Abstract
Currently, porcine circovirus type 2 (PCV2) is considered the major pathogen of porcine circovirus associated-diseases (PCVAD) that causes large economic losses for the swine industry in the world annually, including China. Since the first report of PCV2 in 1998, it has been drawing tremendous attention for the government, farming enterprises, farmers, and veterinary practitioners. Chinese researchers have conducted a number of molecular epidemiological work on PCV2 by molecular approaches in the past several years, which has resulted in the identification of novel PCV2 genotypes and PCV2-like agents as well as the description of new prevalence patterns. Since late 2009, commercial PCV2 vaccines, including the subunit vaccines and inactivated vaccines, have already been used in Chinese swine farms. The aim of this review is to update the insights into the prevalence and control of PCV2 in China, which would contribute to understanding the epidemiology, control measures and design of novel vaccines for PCV2.
Similar content being viewed by others
Introduction
Porcine circoviruses (PCVs), the smallest known animal viruses, including PCV1 and PCV2, are members of the genus Circovirus in the Circoviridae family. PCV1 was first discovered in 1974 as a contaminant of the porcine kidney cell line PK-15 (ATCC CCL-33) and was considered non-pathogenic [1, 2]; Whereas PCV2 was isolated from pigs suffering from post-weaning multisystemic wasting syndrome (PMWS) over 20 years later [3].
Generally, PCV1 genome consists of 1759 nucleotides (NTs), while PCV2 has 1767 or 1768 NTs. PCV1 has only one genome map (Figure 1). For PCV2, it has three kinds of genome organisation, map 1, map 2 and map 3, respectively (Figure 1). PCV2 has three major open reading frames (ORFs), ORF1 (945 NTs, position 51 to 995), ORF2 (702 or 705 NTs, position 1734/1735 to 1030/1033/1034) and ORF3 (315 NTs, position 671 to 357), respectively (Figure 1). The three ORFs encode replicase protein that involves in viral replication (to attain self-replication), the immunogenic capsid protein and the viral pathogenesis-associated protein, respectively [4–6]. One latest study reported that a newly discovered viral protein, ORF4 (180 NTs, position 386 to 565), was not essential for PCV2 replication yet having a role in suppressing caspase activity and regulating CD4(+) and CD8(+) T lymphocytes during PCV2 infection [7].
For life cycle of PCV, firstly, PCV uses glycosaminoglycans (GAG) as attachment receptors. The ssDNA genome is transported into the nucleus and converted by host enzymes into a dsDNA intermediate. The rep and cap mRNAs are transcribed to synthesize proteins, which are then imported from the cytoplasm. Rep/Rep’ then binds to the dsDNA and initiates rolling-circle replication (RCR) by introduction of a nick, which serves as primer [8]. Elongation of the primer by host enzymes leads to replication (Figure 2). Meanwhile, the Rep protein is covalently bound to the DNA and terminates the reaction by introduction of a second cleavage reaction via Tyr-93 (Figure 2). How viruses were assembled and released, have not yet been studied [8].
Life cycles of PCV. Note: The figure was showed in the previous reference [8]. Abbreviations: Glycosaminoglycans (GAG); Rolling-circle replication (RCR); Minimal binding site (MBS).
PCVs, especially PCV2, cause huge hazards to swine industry. At present, PCV2 is associated with a number of diseases, collectively known as porcine circovirus diseases (PCVD) in Europe [9], or porcine circovirus-associated diseases (PCVAD) in North America [10]. Apart from PMWS, PCVD/PCVAD also includes reproductive disorders [11], porcine dermatitis and nephropathy syndrome (PDNS) [12], nervous system lesions [13], porcine respiratory disease complex (PRDC) [14], enteritis [15, 16], and proliferative and necrotizing pneumonia (PNP) [17, 18]. Recently, a new PCV2-related disease syndrome called acute pulmonary edema (APE) that is different from the PCVD/PCVAD syndromes mentioned above was described in U.S. swine herds vaccinated with PCV2 [19].
A number of field and experimental studies that were performed in some countries of North America, Europe and Asia indicated that commercial vaccines (based on genotype PCV2a) against PCV2 were effective in many aspects, including reducing the incidence of PMWS, the number of co-infections, the severity of lesions in lymphoid tissues [20–25], the level of PCV2 viraemia and the severity of microscopically-visible lesions [26–29] as well as improving average daily weight gain and feed conversion ratios [20–25]. However, due to the subsequent possible vaccination pressure, novel variant strains or genotypes (such as PCV1/2, PCV2d) emerged in Canadian and U.S. swine herds [30–32].
In recent years, due to continuous losses caused by PCVD, the government at home and abroad supports scientists’ great deal of work on molecular epidemiology and vaccine development of PCV2. The aim of the review is to update the insights on the prevalence and control of PCV2 in China, which would help in understanding of the epidemiology, adjusting control measures, and the design of novel vaccines against PCV2.
Review
PCV2 Genotypes in China
After the first identification of PCV2 in China in 1998, a comprehensive molecular epidemiological survey was performed in different regions of China between 2001 and 2003 [33]. The results based on full-length ORF2 sequence level identified nine different genotypes (CHN-2A, CHN-2B, CHN-2C, CHN-2D, CHN-2E, CHN-2F, CHN-2G, CHN-2H, and CHN-2I) using restriction fragment length polymorphism (RFLP) analysis. Additional PCV2 genotypes were also reported [34–40]. Subsequently, in order to counteract the current scientific confusion on genotype nomenclature, the EU consortium on PCVD (http://www.pcvd.net/) proposed a unified nomenclature (ORF2 sequences of PCV2 are assigned into different genotypes when the genetic distance among them is at least 0.035) for the PCV2 genotype. The consortium proposed naming the three PCV2 genotypes as PCV-2a, PCV-2b, and PCV-2c [41]. On the other hand, according to the nomenclature of Wang and co-workers [42], Chinese PCV2 strains based on complete genome were classified into PCV2a, PCV2b, PCV2d, PCV2e (Figure 3), and other unidentified genotypes [42–45]. Nevertheless, Segalés et al. [46] had different comments towards the above nomenclatures and argued that PCV2 should still be divided into only three genotypes. In general, these PCV2 strains had three kinds of genome maps and two kinds of genome sizes (Figure 1) [45]. A few studies reported a number of PCV2 variant strains in China [47–50] but the method proposed by EU consortium on PCVD was not utilized. The PCV2c genotype has not been reported so far. In summary, regardless of the classification methods, the above studies demonstrated that various PCV2 genotypes did exist in swine herds of China.
