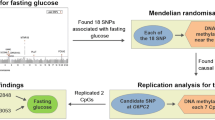
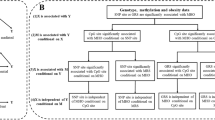
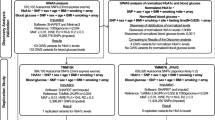
Abstract
Background
Type 2 diabetes mellitus (T2DM) is caused by a combination of environmental, genetic, and epigenetic factors including, fasting blood glucose (FBG), genetic variant rs841853, and cg19693031 methylation. We evaluated the interaction between rs841853 and cg19693031 on the FBG levels of non-diabetic Taiwanese adults.
Methods
We used Taiwan Biobank (TWB) data collected between 2008 and 2016. The TWB data source contains information on basic demographics, personal lifestyles, medical history, methylation, and genotype. The study participants included 1300 people with DNA methylation data. The association of cg19693031 methylation (stratified into quartiles) with rs841853 and FBG was determined using multiple linear regression analysis. The beta-coefficients (β) and p-values were estimated.
Results
The mean ± standard deviation (SD) of FBG in rs841853-CC individuals (92.07 ± 7.78) did not differ significantly from that in the CA + AA individuals (91.62 ± 7.14). However, the cg19693031 methylation levels were significantly different in the two groups (0.7716 ± 0.05 in CC individuals and 0.7631 ± 0.05 in CA + AA individuals (p = 0.002). The cg19693031 methylation levels according to quartiles were β < 0.738592 (< Q1), 0.738592 ≤ 0.769992 (Q1–Q2), 0.769992 ≤ 0.800918 (Q2–Q3), and β ≥ 0.800918 (≥ Q3). FBG increased with decreasing cg19693031 methylation levels in a dose–response manner (ptrend = 0.005). The β-coefficient was − 0.0236 (p = 0.965) for Q2–Q3, 1.0317 (p = 0.058) for Q1–Q2, and 1.3336 (p = 0.019 for < Q1 compared to the reference quartile (≥ Q3). The genetic variant rs841853 was not significantly associated with FBG. However, its interaction with cg19693031 methylation was significant (p-value = 0.036). Based on stratification by rs841853 genotypes, only the CC group retained the inverse and dose–response association between FBG and cg19693031 methylation. The β (p-value) was 0.8082 (0.255) for Q2–Q3, 1.6930 (0.022) for Q1–Q2, and 2.2190 (0.004) for < Q1 compared to the reference quartile (≥ Q3). The ptrend was 0.002.
Conclusion
Summarily, methylation at cg19693031 was inversely associated with fasting blood glucose in a dose-dependent manner. The inverse association was more prominent in rs841853-CC individuals, suggesting that rs841853 could modulate the association between cg19693031 methylation and FBG. Our results suggest that genetic variants may be involved in epigenetic mechanisms associated with FBG, a hallmark of diabetes. Therefore, integrating genetic and epigenetic data may provide more insight into the early-onset of diabetes.
Similar content being viewed by others
Background
Type 2 diabetes mellitus is a serious global public concern with a huge, yet constantly increasing burden. It negatively affects quality of life and greatly increases health care expenditure [1]. Type 2 diabetes affected approximately 462 million people (6.28% of the world’s population) in 2017 [2]. The disease is projected to affect 552 million people by 2030 [3]. The global prevalence rate was approximately 6059 patients per 100,000 in 2017 and is projected to be 7079 and 7862 cases per 100,000 by 2030 and 2040, respectively [2]. It accounted for more than 1 million deaths in 2017, making it the 9th cause of death in the world [2]. The continuous increase in the incidence, prevalence, and mortality of diabetes warrants urgent measures to prevent its occurrence in non-diabetics.
Type 2 diabetes has a multifaceted onset, encompassing a complex interplay of environmental, genetic, and epigenetic processes with poorly explained mechanisms [4,5,6,7,8]. The risk of diabetes is increased by hyperglycemia, increased age, high body mass index (BMI), dyslipidemia, unhealthy diet, and other lifestyle factors such as alcohol consumption and smoking [8,9,10,11,12]. Fasting blood glucose is an important indicator of diabetes, as well as a hallmark of diabetes management [6, 9, 12,13,14]. It is also a predictor of cardiovascular disease risk among diabetics and non-diabetics [15].
