalence of novel porcine circovirus 3 in Korean pig populations. Vet Microbiol. 2017;207:178–80.
Fan S, Ku X, Chen F, Wang Y, Yu X, He Q. Complete genome sequence of a novel porcine circovirus type 3 strain, PCV3/CN/Hubei-618/2016, isolated from China. Genome Announc. 2017;5:e00100–17.
Zheng S, Wu X, Zhang L, Xin C, Liu Y, Shi J, Peng Z, Xu S, Fu F, Yu J, Sun W, Xu S, Li J, Wang J. The occurrence of porcine circovirus 3 without clinical infection signs in Shandong Province. Transbound Emerg Dis. 2017;64:1337–41.
Stadejek T, Wozniak A, Miłek D, Biernacka K. First detection of porcine circovirus type 3 on commercial pig farms in Poland. Transbound Emerg Dis. 2017;64:1350–3.
Nilubol D, Tripipat T, Hoonsuwan T, Tipsombatboon P, Piriyapongsa J. Genetic diversity of the ORF5 gene of porcine reproductive and respiratory syndrome virus (PRRSV) genotypes I and II in Thailand. Arch Virol. 2013;158:943–53.
Fan P, Wei Y, Guo L, Wu H, Huang L, Liu J, Liu C. Synergistic effects of sequential infection with highly pathogenic porcine reproductive and respiratory syndrome virus and porcine circovirus type 2. Virol J. 2013;10:265.
Opriessnig T, Gauger PC, Faaberg KS, Shen H, Beach NM, Meng XJ, Wang C, Halbur PG. Effect of porcine circovirus type 2a or 2b on infection kinetics and pathogenicity of two genetically divergent strains of porcine reproductive and respiratory syndrome virus in the conventional pig model. Vet Microbiol. 2012;158:69–81.
Brunborg IM, Jonassen CM, Moldal T, Bratberg B, Lium B, Koenen F, Schönheit J. Association of myocarditis with high viral load of porcine circovirus type 2 in several tissues in cases of fetal death and high mortality in piglets. A case study. J Vet Diagn Investig. 2007;19:368–75.
Fenaux M, Halbur PG, Haqshenas G, Royer R, Thomas P, Nawagitgul P, Gill M, Toth TE, Meng XJ. Cloned genomic DNA of type 2 porcine circovirus is infectious when injected directly into the liver and lymph nodes of pigs: characterization of clinical disease, virus distribution, and pathologic lesions. J Virol. 2002;76:541–51.
Acknowledgements
We are very grateful to Dr. Zuzhang Wei (Guangxi University, Guangxi, China), Dr. Yu Deng (Xichang College, Sichuan, China), Mr. Hexing Wang (Mengzi Animal Disease Prevention and Control Center, Yunnan, China), Dr. Jianmin Chen, Mr. Shibin Chen and Zhiqiang Hu (anonymous affiliations as requested) for clinical sample collection.
Funding
This study was funded by Guangdong Provincial Department of Science and Technology (Grant nos. 2015A020208008, 2016A040403083 and 2016B020234006) and by South Dakota State University’s agricultural experimental station (Grant no. 3AH-477) to FL and DW.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its Additional file.
Author information
Authors and Affiliations
Contributions
SLZ designed the study, analyzed the data, and wrote the paper; XZ and HZ contributed to sample processing, viral DNA extraction, PCR and subcloning; WKW, QLC, DHL and XHW provided essential reagents and helped to write the paper; BMH, TL and RL provided valuable suggestion. DW and FL revised the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
Not applicable.
Competing interest
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1: Table S1.
The detection results of qPCR for PCV3 in collected samples. (DOC 65 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhai, SL., Zhou, X., Zhang, H. et al. Comparative epidemiology of porcine circovirus type 3 in pigs with different clinical presentations. Virol J 14, 222 (2017). https://doi.org/10.1186/s12985-017-0892-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12985-017-0892-4