Co-Infections of Different PCV2 Genotypes in the Field in China
In 2009, Hesse et al. [51] described natural co-infections of two different PCV2 genotypes in pigs in the field for the first time. A similar molecular epidemiological survey using 118 PCV2-positive samples between 2008 and 2009 was also conducted by our group. Our result indicated that the co-infection rates of PCV2 were 32.2% (38/118) in diseased pigs and 0% (0/41) in asymptomatic pigs, respectively. Sequencing results of the 38 co-infected samples showed that the co-existent genotypes were PCV2a-PCV2b (12/38), PCV2a-PCV2d (15/38), and PCV2e-PCV2d (11/38), respectively. This was the first study demonstrating the co-existence of different PCV2 genotypes or strains in China that could help better understand the molecular epidemiology and the new infection patterns of PCV2 [52]. We believe that the presence of co-infections of different PCV2 genotypes is a more accurate description with regard to the molecular epidemiology, supported by common observations in the field.
Co-Infections of PCV2 with Other Swine Pathogens in China
In addition to co-infections of different PCV2 genotypes, a number of studies demonstrated that PCV2 could co-infect along with by other traditional pathogens (such as porcine reproductive and respiratory syndrome virus, classical swine fever virus, porcine parvovirus, porcine pseudorabies virus, bovine viral diarrhea virus) [53–59] and emerging pathogens (porcine bocavirus, Torque teno sus virus, porcine hokovirus) [60–63].
Novel PCV2 strains in China
PCV2 Strains with nucleotide deletion
In general, the full-length genome of PCV2 is 1767 bp or 1768 bp in size. However, PCV2 strains with gene deletion were identified in China. TZ0601 strain (GenBank accession number EU257511) had one nucleotide (G) deletion in the ORF2 region (Position 1733 to 1026) at position 1059 (Figure 4), resulting in C-terminal truncation of ORF2 [64]. Similarly, YJ strain (HM038032) also had one nucleotide (G) deletion in the ORF2 region (Position 1733 to 1026) (Figure 4), but the deletion occurred at position 1039 [43]. Due to one nucleotide (G) deletion in ORF2 encoding region, for the strains of TZ0601 and YJ, the size of ORF2 is lengthened from 702 bp to 708 bp. In addition, the AH strain (HM038030) and MDJ strain (HM038031) had an identical deletion (C) in the non-coding region at position 39 [43], it suggested that position 39 was a frequent deletion site, despite the fact that these deletions did not affect the ORF1 and ORF2 encoding proteins (Figure 4).
Genome maps of four deletion strains and two insertion strains of PCV2. For TZ0601 and YJ, the nucleotide (G) deletion occurred at position 1059 and 1039, respectively. AH and MDJ had an identical nucleotide (G) deletion at position 39. Moreover, for two insertion strains, 10JS-2 had 11-nt (AGCAGCACCTC) insertion at position 42, while JF2 had one nucleotide (T) insertion at position 1043.
PCV2 Strains with nucleotide insertion
In 2004, a novel PCV2 strain, named Fh16 (AY321993), with gene insertion (11 nt, GGCAGCACCTC) at position 42 (non-coding region from position 1 to 50) was identified in non-PMWS herds in France. However, those herds presented with wasting, necrosis, and proliferous ileitis associated with Lawsonia and Pasteurella multocida. Further genetic analysis showed that Fh16 was belonged to the genotype PCV2b [35]. Furthermore, a Danish PCV2 strain DK475case (EF565360) isolated from non-PMWS herds had the identical 11-nt nucleotide insertion to the Fh16 strain [65].
PCV2 strains with nucleotide insertion also exist in China. Among them, the 10JS-2 strain (JQ806749) had 11-nt insertion (AGCAGCACCTC) at position 42 (Figure 4), which was similar to the strains Fh16 and DK475case [66]. The strain JF2 (HQ402903) had one additional nucleotide insertion (T) at position 1043 comparing to the genotype PCV2a, which led to the changes of ORF2 size (711 bp) (Figure 4) comparing to the previous strains [67]. Notably, phylogenetic analysis suggested that the strains 10JS-2 and JF2 were classified into the genotype of PCV2e and PCV2a, respectively (Figure 3).
PCV2 Recombinant strains
Recombination is an important evolution pathway for viruses. PCV2 is a virus with rapid evolution. Recombination events of PCV2 have been reported from several countries including China since 2007 [30, 39, 51, 68–75]. These recombinant strains did not only arise from intra-genotypic recombination but also inter-genotypic recombination (Figure 5). However, some recombinants’ origins might be doubtful due to artificial genome amplification and/or lack of viral plaque purification tests.
Genome maps of four PCV2 recombinant strains. HN0907 was an intra-genotypic recombinant between PCV2b strains, nucleotides (Position 51 to 413) were from ZhuJi2003 (AY579893, PCV2b) and nucleotides (Position 414 to 995) were from 09CQ (HQ395024, PCV2b). 09HaiN-1 was an inter-genotypic recombinant between PCV2a and PCV2b strains, it exhibited greater similarity to 336 (AY256459, PCV2a) before the breakpoint 309 of ORF2, and shared higher sequence similarity with GSLN-PCV2 (FJ948168, PCV2b) after the breakpoint 309 of ORF2. SXXYA-01 was an inter-genotypic recombinant between PCV2a and PCV2b strains, nucleotides (Position 1034 to 1391) were most from ZhuJi2003 (AY579893) and the remaining sequences were most from DTC (DQ104423). SXXYB-01 was an inter-genotypic recombinant between PCV2a and PCV2b strains, its sequence exhibited higher similarity to DTC (DQ104423, PCV2a) before the breakpoint 1034 nt and higher similarity to ZhuJi2003 (AY579893, PCV2b) after the breakpoint 1034 nt. Note: The recombinant regions were presented using red squares and blue squares.
PCV2-Like viral agents
Porcine circovirus-like agents (P1 and P2) were found in China in 2008. Sequence analyses indicated that the two new agents had 648 nucleotides (P1) and 993 nucleotides (P2) in size (Figure 6), which were closely related to the isolates of PCV2 [76–78]. Epidemiology studies and animal experiments also demonstrated that P1, which caused pallor of the skin and diarrhea in pigs, had a high prevalence in Chinese swine herds [77, 79]. Since then, more porcine circovirus-like strains (Figure 6) were reported [80–82]. However, these PCV2-like strains were reported only by one research team. More epidemiological work should thus be done in order to confirm their actual existence and pathogenicity.