Glucose transporter 1 (or GLUT1), also called the solute carrier family 2, facilitated glucose transporter member 1 (SLC2A1) and thioredoxin-interacting protein (TXNIP) located on chromosome 1 are notable candidate genes for T2D diabetes [16,17,18,19,20,21,22,23,24]. GLUT1 is a well-characterized solute transporter that mainly regulates the cellular uptake of glucose in humans [13, 16]. TXNIP is an important modulator of glucose metabolism and mitochondrial activities associated with changing glucose levels [17,18,19,20]. TXNIP controls glucose uptake in cells, partly by binding to GLUT1, serving as a glucose-sensitive switch [17,18,19,20]. Its expression is a key element in glucose uptake mediated by GLUT1 [19].
Association studies and meta-analyses identified GLUT1 rs841853 single nucleotide polymorphism (SNP), also known as GLUT1 XbaI polymorphism as one of the genetic variants associated with diabetes and diabetic nephropathy, a diabetes-related complication [24,25,26,27,28,29,30,31,32,33,34,35].
DNA methylation, the most studied gene regulatory epigenetic process, is affected by environmental [6, 36, 37] and genetic factors [10, 38,39,40]. Perturbed DNA methylation influences gene expression [41, 42]. Several epigenome-wide association studies (EWAS) on methylation have identified TXNIP cg19693031 as the top diabetes-related methylation site [4, 22, 23, 43,44,45,46,47,48]. This site has also been associated with fasting blood glucose [22, 49].
DNA methylation is a strong disease marker that appears early during disease onset, especially cancer [50,51,52]. Genetic variants are population- and disease-specific. Hence, the identification of variants and biomarkers specific to certain diseases could be helpful in targeted therapy [53]. The genome intertwines with the epigenome [54] and there is a high probability that genomic variations cause diseases by affecting DNA methylation [55]. Therefore, the integration of genetic and methylation data could expand our understanding of disease etiology and prognosis. However, this area of research is lagging [54]. To our knowledge, no study has investigated the joint effect of genetic and epigenetic factors on diabetes and or FBG. In this regard and considering the important individual roles of TXNIP and GLUT1 in diabetes susceptibility and the direct interplay of both genes in glucose metabolism [17,18,19,20], we evaluated the interaction between the genetic variant (GLUT1 rs841853) and the epigenetic aberration (TXNIP cg19693031 methylation) on the fasting blood glucose levels of Taiwanese without type 2 diabetes. We hypothesized that the association between FBG and TXNIP cg19693031 methylation among non-diabetics may differ based on GLUT1 rs841853 genotypes.
Materials and methods
Study participants
We used TWB data collected between 2008 and 2016 for this study. The biobank contains data on basic demographics, personal lifestyle activities, dietary status, personal and family medical history, physical examination, methylation, and genotype. Taiwanese men and women aged 30 to 70 who have never had cancer were eligible to participate in the TWB project as explained previously [56, 57]. More information on the TWB can be found at https://www.twbiobank.org.tw/new_web_en/about-export.php. Data collection in the TWB was done following standard procedures. Initially, we recruited 1442 participants with data on methylation. However, we excluded people with diabetes (n = 115) and those with incomplete genotypes (n = 27). The final study participants included 1300 individuals. All participants provided written informed consents. This study was carried out in accordance with the Declaration of Helsinki and was approved by the Institutional Review Board of Chung Shan Medical University Hospital (CS1-2212).
Definition of variables
In our study, diabetes was defined by (1) fasting blood glucose levels above 126 mg/dl, (2) hemoglobin A1c (HbA1c) levels of at least 6.5% [58], and (3) self-report of a previous clinical diagnosis with diabetes. We defined hypertension using the following criteria: (1) systolic blood pressure 140 mmHg; (2) diastolic blood pressure 90 mmHg) [59] or (3) self-report of prior hypertension diagnosis. The Hitachi LST008 analyzer was used to determine fasting blood glucose, high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), and triglycerides. BMI was calculated using the standard formula: weight in kilograms divided by height in meters squared (kg/m2). Other variables were self-reported and were defined as follows: (1) cigarette smoking: habitual smoking for over 6 months; (2) alcohol drinking: habitual alcohol consumption of over 150 ml per week for the past 6 months; (3) regular exercise: any physical activity lasting over 30 min a day and taken at least three times a week; (4) tea drinking: habitual daily consumption of tea; (5) coffee drinking: consumption of coffee at least three times a week; (6) vegetarian: maintained a vegetarian diet for the past 6 months.