Genome maps of eight PCV2-like agents. For P1, twenty-two nucleotides (GGATCCACTAGTAACGGCCGCC) from 5'-terminal region might originate from porcine endogenous retroviruses and the rest of the sequence shared 98.42% identity with ORF2 of PCV2 genome (AF381175). For P2, its ORF3 sequences shared 64.7% ~ 97.4% identity with ORF2 of PCV genome. For JSTZ, ZJQDH1 and ZJQDH2, online Blastn results showed they shared hightest nucleotide identity (98% ~ 100%) with the partial sequences of PCV2 (the strain of GZ-CS1, JQ809462). For JSHM, online Blastn results showed it shared hightest nucleotide identity (98% ~ 100%) with the partial sequences of PCV2 (the strain of CQWZ12, KF742551). For CH-IVT1, online Blastn results showed it shared hightest nucleotide identity (100%) with the partial sequences of PCV2 (the strain of BF, AF381175). For BIV, online Blastn results showed it shared hightest nucleotide identity (99%) with the partial sequences of PCV2 (the strain of 10JS-2, JQ806749). Note: Complete genomes of P1 and P2 were showed in reference [78] and reference [76], respectively.
PCV2 DNA in Chinese commercial swine vaccines
PCV1 is non-pathogenic to pigs, and it is often detected in cell lines, pepsin, and commercial swine vaccines [83–87]. However, some studies reported that PCV2 DNA also existed in human stool, human vaccines, beef, calf bone marrow, rodents, and even commercial swine vaccines in China [88–93]. Infections of pigs and PCV-associated diseases by vaccination (Figure 7) would have occurred if PCV2 retained its infectivity in those commercial swine vaccines. To some extent, it has warned the vaccine manufacturers that good quality control measures should be taken in order to avoid the spread of PCV2 through contaminated vaccines, especially live vaccines.
PCV2 DNA in mosquitoes collected from China
PCV2 has a high prevalence in swine herds in the world, implying that there might be other transmission routes (Figure 7). Recently, extensive PCV2 DNA was detected from water samples in Brazil, farm air in Canada, house flies in UK, and even in Culex (a kind of mosquito) (6.78%, 4/59 sampling) collected in China [94–97]. In general, mosquito is considered as an important vector for many zoonotic infectious diseases, such as Japanese encephalitis B and dengue fever. The control and killing of this vector (such as Culex and house flies) is usually overlooked under the poor swine farm conditions in China, which might exacerbate the spread of PCV2 in pig farms.
Control of PCV2 in China
Vaccine
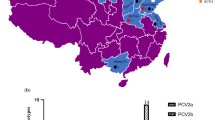
Safe and effective vaccines are considered as the best control measure for viral diseases. Commercial PCV2 vaccines (including the subunit vaccine, inactivated PCV1-2 chimera vaccine and inactivated PCV2 vaccine) have been proven to be effective in the EU, North America, and South Korea [98]. In recent years, PCV2 is given more attention by farming enterprises and the government in China. Since the first imported vaccine (Ingelvac circoflex, Boehringer Ingelheim, Ltd.) being available in China at the end of 2009, there have been at least five commercial vaccines (one subunit vaccine and four inactivated vaccines) from 16 manufacturers (Table 1). Among these vaccines, one vaccine is based on PCV2d genotype, two vaccines are based on PCV2a genotype and three vaccines are based on PCV2b genotype. In general, PCV2 vaccines induced a humoral immune response, characterized by producing neutralizing antibodies (NA) that are cross-protective against both predominant PCV2 genotypes in conventional pigs [26]. Besides, the infection tests based on SPF pigs and colostrum-deprived pigs also demonstrated that cross-protection was present between PCV2a and PCV2b [99, 100]. However, one recent study reported that a PCV2 vaccine based on genotype PCV2b was more effective in protecting pigs against the effects of PCV2b than those based on the genotype PCV2a [101]. The result could act as a further support for the antigenic variability of PCV2 [64, 102]. Moreover, sequence varieties between PCV2 isolates could translate to functional antigenic differences in viral neutralization in vivo [103].
Other measures
In addition to vaccination, other comprehensive measures have been taken in China, which mainly followed the recommendations in the developed countries, such as “Madec’s 20-point plan”, including “all-in”–“all-out” procedures, disinfection, limiting animal contact, preventing mixing of batches and cross-fostering and the isolation or euthanasia of diseased pigs; It also included the environmental factors such as the maintenance of appropriate temperature, air-flow, and space within pens; and furthermore, appropriate use of anti-parasitic treatments and vaccination [104], co-infections control [105–107], breeding and semen quality control [108–110], and herd nutrition improvement [111–113] are included as well. Nevertheless, autogenous vaccines were also considered as an effective method to prevent and control PCV2 on some pig farms from our experience.
Conclusions
In summary, PCV2 infection is very common in Chinese swine herds. There are various PCV2 genotypes, PCV2-like strains, or mutants circulating in China. Together with the potential cross-species transmission of PCV2 (Figure 7), these factors lead to great challenges for the control of PCV2 despite the availability of vaccines. Furthermore, although cross-protection within inter-genotypes of PCV2 is present, NA only neutralizes certain PCV2 genotype but not all PCV2 genotypes [103]. At present, PCV2 infection status can be quite complex clinically, if it is not mastered well by veterinarians or practitioners. It might hinder choosing the right choice of PCV2 vaccines and thus not conducive to the prevention of PCV2. To overcome this problem, it is necessary to perform the detection and sequencing of PCV2 antigen. In addition to effective comprehensive measures, the development of polyvalent vaccines (such as PCV2a + PCV2b + PCV2d) or universal vaccines against all PCV2 genotypes are needed in the future.
References
Tischer I, Rasch R, Tochtermann G: Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralbl Bakteriol Orig A. 1974, 226: 153-167.
Tischer I, Mields W, Wolff D, Vagt M, Griem W: Studies on epidemiology and pathogenicity of porcine circovirus. Arch Virol. 1986, 91: 271-276. 10.1007/BF01314286.
Allan GM, Ellis JA: Porcine circoviruses: a review. J Vet Diagn Invest. 2000, 12: 3-14. 10.1177/104063870001200102.
Mankertz A, Mankertz J, Wolf K, Buhk HJ: Identification of a protein essential for replication of porcine circovirus. J Gen Virol. 1998, 79: 381-384.
Nawagitgul P, Morozov I, Bolin SR, Harms PA, Sorden SD, Paul PS: Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J Gen Virol. 2000, 81: 2281-2287.
Liu J, Chen I, Du Q, Chua H, Kwang J: The ORF3 protein of porcine circovirus type 2 is involved in viral pathogenesis in vivo. J Virol. 2006, 80: 5065-5073. 10.1128/JVI.80.10.5065-5073.2006.
He J, Cao J, Zhou N, Jin Y, Wu J, Zhou J: Identification and functional analysis of the novel ORF4 protein encoded by porcine circovirus type 2. J Virol. 2013, 87: 1420-1429. 10.1128/JVI.01443-12.
Finsterbusch T, Mankertz A: Porcine circoviruses—small but powerful. Virus Res. 2009, 143: 177-183. 10.1016/j.virusres.2009.02.009.
Segalés J: Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology and laboratory diagnosis. Virus Res. 2012, 164: 10-19. 10.1016/j.virusres.2011.10.007.