SNP and DNA methylation assessments
We chose GLUT1 rs841853 and TXNIP cg19693031 due to their well-established links with T2DM. DNA methylation and SNP genotyping were conducted at Academia Sinica. Genotyping was done using custom TWB chips (versions 1 and 2) and imputed using SHAPEIT2 (v2.r790) and IMPUTE2 (v2.3.1) software. SNP genotyping and imputation details have been described elsewhere [60]. Briefly, the TWB version 1 chip contains about 650,000 SNPs for GWAS typed on the Axiom genome-wide CHB 1 Array plate (Affymetrix, Inc., Santa Clara, CA, USA). The TWB version 2 chip (Thermo Fisher Scientific, Inc., Santa Clara, CA, USA) can detect approximately 714,431 SNPs. GLUT1 rs841853 passed quality control tests: call rate > 95%, the Hardy–Weinberg equilibrium (p-value > 1.0 × 10–6), and a minor allele frequency > 0.01. DNA methylation was assessed with the Infinium MethylationEPIC BeadChipEPIC array (Illumina Inc. San Diego, CA, USA) using whole blood DNA treated with sodium bisulfite. The R software with the ChAMP package was used to process methylation data, including normalization and correction for batch effect and cell-type heterogeneity.
Statistical analyses
We divided the TXNIP cg19693031 methylation levels into quartiles. The cut-offs were 0.738592 (25th percentile or Q1), 0.769992 (50th percentile or Q2), and 0.800918 (75th percentile or Q3). For GLUT1 rs841853, we used the dominant model and classified the genotypes into CC and CA + AA. Differences between discrete and continuous variables stratified by the GLUT1 rs841853 genotypes (CC and CA + AA) were compared using the Chi-square test and the t-test, respectively. The association of cg19693031 methylation, rs841853, and other variables with fasting blood glucose, as well as the interaction between cg19693031 methylation and rs841853 was determined by multiple linear regression analysis. We used ≥ Q3 as the reference group. Plink 1.9 and SAS 9.4 (SAS Institute, Cary, NC, USA) were used for data analyses.
Results
Table 1 shows a summary of the demographic characteristics of the study participants classified into the rs841853 CC and CA + AA genotypes. There were 735 and 565 participants with CC and CA + AA genotypes, respectively. FBG levels (mean ± SD) between the genotypes (92.07 ± 7.78 for CC and 91.62 ± 7.14 for CA + AA) did not differ significantly. However, the cg19693031 methylation levels were significantly different in the two groups (0.7716 ± 0.05 in CC individuals and 0.7631 ± 0.05 in CA + AA individuals (p = 0.002). The cg19693031 methylation levels according to quartiles were β < 0.738592 (< Q1), 0.738592 ≤ 0.769992 (Q1–Q2), 0.769992 ≤ 0.800918 (Q2–Q3), and β ≥ 0.800918 (≥ Q3). Demographic characteristics of the study participants grouped into GLUT1 rs841853 genotype (CC, CA, and AA) are shown in Additional file 1: Table S1.
Table 2 shows the association of cg19693031 methylation and the rs841853 variant with FBG. FBG increased with decreasing cg19693031 methylation levels in a dose–response manner (ptrend = 0.005). The β (p-value) was − 0.0236 (0.965) for Q2–Q3, 1.0317 (0.058) for Q1–Q2, and 1.3336 (0.019) for < Q1 compared to the reference quartile (≥ Q3). The rs841853 variant was not significantly associated with fasting blood glucose (β = − 0.4576, p = 0.232). However, its interaction with cg19693031 methylation (i.e., rs841853 genotypes*cg19693031 quartiles) was significant (p = 0.036) as shown in Table 3. Additionally, the additive model revealed a significant association between the AA genotype (reference: CC) and FBG (β = − 1.7643, p-value = 0.036) as shown in Additional file 1: Table S2.
When we examined the association between cg19693031 and FBG based on rs841853 genotypes (Table 3), the cg19693031 methylation levels were inversely associated with FBG in both groups but only the CC genotype showed significant results (β; p-value = 0.8082; 0.255 for Q2–Q3, 1.6930; 0.022 for Q1–Q2, and 2.2190; 0.004 for < Q1) compared to the reference quartile (≥ Q3). The trend test was significant only for the CC genotype (ptrend = 0.002).
When we combined the rs841853 genotypes and cg19693031 methylation quartiles using CC and ≥ Q3 as the reference group (Table 4), FBG levels were significantly higher among individuals carrying the CC genotype in the Q1–Q2 (β = 1.7709, p-value = 0.013) and < Q1 (β = 2.3116, p-value = 0.001) quartiles.
Discussion
The heritability of diabetes is estimated at 20–80% [61, 62]. However, only 5–15% of this heritability has been explained [63]. Some methylation sites are believed to be heritable [64, 65]. Therefore, SNPs alone cannot fully delineate genetic heritability [6]. To our knowledge, this is the first study on blood sugar levels based on a genetic variant (rs841853) and an epigenetic modification (cg19693031 methylation) among non-diabetics.