Opriessnig T, Meng XJ, Halbur PG: Porcine circovirus type 2 associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J Vet Diagn Invest. 2007, 19: 591-615. 10.1177/104063870701900601.
West KH, Bystrom JM, Wojnarowicz C, Shantz N, Jacobson M, Allan GM, Haines DM, Clark EG, Krakowka S, McNeilly F, Konoby C, Martin K, Ellis JA: Myocarditis and abortion associated with intrauterine infection of sows with porcine circovirus 2. J Vet Diagn Invest. 1999, 11: 530-532. 10.1177/104063879901100608.
Rosell C, Segalés J, Ramos-Vara JA, Folch JM, Rodríguez-Arrioja GM, Duran CO, Balasch M, Plana-Durán J, Domingo M: Identification of porcine circovirus in tissues of pigs with porcine dermatitis and nephropathy syndrome. Vet Rec. 2000, 146: 40-43. 10.1136/vr.146.2.40.
Stevenson GW, Kiupel M, Mittal SK, Choi J, Latimer KS, Kanitz CL: Tissue distribution and genetic typing of porcine circoviruses in pigs with naturally occurring congenital tremors. J Vet Diagn Invest. 2001, 13: 57-62. 10.1177/104063870101300111.
Kim J, Chung HK, Chae C: Association of porcine circovirus 2 with porcine respiratory disease complex. Vet J. 2003, 166: 251-256. 10.1016/S1090-0233(02)00257-5.
Kim J, Ha Y, Jung K, Choi C, Chae C: Enteritis associated with porcine circovirus 2 in pigs. Can J Vet Res. 2004, 68: 218-221.
Jensen TK, Vigre H, Svensmark B, Bille-Hansen V: Distinction between porcine circovirus type 2 enteritis and porcine proliferative enteropathy caused by Lawsonia intracellularis. J Comp Pathol. 2006, 135: 176-182. 10.1016/j.jcpa.2006.08.006.
Drolet R, Larochelle R, Morin M, Delisle B, Magar R: Detection rates of porcine reproductive and respiratory syndrome virus, porcine circovirus type 2, and swine influenza virus in porcine proliferative and necrotizing pneumonia. Vet Pathol. 2003, 40: 143-148. 10.1354/vp.40-2-143.
Grau-Roma L, Segalés J: Detection of porcine reproductive and respiratory syndrome virus, porcine circovirus type 2, swine influenza virus and Aujeszky’ s disease virus in cases of porcine proliferative and necrotizing pneumonia (PNP) in Spain. Vet Microbiol. 2007, 119: 144-151. 10.1016/j.vetmic.2006.09.009.
Cino-Ozuna AG, Henry S, Hesse R, Nietfeld JC, Bai J, Scott HM, Rowland RR: Characterization of a new disease syndrome associated with porcine circovirus type 2 in previously vaccinated herds. J Clin Microbiol. 2011, 49: 2012-2016. 10.1128/JCM.02543-10.
Fachinger V, Bischoff R, Jedidia SB, Saalmuller A, Elbers K: The effect of vaccination against porcine circovirus type 2 in pigs suffering from porcine respiratory disease complex. Vaccine. 2008, 26: 1488-1499. 10.1016/j.vaccine.2007.11.053.
Horlen KP, Dritz SS, Nietfeld JC, Henry SC, Hesse RA, Oberst R, Hays M, Anderson J, Rowland RR: A field evaluation of mortality rate and growth performance in pigs vaccinated against porcine circovirus type 2. J Am Vet Med Assoc. 2008, 232: 906-912. 10.2460/javma.232.6.906.
Kixmöller M, Ritzmann M, Eddicks M, Saalmüller A, Elbers K, Fachinger V: Reduction of PMWS-associated clinical signs and co-infections by vaccination against PCV2. Vaccine. 2008, 26: 3443-3451. 10.1016/j.vaccine.2008.04.032.
Pejsak Z, Podgórska K, Truszczyński M, Karbowiak P, Stadejek T: Efficacy of different protocols of vaccination against porcine circovirus type 2 (PCV2) in a farm affected by postweaning multisystemic wasting syndrome (PMWS). Comp Immunol Microbiol Infect Dis. 2010, 33: e1-e5. 10.1016/j.cimid.2009.09.006.
Segalés J, Urniza A, Alegre A, Bru T, Crisci E, Nofrarías M, López-Soria S, Balasch M, Sibila M, Xu Z, Chu HJ, Fraile L, Plana-Duran J: A genetically engineered chimeric vaccine against porcine circovirus type 2 (PCV2) improves clinical, pathological and virological outcomes in postweaning multisystemic wasting syndrome affected farms. Vaccine. 2009, 27: 7313-7321. 10.1016/j.vaccine.2009.09.084.
Lyoo K, Joo H, Caldwell B, Kim H, Davies PR, Torrison J: Comparative efficacy of three commercial PCV2 vaccines in conventionally reared pigs. Vet J. 2011, 189: 58-62. 10.1016/j.tvjl.2010.06.015.
Fort M, Sibila M, Allepuz A, Mateu E, Roerink F, Segalés J: Porcine circovirus type 2 (PCV2) vaccination of conventional pigs prevents viremia against PCV2 isolates of different genotypes and geographic origins. Vaccine. 2008, 26: 1063-1071. 10.1016/j.vaccine.2007.12.019.
Fort M, Sibila M, Pérez-Martín E, Nofrarías M, Mateu E, Segalés J: One dose of a porcine circovirus 2 (PCV2) sub-unit vaccine administered to 3-week-old conventional piglets elicits cell-mediated immunity and significantly reduces PCV2 viremia in an experimental model. Vaccine. 2009, 27: 4031-4037. 10.1016/j.vaccine.2009.04.028.
Opriessnig T, Patterson AR, Elsener J, Meng XJ, Halbur PG: Influence of maternal antibodies on efficacy of porcine circovirus type 2 (PCV2) vaccination to protect pigs from experimental infection with PCV2. Clin Vaccine Immunol. 2008, 15: 397-401. 10.1128/CVI.00416-07.
Opriessnig T, Patterson AR, Madson DM, Pal N, Halbur PG: Comparison of efficacy of commercial one dose and two dose PCV2 vaccines using a mixed PRRSV–PCV2–SIV clinical infection model 2–3-months post vaccination. Vaccine. 2009, 27: 1002-1007. 10.1016/j.vaccine.2008.11.105.
Gagnon CA, Music N, Fontaine G, Tremblay D, Harel J: Emergence of a new type of porcine circovirus in swine (PCV): a type 1 and type 2 PCV recombinant. Vet Microbiol. 2010, 144: 18-23. 10.1016/j.vetmic.2009.09.072.