In our study, we found significant differences in baseline TXNIP cg19693031 methylation levels between GLUT1 rs841853 genotypes (CC and CA + AA). There were no differences in FBG between these genotypes. Multiple linear regression analyses showed an inverse association between FBG and cg19693031 methylation levels in a dose–response manner. These results are consistent with those from numerous studies in which diabetes patients (higher FBG) had lower levels of TXNIP methylation [4, 22, 23, 43,44,45,46,47,48,49].
FBG did not differ between the GLUT1 rs841853 genotypes as previously reported [24]. The dominant model showed no significant association between rs841853-CA + AA genotype and FBG in our study. However, the additive model revealed a significant association between the rs841853-AA genotype and FBG. Previous literature contains contradictory findings regarding rs841853 and diabetes. For instance, rs841853 was not significantly associated with diabetes in several studies [24, 66,67,68]. Contrarily, the variant was confirmed as a diabetes-related SNP in Japanese [33, 69], Tunisian [34], and Bangladeshi [35] women. Some meta-analyses reported significant associations between rs841853 and diabetes [26, 30]. However, a meta-analysis found an association only among Asians, not Blacks or Caucasians. This suggests that the effect of the variant on T2DM varies across races [25]. Additional investigations of other diabetes-associated SNPs, including the recently reported polymorphism rs1800977 (C69T) within the ATP-binding cassette transporter A1 (ABCA1) [70] gene are necessary.
Even though the dominant model suggested that the rs841853-CA + AA genotype might not be independently associated with FBG in our study, the additive model showed a significant association between the AA genotype and FBG. Furthermore, the interaction between rs841853 and cg19693031 methylation was significant. The presence of a significant interaction between the genetic variant and the epigenetic process implies that rs841853 might be involved in the epigenetic mechanism (cg19693031 methylation) underlying diabetes. When we stratified the participants by the rs841853 genotypes, the dose–response and inverse association between FBG and cg19693031 methylation was retained only in the CC genotype. That is, in the presence of the CC genotype, lower levels of methylation (i.e., β < 0.738592 and 0.738592 ≤ 0.769992 corresponding to Q1–Q2 and < Q1) were associated with an increase (1.6930 for Q1–Q2 and 2.2190 for < Q1) in FBG levels. The increase in FBG (a hallmark of diabetes) indicates a higher probability of exposure to diabetes among non-diabetics with the rs841853 CC genotype who had lower levels of cg19693031 methylation. This could also imply that the GLUT1 rs841853 CC genotype and cg19693031 methylation might jointly influence the expression of TXNIP. However, we cannot clearly state the underlying mechanism. Notwithstanding, TXNIP is a gatekeeper for glucose metabolism which enhances glucose toxicity and pancreatic β cell apoptosis when highly expressed [17, 18, 20, 71, 72]. This gene is highly expressed in diabetes, which is characterized by impaired glucose-induced insulin production [71] and its inhibition could reduce glucotoxicity-related β-cell loss [73]. It is regarded as the main regulatory channel and an endocytosis adaptor for GLUT1 in glucose metabolism and the resulting mitochondrial actions in response to fluctuating glucose levels. That is, TXNIP is a signal regulation channel in glucose metabolism where it reduces glucose uptake by promoting GLUT1 endocytosis [18, 20].
An increase in GLUT1 mRNA expression is associated with an increase in glucose uptake [18]. However, TXNIP expression appears to be negatively associated with glucose levels and GLUT1. In the brain, GLUT1 is overly expressed, while TXNIP expression is very low [20]. TXNIP degradation resulting from glucose uptake was associated with the release of GLUT1 from endocytosis [17]. Furthermore, its overexpression in cultured adipocytes was associated with inhibited glucose uptake and vice versa [18]. TXNIP inhibits glucose influx directly or indirectly. The indirect mechanism involves the promotion of GLUT1 endocytosis by TXNIP that is transcriptionally induced by glucose [17, 18, 20]. The direct mechanism involves the binding of TXNIP to GLUT1 which inhibits the transport of glucose by GLUT1 at the plasma membrane [17]. In diabetes pathogenesis, slightly elevated blood sugar levels early in the disease onset enhance TXNIP expression and suppress glucose uptake by cells. This leads to increased blood sugar levels and subsequent overexpression of TXNIP, which down-regulates GLUT1 function and reduces glucose uptake in the periphery [18].
Our study was limited to participants without diabetes. However, when we included an additional model to determine FBG levels in diabetic patients (n = 114) based on GLUT1 TXNIP cg19693031 and variant rs841853 (data not shown), we found that (1) FBG levels decreased significantly with increasing methylation levels (β = − 377.4484, p < 0.001); (2) Compared to CC homozygotes, FBG levels were higher in patients with CA genotype (β = 11.0338) but lower in those with AA genotype (β = − 25.9662) even though these results were not significant (p > 0.05). Despite these, selection bias cannot be ruled out due to the retrospective nature of our study.
DNA methylation is a strong disease marker that appears early during disease onset, especially cancer [50,51,52]. Genetic variants are population and disease-specific. Therefore, identifying specific variants and biomarkers for certain diseases could be useful in targeted therapy [53]. Therefore, monitoring the methylation patterns of diabetes-related genes in non-diabetics with a specific genetic variation could help in the identification of individuals at risk of diabetes.
Conclusion
TXNIP cg19693031 methylation was inversely associated with FBG in a dose-dependent manner. The rs841853 variant was not independently associated with fasting blood glucose but had a significant interaction with cg19693031 methylation. The inverse association between TXNIP cg19693031 methylation and FBG became more prominent in the presence of the GLUT1 rs841853 CC genotype, suggesting that rs841853 could modulate the association between cg19693031 methylation and FBG. Both genetic variants and epigenetic changes associated with FBG could help in the early identification of individuals at risk of T2DM. Hence, considering the methylation patterns of diabetes-related genes in non-diabetics with specific genetic variants is essential when investigating the pathogenesis of T2DM.
Availability of data and materials
The data that support the findings of this study are available from Taiwan Biobank but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are, however, available from the authors upon reasonable request and with permission of Taiwan Biobank.
References
Ramtahal R, Khan C, Maharaj-Khan K, Nallamothu S, Hinds A, Dhanoo A, et al. Prevalence of self-reported sleep duration and sleep habits in type 2 diabetes patients in South Trinidad. J Epidemiol Global Health. 2015;5 (4):S35–43.
Khan MAB, Hashim MJ, King JK, Govender RD, Mustafa H, Al KJ. Epidemiology of type 2 diabetes–global burden of disease and forecasted trends. J Epidemiol Global Health. 2020;10 (1):107.
Whiting DR, Guariguata L, Weil C, Shaw J. IDF diabetes atlas: global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Res Clin Pract. 2011;94 (3):311–21.
Meeks KA, Henneman P, Venema A, Addo J, Bahendeka S, Burr T, et al. Epigenome-wide association study in whole blood on type 2 diabetes among sub-Saharan African individuals: findings from the RODAM study. Int J Epidemiol. 2019;48 (1):58–70.
Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40 (5):638–45.
Drong A, Lindgren C, McCarthy M. The genetic and epigenetic basis of type 2 diabetes and obesity. Clin Pharmacol Ther. 2012;92 (6):707–15.
Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42 (7):579–89.
Oluyombo R, Olamoyegun MA, Olaifa O, Iwuala SO, Babatunde OA. Cardiovascular risk factors in semi-urban communities in southwest Nigeria: patterns and prevalence. J Epidemiol Global Health. 2015;5 (2):167–74.
Lone S, Lone K, Khan S, Pampori RA. Assessment of metabolic syndrome in Kashmiri population with type 2 diabetes employing the standard criteria’s given by WHO, NCEPATP III and IDF. J Epidemiol Global Health. 2017;7 (4):235–9.
Mahanta TG, Joshi R, Mahanta BN, Xavier D. Prevalence of modifiable cardiovascular risk factors among tea garden and general population in Dibrugarh, Assam, India. J Epidemiol Global Health. 2013;3 (3):147–56.
Bo A, Thomsen R, Nielsen J, Nicolaisen SK, Beck-Nielsen H, Rungby J, et al. Early-onset type 2 diabetes: age gradient in clinical and behavioural risk factors in 5115 persons with newly diagnosed type 2 diabetes—results from the DD2 study. Diabetes/Metab Res Rev. 2018;34 (3):e2968.
Pippitt K, Li M, Gurgle HE. Diabetes mellitus: screening and diagnosis. Am Fam Physician. 2016;93 (2):103–9.
Al Slail FY, Abid O, Assiri AM, Memish ZA, Ali MK. Cardiovascular risk profiles of adults with type-2 diabetes treated at urban hospitals in Riyadh, Saudi Arabia. J Epidemiol Global Health. 2016;6 (1):29–36.
Fonseca VA. Defining and characterizing the progression of type 2 diabetes. Diabetes Care. 2009;32 (suppl 2):S151–6.
Levitan EB, Song Y, Ford ES, Liu S. Is nondiabetic hyperglycemia a risk factor for cardiovascular disease?: a meta-analysis of prospective studies. Arch Intern Med. 2004;164 (19):2147–55.