Opriessnig T, Xiao CT, Gerber PF, Halbur PG: Emergence of a novel mutant PCV2b variant associated with clinical PCVAD in two vaccinated pig farms in the U.S. concurrently infected with PPV2. Vet Microbiol. 2013, 163: 177-183. 10.1016/j.vetmic.2012.12.019.
Xiao CT, Halbur PG, Opriessnig T: Complete genome sequence of a novel porcine circovirus type 2b variant present in cases of vaccine failures in the United States. J Virol. 2012, 86: 12469-10.1128/JVI.02345-12.
Wen LB, Guo X, Yang HC: Genotyping of porcine circovirus type 2 from a variety of clinical conditions in China. Vet Microbiol. 2005, 110: 141-146. 10.1016/j.vetmic.2005.07.003.
Carman S, Cai HY, DeLay J, Youssef SA, McEwen BJ, Gagnon CA, Tremblay D, Hazlett M, Lusis P, Fairles J, Alexander HS, van Dreumel T: The emergence of a new strain of porcine circovirus-2 in Ontario and Quebec swine and its association with severe porcine circovirus associated disease–2004–2006. Can J Vet Res. 2008, 72: 259-268.
de Boisséson C, Béven V, Bigarré L, Thiéry R, Rose N, Eveno E, Madec F, Jestin A: Molecular characterization of Porcine circovirus type 2 isolates from post-weaning multisystemic wasting syndrome-affected and non-affected pigs. J Gen Virol. 2004, 85: 293-304. 10.1099/vir.0.19536-0.
Grau-Roma L, Crisci E, Sibila M, López-Soria S, Nofrarias M, Cortey M, Fraile L, Olvera A, Segalés J: A proposal on porcine circovirus type 2 (PCV2) genotype definition and their relation with postweaning multisystemic wasting syndrome (PMWS) occurrence. Vet Microbiol. 2008, 128: 23-35. 10.1016/j.vetmic.2007.09.007.
Larochelle R, Magar R, D’Allaire S: Genetic characterization and phylogenetic analysis of porcine circovirus type 2 (PCV2) strains from cases presenting various clinical conditions. Virus Res. 2002, 90: 101-112. 10.1016/S0168-1702(02)00141-7.
de Castro AM MG, Cortez A, Heinemann MB, Brandão PE, Richtzenhain LJ: Genetic diversity of Brazilian strains of porcine circovirus type 2 (PCV-2) revealed by analysis of the cap gene (ORF-2). Arch Virol. 2007, 52: 1435-1445.
Olvera A, Cortey M, Segalés J: Molecular evolution of porcine circovirus type 2 genomes: phylogeny and clonality. Virology. 2007, 357: 175-185. 10.1016/j.virol.2006.07.047.
Timmusk S, Wallgren P, Brunborg IM, Wikström FH, Allan G, Meehan B, McMenamy M, McNeilly F, Fuxler L, Belák K, Põdersoo D, Saar T, Berg M, Fossum C: Phylogenetic analysis of porcine circovirus type 2 (PCV2) pre- and post-epizootic postweaning multisystemic wasting syndrome (PMWS). Virus Genes. 2008, 36: 509-520. 10.1007/s11262-008-0217-1.
Segalés J, Olvera A, Grau-Roma L, Charreyre C, Nauwynck H, Larsen L, Dupont K, McCullough K, Ellis J, Krakowka S, Mankertz A, Fredholm M, Fossum C, Timmusk S, Stockhofe-Zurwieden N, Beattie V, Armstrong D, Grassland B, Baekbo P, Allan G: PCV-2 genotype definition and nomenclature. Vet Rec. 2008, 162: 867-868. 10.1136/vr.162.26.867.
Wang F, Guo X, Ge X, Wang Z, Chen Y, Cha Z, Yang H: Genetic variation analysis of Chinese strains of porcine circovirus type 2. Virus Res. 2009, 145: 151-156. 10.1016/j.virusres.2009.05.015.
Guo LJ, Lu YH, Wei YW, Huang LP, Liu CM: Porcine circovirus type 2 (PCV2): genetic variation and newly emerging genotypes in China. Virol J. 2010, 7: 273-10.1186/1743-422X-7-273.
Zhao GH, Cheng W, Zhang PJ, Han YS, Chen DK: Novel genotypes of type 2 porcine circovirus (PCV2) in PMWS pigs in China between 2008 and 2009. J Anim Vet Adv. 2010, 9: 3083-3091.
Zhai SL, Chen SN, Zhang JW, Wei ZZ, Long JX, Yuan SS, Wei WK, Chen QL, Xuan H, Wu DC: Dissection of the possible routes on porcine circoviruses infecting human. J Anim Vet Adv. 2012, 11: 1281-1286. 10.3923/javaa.2012.1281.1286.
Cortey M, Olvera A, Grau-Roma L, Segalés J: Further comments on porcine circovirus type 2 (PCV2) genotype definition and nomenclature. Vet Microbiol. 2011, 149: 522-523. 10.1016/j.vetmic.2010.11.009.
Li W, Wang X, Ma T, Feng Z, Li Y, Jiang P: Genetic analysis of porcine circovirus type 2 (PCV2) strains isolated between 2001 and 2009: genotype PCV2b predominate in postweaning multisystemic wasting syndrome occurrences in eastern China. Virus Genes. 2010, 40: 244-251. 10.1007/s11262-009-0438-y.
Mu C, Yang Q, Zhang Y, Zhou Y, Zhang J, Martin DP, Xia P, Cui B: Genetic variation and phylogenetic analysis of porcine circovirus type 2 infections in central China. Virus Genes. 2012, 45: 463-473. 10.1007/s11262-012-0789-7.
Wang C, Pang VF, Lee F, Huang TS, Lee SH, Lin YJ, Lin YL, Lai SS, Jeng CR: Prevalence and genetic variation of porcine circovirus type 2 in Taiwan from 2001 to 2011. Res Vet Sci. 2013, 94: 789-795. 10.1016/j.rvsc.2012.11.019.
Wei C, Zhang M, Chen Y, Xie J, Huang Z, Zhu W, Xu T, Cao Z, Zhou P, Su S, Zhang G: Genetic evolution and phylogenetic analysis of porcine circovirus type 2 infections in southern China from 2011 to 2012. Infect Genet Evol. 2013, 17: 87-92.
Hesse R, Kerrigan M, Rowland RR: Evidence for recombination between PCV2a and PCV2b in the field. Virus Res. 2008, 132: 201-207. 10.1016/j.virusres.2007.10.013.
Zhai SL, Chen SN, Wei ZZ, Zhang JW, Huang L, Lin T, Yue C, Ran DL, Yuan SS, Wei WK, Long JX: Co-existence of multiple strains of porcine circovirus type 2 in the same pig from China. Virol J. 2011, 8: 517-10.1186/1743-422X-8-517.