Galochkina T, Chong MNF, Challali L, Abbar S, Etchebest C. New insights into GluT1 mechanics during glucose transfer. Sci Rep. 2019;9 (1):1–14.
Wu N, Zheng B, Shaywitz A, Dagon Y, Tower C, Bellinger G, et al. AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Mol Cell. 2013;49 (6):1167–75.
Parikh H, Carlsson E, Chutkow WA, Johansson LE, Storgaard H, Poulsen P, et al. TXNIP regulates peripheral glucose metabolism in humans. PLoS Med. 2007;4 (5):e158.
Beg M, Zhang W, McCourt AC, Enerbäck S. ATGL activity regulates GLUT1-mediated glucose uptake and lactate production via TXNIP stability in adipocytes. J Biol Chem. 2021;296:100332.
Waldhart AN, Dykstra H, Peck AS, Boguslawski EA, Madaj ZB, Wen J, et al. Phosphorylation of TXNIP by AKT mediates acute influx of glucose in response to insulin. Cell Rep. 2017;19 (10):2005–13.
Wondafrash DZ, Nire’a AT, Tafere GG, Desta DM, Berhe DA, Zewdie KA. Thioredoxin-interacting protein as a novel potential therapeutic target in diabetes mellitus and its underlying complications. Diabetes Metab Syndr Obesity Targets Therapy. 2020;13:43.
Soriano-Tárraga C, Jiménez-Conde J, Giralt-Steinhauer E, Mola-Caminal M, Vivanco-Hidalgo RM, Ois A, et al. Epigenome-wide association study identifies TXNIP gene associated with type 2 diabetes mellitus and sustained hyperglycemia. Hum Mol Genet. 2016;25 (3):609–19.
Walaszczyk E, Luijten M, Spijkerman AM, Bonder MJ, Lutgers HL, Snieder H, et al. DNA methylation markers associated with type 2 diabetes, fasting glucose and HbA 1c levels: a systematic review and replication in a case–control sample of the Lifelines study. Diabetologia. 2018;61 (2):354–68.
Ramadan RA, Zaki AM, Magour GM, Zaki MA, Aglan SA, Madkour MA, et al. Association of XbaI GLUT1 polymorphism with susceptibility to type 2 diabetes mellitus and diabetic nephropathy. Am J Mol Biol. 2016;6:71–8.
Du B, Liu S, Cui C, Wang S, Cui W. Association between glucose transporter 1 rs841853 polymorphism and type 2 diabetes mellitus risk may be population specific (葡萄糖转移酶 1 的 rs841853 基因多态性与 2 型糖尿发生风险的关系可能具有人群特异性). J Diabetes. 2013;5 (3):291–9.
Stefanidis I, Tziastoudi M, Tsironi E, Dardiotis E, Tachmitzi S, Fotiadou A, et al. The contribution of genetic variants of SLC2A1 gene in T2DM and T2DM-nephropathy: association study and meta-analysis. Ren Fail. 2018;40 (1):561–76.
Mooyaart AL. Diabetic nephropathy: pathology, genetics and carnosine metabolism. Leiden: Leiden University; 2011.
Liu Z-H, Guan T-J, Chen Z-H, Li L-S. Glucose transporter (GLUT1) allele (XbaI–) associated with nephropathy in non-insulin-dependent diabetes mellitus. Kidney Int. 1999;55 (5):1843–8.
Grzeszczak W, Moczulski DK, Zychma M, Zukowska-Szczechowska E, Trautsolt W, Szydlowska I. Role of GLUT1 gene in susceptibility to diabetic nephropathy in type 2 diabetes. Kidney Int. 2001;59 (2):631–6.
Stefanidis I, Kytoudis K, Papathanasiou AA, Zaragotas D, Melistas L, Kitsios GD, et al. XbaI GLUT1 gene polymorphism and the risk of type 2 diabetes with nephropathy. Dis Markers. 2009;27 (1):29–35.
Zintzaras E, Stefanidis I. Association between the GLUT1 gene polymorphism and the risk of diabetic nephropathy: a meta-analysis. J Hum Genet. 2005;50 (2):84–91.
Cui W, Du B, Zhou W, Jia Y, Sun G, Sun J, et al. Relationship between five GLUT1 gene single nucleotide polymorphisms and diabetic nephropathy: a systematic review and meta-analysis. Mol Biol Rep. 2012;39 (8):8551–8.
Tao T, Tanizawa Y, Matsutani A, Matsubara A, Kaneko T, Kaku K. HepG2/erythrocyte glucose transporter (GLUT1) gene in NIDDM: a population association study and molecular scanning in Japanese subjects. Diabetologia. 1995;38 (8):942–7.
Makni K, Mnif F, Boudawara M, Hamza N, Rekik N, Abid M, et al. Association of glucose transporter 1 polymorphisms with type 2 diabetes in the Tunisian population. Diabetes Metab Res Rev. 2008;24 (7):544–8.
Bappy HJA, Goswami A, Huda N, Hosen MI, Nabi AN. Gender specific association of missense variant rs1805097 of IRS-2 and noncoding variant rs841853 of GLUT-1 genes with susceptibility to type 2 diabetes in Bangladeshi population. Gene Rep. 2020;21:100866.
Liu H, Li S, Wang X, Zhu J, Wei Y, Wang Y, et al. DNA methylation dynamics: identification and functional annotation. Brief Funct Genomics. 2016;15 (6):470–84.
Tantoh DM, Lee K-J, Nfor ON, Liaw Y-C, Lin C, Chu H-W, et al. Methylation at cg05575921 of a smoking-related gene (AHRR) in non-smoking Taiwanese adults residing in areas with different PM 2.5 concentrations. Clin Epigenet. 2019;11 (1):1–8.
Fraser HB, Lam LL, Neumann SM, Kobor MS. Population-specificity of human DNA methylation. Genome Biol. 2012;13 (2):1–12.
Gibbs JR, Van Der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai S-L, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6 (5):e1000952.
Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12 (1):1–13.
Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38 (1):23–38.
Jin Z, Liu Y. DNA methylation in human diseases. Genes Diseases. 2018;5:1–8.
Chambers JC, Loh M, Lehne B, Drong A, Kriebel J, Motta V, et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol. 2015;3 (7):526–34.
Kulkarni H, Kos MZ, Neary J, Dyer TD, Kent JW Jr, Göring HH, et al. Novel epigenetic determinants of type 2 diabetes in Mexican-American families. Hum Mol Genet. 2015;24 (18):5330–44.
Florath I, Butterbach K, Heiss J, Bewerunge-Hudler M, Zhang Y, Schoettker B, et al. Type 2 diabetes and leucocyte DNA methylation: an epigenome-wide association study in over 1,500 older adults. Diabetologia. 2016;59 (1):130–8.
Al Muftah WA, Al-Shafai M, Zaghlool SB, Visconti A, Tsai P-C, Kumar P, et al. Epigenetic associations of type 2 diabetes and BMI in an Arab population. Clin Epigenet. 2016;8 (1):1–10.
Juvinao-Quintero DL, Marioni RE, Ochoa-Rosales C, Russ TC, Deary IJ, Van Meurs JB, et al. DNA methylation of blood cells is associated with prevalent type 2 diabetes in a meta-analysis of four European cohorts. Clin Epigenet. 2021;13 (1):1–14.
Cardona A, Day FR, Perry JR, Loh M, Chu AY, Lehne B, et al. Epigenome-wide association study of incident type 2 diabetes in a British population: EPIC-Norfolk study. Diabetes. 2019;68 (12):2315–26.
Nuotio M-L, Pervjakova N, Joensuu A, Karhunen V, Hiekkalinna T, Milani L, et al. An epigenome-wide association study of metabolic syndrome and its components. Sci Rep. 2020;10 (1):1–12.
Diaz-Lagares A, Mendez-Gonzalez J, Hervas D, Saigi M, Pajares MJ, Garcia D, et al. A novel epigenetic signature for early diagnosis in lung cancer. Clin Cancer Res. 2016;22 (13):3361–71.
Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4 (9):707.
Mikeska T, Bock C, Do H, Dobrovic A. DNA methylation biomarkers in cancer: progress towards clinical implementation. Expert Rev Mol Diagn. 2012;12 (5):473–87.
Krzyszczyk P, Acevedo A, Davidoff EJ, Timmins LM, Marrero-Berrios I, Patel M, et al. The growing role of precision and personalized medicine for cancer treatment. Technology. 2018;6 (03no4):79–100.
Romanowska J, Haaland ØA, Jugessur A, Gjerdevik M, Xu Z, Taylor J, et al. Gene–methylation interactions: discovering region-wise DNA methylation levels that modify SNP-associated disease risk. Clin Epigenet. 2020;12 (1):1–18.
Shilpi A, Bi Y, Jung S, Patra SK, Davuluri RV. Identification of genetic and epigenetic variants associated with breast cancer prognosis by integrative bioinformatics analysis. Cancer informatics. 2017. https://doi.org/10.4137/CIN.S39783.
Fan C-T, Lin J-C, Lee C-H. Taiwan Biobank: a project aiming to aid Taiwan’s transition into a biomedical island. 2008.