Yue F, Cui S, Zhang C, Yoon KJ: A multiplex PCR for rapid and simultaneous detection of porcine circovirus type 2, porcine parvovirus, porcine pseudorabies virus, and porcine reproductive and respiratory syndrome virus in clinical specimens. Virus Genes. 2009, 38: 392-397. 10.1007/s11262-009-0333-6.
Cao S, Chen H, Zhao J, Lü J, Xiao S, Jin M, Guo A, Wu B, He Q: Detection of porcine circovirus type 2, porcine parvovirus and porcine pseudorabies virus from pigs with postweaning multisystemic wasting syndrome by multiplex PCR. Vet Res Commun. 2005, 29: 263-269.
Jiang Y, Shang H, Xu H, Zhu L, Chen W, Zhao L, Fang L: Simultaneous detection of porcine circovirus type 2, classical swine fever virus, porcine parvovirus and porcine reproductive and respiratory syndrome virus in pigs by multiplex polymerase chain reaction. Vet J. 2010, 183: 172-175. 10.1016/j.tvjl.2008.11.016.
Xu XG, Chen GD, Huang Y, Ding L, Li ZC, Chang CD, Wang CY, Tong DW, Liu HJ: Development of multiplex PCR for simultaneous detection of six swine DNA and RNA viruses. J Virol Methods. 2012, 183: 69-74. 10.1016/j.jviromet.2012.03.034.
Liu JK, Wei CH, Yang XY, Dai AL, Li XH: Multiplex PCR for the simultaneous detection of porcine reproductive and respiratory syndrome virus, classical swine fever virus, and porcine circovirus in pigs. Mol Cell Probes. 2013, 27: 149-152. 10.1016/j.mcp.2013.03.001.
Zheng LL, Wang YB, Li MF, Chen HY, Guo XP, Geng JW, Wang ZY, Wei ZY, Cui BA: Simultaneous detection of porcine parvovirus and porcine circovirus type 2 by duplex real-time PCR and amplicon melting curve analysis using SYBR Green. J Virol Methods. 2013, 187: 15-19. 10.1016/j.jviromet.2012.06.024.
Deng Y, Sun CQ, Cao SJ, Lin T, Yuan SS, Zhang HB, Zhai SL, Huang L, Shan TL, Zheng H, Wen XT, Tong GZ: High prevalence of bovine viral diarrhea virus 1 in Chinese swine herds. Vet Microbiol. 2012, 159: 490-493. 10.1016/j.vetmic.2012.04.023.
Zhai S, Yue C, Wei Z, Long J, Ran D, Lin T, Deng Y, Huang L, Sun L, Zheng H, Gao F, Zheng H, Chen S, Yuan S: High prevalence of a novel porcine bocavirus in weanling piglets with respiratory tract symptoms in China. Arch Virol. 2010, 155: 1313-1317. 10.1007/s00705-010-0698-9.
Zhang HB, Huang L, Liu YJ, Lin T, Sun CQ, Deng Y, Wei ZZ, Cheung AK, Long JX, Yuan SS: Porcine bocaviruses: genetic analysis and prevalence in Chinese swine population. Epidemiol Infect. 2011, 139: 1581-1586. 10.1017/S0950268811000847.
Zhai SL, Long JX, Wei WK, Chen QL, Luo ML, Lv DH, Wu DC, Gao F, Yuan SS, Tong GZ, Wei ZZ: High prevalence of torque teno sus virus in China and genetic diversity of the 5' non-coding region. Arch Virol. 2013, 158: 1567-1573. 10.1007/s00705-013-1644-4.
Li S, Wei Y, Liu J, Tang Q, Liu C: Prevalence of porcine hokovirus and its co-infection with porcine circovirus 2 in China. Arch Virol. 2013, 158: 1987-1991. 10.1007/s00705-013-1690-y.
Shang SB, Jin YL, Jiang XT, Zhou JY, Zhang X, Xing G, He JL, Yan Y: Fine mapping of antigenic epitopes on capsid proteins of porcine circovirus, and antigenic phenotype of porcine circovirus type 2. Mol Immunol. 2009, 46: 327-334. 10.1016/j.molimm.2008.10.028.
Dupont K, Nielsen EO, Baekbo P, Larsen LE: Genomic analysis of PCV2 isolates from Danish archives and a current PMWS case–control study supports a shift in genotypes with time. Vet Microbiol. 2008, 128: 56-64. 10.1016/j.vetmic.2007.09.016.
Cai L, Han X, Hu D, Li X, Wang B, Ni J, Zhou Z, Yu X, Zhai X, Tian K: A novel porcine circovirus type 2a strain, 10JS-2, with eleven-nucleotide insertions in the origin of genome replication. J Virol. 2012, 86: 7017-10.1128/JVI.00760-12.
Huang LP, Lu YH, Wei YW, Guo LJ, Liu CM: Identification of one critical amino acid that determines a conformational neutralizing epitope in the capsid protein of porcine circovirus type 2. BMC Microbiol. 2011, 11: 188-10.1186/1471-2180-11-188.
Cheung AK: Homologous recombination within the capsid gene of porcine circovirus type 2 subgroup viruses via natural co-infection. Arch Virol. 2009, 154: 531-534. 10.1007/s00705-009-0329-5.
Lefebvre DJ, Van Doorsselaere J, Delputte PL, Nauwynck HJ: Recombination of two porcine circovirus type 2 strains. Arch Virol. 2009, 154: 875-879. 10.1007/s00705-009-0379-8.
Kim HK, Luo Y, Moon HJ, Park SJ, Keum HO, Rho S, Park BK: Phylogenetic and recombination analysis of genomic sequences of PCV2 isolated in Korea. Virus Genes. 2009, 39: 352-358. 10.1007/s11262-009-0395-5.
Cadar D, Cságola A, Lorincz M, Tombácz K, Spînu M, Tuboly T: Detection of natural inter- and intra-genotype recombination events revealed by cap gene analysis and decreasing prevalence of PCV2 in wild boars. Infect Genet Evol. 2012, 12: 420-427. 10.1016/j.meegid.2012.01.014.
Ma CM, Hon CC, Lam TY, Li VY, Wong CK, de Oliveira T, Leung FC: Evidence for recombination in natural populations of porcine circovirus type 2 in Hong Kong and mainland China. J Gen Virol. 2007, 88: 1733-1737. 10.1099/vir.0.82629-0.
Cai L, Han X, Ni J, Yu X, Zhou Z, Zhai X, Chen X, Tian K: Natural recombinants derived from different patterns of recombination between two PCV2b parental strains. Virus Res. 2011, 158: 281-288. 10.1016/j.virusres.2011.03.021.
Cai L, Ni J, Xia Y, Zi Z, Ning K, Qiu P, Li X, Wang B, Liu Q, Hu D, Yu X, Zhou Z, Zhai X, Han X, Tian K: Identification of an emerging recombinant cluster in porcine circovirus type 2. Virus Res. 2012, 165: 95-102. 10.1016/j.virusres.2012.01.008.
Huang Y, Shao M, Xu X, Zhang X, Du Q, Zhao X, Zhang W, Lyu Y, Tong D: Evidence for different patterns of natural inter-genotype recombination between two PCV2 parental strains in the field. Virus Res. 2013, 175: 78-86. 10.1016/j.virusres.2013.03.014.
Wen L, He K, Yang H, Ni Y, Zhang X, Guo R, Pan Q: Complete nucleotide sequence of a novel porcine circovirus-like agent and its infectivity in vitro. Sci China C Life Sci. 2008, 51: 453-458. 10.1007/s11427-008-0053-5.
Wen L, He K, Yang H, Yu Z, Mao A, Zhong S, Ni Y, Zhang X, Li B, Wang X, Zhou J, Guo R, Lv L, Jiang J: Prevalence of porcine circovirus-like agent P1 in Jiangsu China. Virol J. 2011, 8: 543-10.1186/1743-422X-8-543.
Wen L, He K, Yu Z, Mao A, Ni Y, Zhang X, Guo R, Li B, Wang X, Zhou J, Lv L: Complete genome sequence of a novel porcine circovirus-like agent. J Virol. 2012, 86: 639-10.1128/JVI.06469-11.
Wen L, He K, Xiao Q, Yu Z, Mao A, Ni Y, Zhang X, Li B, Wang X, Guo R, Zhou J, Lv L, Jiang J: A novel porcine circovirus-like agent P1 is associated with wasting syndromes in pigs. PLoS One. 2012, 7: e41565-10.1371/journal.pone.0041565.
Wen L, He K, Ni Y, Zhang X, Li B, Wang X, Guo RL, Yu Z, Mao A, Zhou J, Lv L, Jiang J: Complete genome sequence of the rearranged porcine circovirus type 2. J Virol. 2012, 86: 5963-10.1128/JVI.00494-12.
Wen L, He K, Ni Y, Zhang X, Li B, Wang X, Guo RL, Yu Z, Mao A, Zhou J, Lv L, Jiang J: In vitro intragenomic rearrangement of porcine circovirus type 2. J Virol. 2012, 86: 8911-10.1128/JVI.01377-12.
Wen L, He K, Li B, Wang X, Guo RL, Yu Z, Mao A, Ni Y, Zhang X, Zhou J, Lv L, Jiang J: In vitro and in vivo isolation of a novel rearranged porcine circovirus type 2. J Virol. 2012, 86: 13120-10.1128/JVI.02392-12.
Ma H, Shaheduzzaman S, Willliams DK, Gao Y, Khan AS: Investigations of porcine circovirus type 1 (PCV1) in vaccine-related and other cell lines. Vaccine. 2011, 29: 8429-8437. 10.1016/j.vaccine.2011.07.123.
Wang C, Pang VF, Jeng CR, Lee F, Huang YW, Lin YL, Hsiao SH, Lai SS: Detection of porcine circovirus type 1 in commercial porcine vaccines by loop-mediated isothermal amplification. Folia Microbiol (Praha). 2011, 56: 483-489. 10.1007/s12223-011-0072-7.
Fenaux M, Opriessnig T, Halbur PG, Xu Y, Potts B, Meng XJ: Detection and in vitro and in vivo characterization of porcine circovirus DNA from a porcine-derived commercial pepsin product. J Gen Virol. 2004, 85: 3377-3382. 10.1099/vir.0.80429-0.
Baylis SA, Finsterbusch T, Bannert N, Blümel J, Mankertz A: Analysis of porcine circovirus type 1 detected in Rotarix vaccine. Vaccine. 2011, 29: 690-697. 10.1016/j.vaccine.2010.11.028.
Victoria JG, Wang C, Jones MS, Jaing C, McLoughlin K, Gardner S, Delwart EL: Viral nucleic acids in live-attenuated vaccines: detection of minority variants and an adventitious virus. J Virol. 2010, 84: 6033-6040. 10.1128/JVI.02690-09.
Li L, Kapoor A, Slikas B, Bamidele O, Wang C, Shaukat S, Masroor M, Wilson M, Ndjango J, Peeters M, Gross-Camp N, Muller M, Hahn B, Wolfe N, Triki H, Bartkus J, Zaidi S, Delwart E: Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol. 2010, 84: 1674-1682. 10.1128/JVI.02109-09.
Li L, Shan T, Soji OB, Alam MM, Kunz TH, Zaidi SZ, Delwart E: Possible cross-species transmission of circoviruses and cycloviruses among farm animals. J Gen Virol. 2011, 92: 768-772. 10.1099/vir.0.028704-0.
Nayar GP, Hamel AL, Lin L, Sachvie C, Grudeski E, Spearman G: Evidence for circovirus in cattle with respiratory disease and from aborted bovine fetuses. Can Vet J. 1999, 40: 277-278.
Kappe EC, Halami MY, Schade B, Alex M, Hoffmann D, Gangl A, Meyer K, Dekant W, Schwarz BA, Johne R, Buitkamp J, Böttcher J, Müller H: Bone marrow depletion with haemorrhagic diathesis in calves in Germany: characterization of the disease and preliminary investigations on its aetiology. Berl Munch Tierarztl Wochenschr. 2010, 123: 31-41.
Lrincz M, Csagola A, Biksi I, Szeredi L, Dan A, Tuboly T: Detection of porcine circovirus in rodents-short communication. Acta Vet Hung. 2010, 58: 265-268. 10.1556/AVet.58.2010.2.12.
Ye ZM: Contaminated investigation of bovine viral diarrhea virus and porcine circoviruses in commercial swine vaccines. Swine Prod. 2011, 6: 101-102. (In Chinese)
Garcia LA, Viancelli A, Rigotto C, Pilotto MR, Esteves PA, Kunz A, Barardi CR: Surveillance of human and swine adenovirus, human norovirus and swine circovirus in water samples in Santa Catarina, Brazil. J Water Health. 2012, 10: 445-452. 10.2166/wh.2012.190.
Verreault D, Létourneau V, Gendron L, Massé D, Gagnon CA, Duchaine C: Airborne porcine circovirus in Canadian swine confinement buildings. Vet Microbiol. 2010, 141: 224-230. 10.1016/j.vetmic.2009.09.013.
Blunt R, McOrist S, McKillen J, McNair I, Jiang T, Mellits K: House fly vector for porcine circovirus 2b on commercial pig farms. Vet Microbiol. 2011, 149: 452-455. 10.1016/j.vetmic.2010.11.019.
Yang X, Hou L, Ye J, He Q, Cao S: Detection of porcine circovirus type 2 (PCV2) in mosquitoes from pig farms by PCR. Pak Vet J. 2012, 32: 134-135.