Lin J-C, Fan C-T, Liao C-C, Chen Y-S. Taiwan Biobank: making cross-database convergence possible in the Big Data era. Gigascience. 2018;7 (1):gix110.
Association AD. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2014;37 (Supplement 1):S81–90.
Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL Jr, et al. Seventh report of the joint national committee on prevention, detection, evaluation, and treatment of high blood pressure. Hypertension. 2003;42 (6):1206–52.
Wei C-Y, Yang J-H, Yeh E-C, Tsai M-F, Kao H-J, Lo C-Z, et al. Genetic profiles of 103,106 individuals in the Taiwan Biobank provide insights into the health and history of Han Chinese. NPJ Genom Med. 2021;6 (1):1–10.
Meigs JB, Cupples LA, Wilson P. Parental transmission of type 2 diabetes: the Framingham Offspring Study. Diabetes. 2000;49 (12):2201–7.
Poulsen P, Kyvik KO, Vaag A, Beck-Nielsen H. Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance–a population-based twin study. Diabetologia. 1999;42 (2):139–45.
Maher B. Personal genomes: the case of the missing heritability. Nat News. 2008;456 (7218):18–21.
Boks MP, Derks EM, Weisenberger DJ, Strengman E, Janson E, Sommer IE, et al. The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PLoS ONE. 2009;4 (8):e6767.
Kaminsky ZA, Tang T, Wang S-C, Ptak C, Oh GH, Wong AH, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. 2009;41 (2):240–5.
Baroni MG, Alcolado JC, Gragnoli C, Franciosi AM, Cavallo MG, Fiore V, et al. Affected sib-pair analysis of the GLUT1 glucose transporter gene locus in non-insulin-dependent diabetes mellitus (NIDDM): evidence for no linkage. Hum Genet. 1994;93 (6):675–80.
Alcolado J, Baroni M. Restriction fragment length polymorphisms at the GLUT4 and GLUT1 gene loci in type 2 diabetes. Diabet Med. 1992;9 (1):58–60.
Elbein SC, Hoffman MD, Matsutani A, Permutt MA. Linkage analysis of GLUT1 (HepG2) and GLUT2 (liver/islet) genes in familial NIDDM. Diabetes. 1992;41 (12):1660–7.
Li S, Oelbaum R, Baroni M, Stock J, Galton D. Association of genetic variant of the glucose transporter with non-insulin-dependent diabetes mellitus. Lancet. 1988;332 (8607):368–70.
Ghafar MTA, Shalaby KH, Okda HI, Rizk FH. Association of ABCA1 (C69T) gene polymorphism with dyslipidemia and type 2 diabetes among the Egyptian population. Meta Gene. 2020;25:100714.
Minn AH, Pise-Masison CA, Radonovich M, Brady JN, Wang P, Kendziorski C, et al. Gene expression profiling in INS-1 cells overexpressing thioredoxin-interacting protein. Biochem Biophys Res Commun. 2005;336 (3):770–8.
Mohammad Alhawiti N, Al Mahri S, Azhar Aziz M, Shafi Malik S, Mohammad S. TXNIP in metabolic regulation: physiological role and therapeutic outlook. Curr Drug Targets. 2017;18 (9):1095–103.
Chen J, Saxena G, Mungrue IN, Lusis AJ, Shalev A. Thioredoxin-interacting protein: a critical link between glucose toxicity and β-cell apoptosis. Diabetes. 2008;57 (4):938–44.
Acknowledgements
Not applicable.
Funding
This work was supported by funds from Chung Shan Medical University Hospital (CSH-2021-D-006) and the Ministry of Science and Technology (MOST), Taiwan (MOST 109-2121-M-040-002; MOST 110-2121-M-040-002; MOST 109-2811-M-040-500; MOST 110-2811-M-040-001).
Author information
Authors and Affiliations
Contributions
HHT,CYS, , SYH, DMT, ONN, , YHC, and YPL conceived the study. All authors designed the study. SYH and YPL analyzed the data. All authors interpreted the data. DMT and HHT drafted the manuscript. All authors reviewed, read, and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Institutional Review Board of Chung Shan Medical University Hospital (CS1-20212).
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationship that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Demographic characteristics of the study participants grouped by GLUT1 rs841853 genotypes (CC, CA, and AA). Table S2. Association of TXNIP cg19693031 methylation and GLUT1 rs841853 genotype with fasting blood glucose (additive model).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Tsai, HH., Shen, CY., Ho, CC. et al. Interaction between a diabetes-related methylation site (TXNIP cg19693031) and variant (GLUT1 rs841853) on fasting blood glucose levels among non-diabetics. J Transl Med 20, 87 (2022). https://doi.org/10.1186/s12967-022-03269-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-022-03269-y