Grau-Roma L, Fraile L, Segalés J: Recent advances in the epidemiology, diagnosis and control of diseases caused by porcine circovirus type 2. Vet J. 2011, 187: 23-32. 10.1016/j.tvjl.2010.01.018.
Opriessnig T, Ramamoorthy S, Madson DM, Patterson AR, Pal N, Carman S, Meng XJ, Halbur PG: Differences in virulence among porcine circovirus type 2 isolates are unrelated to cluster type 2a or 2b and prior infection provides heterologous protection. J Gen Virol. 2008, 89: 2482-2491. 10.1099/vir.0.2008/001081-0.
Opriessnig T, Gerber PF, Xiao CT, Mogler M, Halbur P: GA commercial vaccine based on PCV2a and an experimental vaccine based on a variant mPCV2b are both effective in protecting pigs against challenge with a 2013 U.S. variant mPCV2b strain. Vaccine. 2014, 32: 230-237. 10.1016/j.vaccine.2013.11.010.
Opriessnig T, O’Neill K, Gerber PF, de Castro AM, Gimenéz-Lirola LG, Beach NM, Zhou L, Meng XJ, Wang C, Halbur PG: A PCV2 vaccine based on genotype 2b is more effective than a 2a-based vaccine to protect against PCV2b or combined PCV2a/2b viremia in pigs with concurrent PCV2, PRRSV and PPV infection. Vaccine. 2013, 31: 487-494. 10.1016/j.vaccine.2012.11.030.
Lefebvre DJ, Costers S, Van Doorsselaere J, Misinzo G, Delputte PL, Nauwynck HJ: Antigenic differences among porcine circovirus type 2 strains, as demonstrated by the use of monoclonal antibodies. J Gen Virol. 2008, 89: 177-187. 10.1099/vir.0.83280-0.
Kurtz S, Grau-Roma L, Cortey M, Fort M, Rodríguez F, Sibila M, Segalés J: Pigs naturally exposed to porcine circovirus type 2 (PCV2) generate antibody responses capable to neutralise PCV2 isolates of different genotypes and geographic origins. Vet Res. 2014, 45: 29-10.1186/1297-9716-45-29.
Madec F, Eveno E, Morvan P, Hamon L, Blanchard P, Cariolet R, Amenna N, Truong C, Mahé D, Albina E, Jestin A: Post-weaning multisystemic wasting syndrome (PMWS) in pigs in France: clinical observations from followup studies on affected farms. Livest Prod Sci. 2000, 63: 223-233. 10.1016/S0301-6226(99)00138-4.
Opriessnig T, Yu S, Gallup JM, Evans RB, Fenaux M, Pallares F, Thacker EL, Brockus CW, Ackermann MR, Thomas P, Meng XJ, Halbur PG: Effect of vaccination with selective bacterins on conventional pigs infected with type 2 porcine circovirus. Vet Pathol. 2003, 40: 521-529. 10.1354/vp.40-5-521.
Opriessnig T, Fenaux M, Yu S, Evans RB, Cavanaugh D, Gallup JM, Pallares FJ, Thacker EL, Lager KM, Meng XJ, Halbur PG: Effect of porcine parvovirus vaccination on the development of PMWS in segregated early weaned pigs coinfected with type 2 porcine circovirus and porcine parvovirus. Vet Microbiol. 2004, 98: 209-220. 10.1016/j.vetmic.2003.11.006.
Opriessnig T, Thacker EL, Yu S, Fenaux M, Meng XJ, Halbur PG: Experimental reproduction of postweaning multisystemic wasting syndrome in pigs by dual infection with Mycoplasma hyopneumoniae and porcine circovirus type 2. Vet Pathol. 2004, 41: 624-640. 10.1354/vp.41-6-624.
Opriessnig T, Fenaux M, Thomas P, Hoogland MJ, Rothschild MF, Meng XJ, Halbur PG: Evidence of breed-dependent differences in susceptibility to porcine circovirus type-2-associated disease and lesions. Vet Pathol. 2006, 43: 281-293. 10.1354/vp.43-3-281.
Madson D, Patterson A, Ramamoorthy S, Pal N, Meng XJ, Opriessnig T: Reproductive failure experimentally induced in sows via artificial insemination with semen spiked with porcine circovirus type 2 (PCV2). Vet Pathol. 2009, 46: 707-716. 10.1354/vp.08-VP-0234-O-FL.
Madson DM, Ramamoorthy S, Kuster C, Pal N, Meng XJ, Halbur PG, Opriessnig T: Infectivity of porcine circovirus type 2 DNA in semen from experimentally-infected boars. Vet Res. 2009, 40: 10-10.1051/vetres:2008048.
Bassaganya-Riera J, Pogranichniy RM, Jobgen SC, Halbur PG, Yoon KJ, O’Shea M, Mohede I, Hontecillas R: Conjugated linoleic acid ameliorates viral infectivity in a pig model of virally induced immunosuppression. J Nutr. 2003, 133: 3204-3214.
Fraile L, Crisci E, Weenberg J, Armadans M, Mendoza L, Ruiz L, Bernaus S, Montoya M: Effect of treatment with phytosterols in three herds with porcine respiratory disease complex. J Swine Health Prod. 2009, 17: 32-41.
Jung BG, Toan NT, Cho SJ, Ko JH, Jung YK, Lee BJ: Dietary aluminosilicate supplement enhances immune activity in mice and reinforces clearance of porcine circovirus type 2 in experimentally infected pigs. Vet Microbiol. 2010, 143: 117-125. 10.1016/j.vetmic.2009.11.009.
Acknowledgments
This review is part of the work supported by grants (No. 2011A090200117, No. 2012A020602075 and No. 2012224–26) funded by Guangdong Provincial Department of Science and Technology and Guangzhou Science and Technology and Information Bureau, respectively. We also thank Dr. Susanna KP. Lau (Department of Microbiology, The University of Hong Kong), Prof. Yao-Wei Huang (College of Animal Sciences, Zhejiang University & College of Veterinary Medicine, Virginia Polytechnic Institute and State University), Dr. Shanzhong Gong (The University of Texas at Austin) for improving the language quality of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that there is no competing interests regarding the publication of this article.
Authors’ contribution
SLZ structured the review and prepared the draft of the manuscript. SNC collected the literatures and prepared the manuscript. BBS, SFD and JH prepared the tables and figures. ZHX, MHT, FGW, XJL, DHL, XHW, JY, WKW and MLL revised the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Zhai, SL., Chen, SN., Xu, ZH. et al. Porcine circovirus type 2 in China: an update on and insights to its prevalence and control. Virol J 11, 88 (2014). https://doi.org/10.1186/1743-422X-11-88
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1743-422X-11-88