Abstract
Aims
Hemoglobin A1c (HbA1c) levels are widely employed to diagnose diabetes. However, estimates of the heritability of HbA1c and glucose levels are different. Therefore, we explored HbA1c- and blood glucose-associated loci in a non-diabetic Japanese population.
Methods
We conducted a two-stage genome-wide association study (GWAS) on variants associated with HbA1c and blood glucose levels in a Japanese population. In the initial stage, data of 4911 participants of the Japan Multi-Institutional Collaborative Cohort (J-MICC) were subjected to discovery analysis. In the second stage, two datasets from the Tohoku Medical Megabank project, with 8175 and 40,519 participants, were used for the replication study. Association of the imputed variants with HbA1c and blood glucose levels was determined via linear regression analyses adjusted for age, sex, body mass index (BMI), smoking, and genetic principal components (PC1–PC10). Moreover, we performed a BMI-stratified GWAS on HbA1c levels in the J-MICC. The discovery analysis and BMI-stratified GWAS results were validated with re-analyses of normalized HbA1c levels adjusted for site in addition to the above, and blood glucose adjusted for fasting time as an additional covariate.
Results
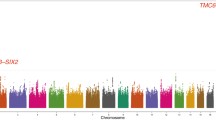
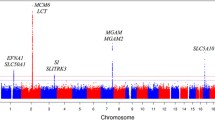
Genetic variants associated with HbA1c levels were identified in KCNQ1 and TMC6. None of the genetic variants associated with blood glucose levels in the discovery analysis were replicated. Association of rs2299620 in KCNQ1 with HbA1c levels showed heterogeneity between individuals with BMI ≥ 25 kg/m2 and BMI < 25 kg/m2.
Conclusions
The variant rs2299620 in KCNQ1 might affect HbA1c levels differentially based on BMI grouping in the Japanese population.

Similar content being viewed by others
Data availability
Data are available upon reasonable request. Details can be found on the J-MICC Study website (http://www.jmicc.com/).
References
American Diabetes Association. Standards of medical care in diabetes–2010. Diabetes Care. 2010;33(Suppl 1):S11-61. https://doi.org/10.2337/dc10-S011.
World Health Organization. Use of Glycated Haemoglobin (HbA1c) in the Diagnosis of Diabetes Mellitus: Abbreviated Report of a WHO Consultation. 2011.
Soranzo N, Sanna S, Wheeler E, et al. Common variants at 10 genomic loci influence hemoglobin A1(C) levels via glycemic and nonglycemic pathways. Diabetes. 2010;59(12):3229–39. https://doi.org/10.2337/db10-0502.
Chen P, Takeuchi F, Lee JY, et al. Multiple nonglycemic genomic loci are newly associated with blood level of glycated hemoglobin in East Asians. Diabetes. 2014;63(7):2551–62. https://doi.org/10.2337/db13-1815.
Hachiya T, Komaki S, Hasegawa Y, et al. Genome-wide meta-analysis in Japanese populations identifies novel variants at the TMC6-TMC8 and SIX3-SIX2 loci associated with HbA1c. Sci Rep. 2017;7(1):16147. https://doi.org/10.1038/s41598-017-16493-0.
Sakaue S, Kanai M, Tanigawa Y, et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat Genet. 2021;53(10):1415–24. https://doi.org/10.1038/s41588-021-00931-x.
Soranzo N. Genetic determinants of variability in glycated hemoglobin (HbA(1c)) in humans: review of recent progress and prospects for use in diabetes care. Curr Diab Rep. 2011;11(6):562–9. https://doi.org/10.1007/s11892-011-0232-9.
Ryu J, Lee C. Association of glycosylated hemoglobin with the gene encoding CDKAL1 in the Korean association resource (KARE) study. Hum Mutat. 2012;33(4):655–9. https://doi.org/10.1002/humu.22040.
Li J, Glessner JT, Zhang H, et al. GWAS of blood cell traits identifies novel associated loci and epistatic interactions in Caucasian and African-American children. Hum Mol Genet. 2013;22(7):1457–64. https://doi.org/10.1093/hmg/dds534.
van der Harst P, Zhang W, Mateo Leach I, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492(7429):369–75. https://doi.org/10.1038/nature11677.
Guillem F, Lawson S, Kannengiesser C, et al. Two nonsense mutations in the TMPRSS6 gene in a patient with microcytic anemia and iron deficiency. Blood. 2008;112(5):2089–91. https://doi.org/10.1182/blood-2008-05-154740.
Delpierre G, Collard F, Fortpied J, et al. Fructosamine 3-kinase is involved in an intracellular deglycation pathway in human erythrocytes. Biochem J. 2002;365(Pt 3):801–8. https://doi.org/10.1042/BJ20020325.
Pilia G, Chen WM, Scuteri A, et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2006;2(8):e132. https://doi.org/10.1371/journal.pgen.0020132.
jMorp. TGA000007 > HbA1c (NGSP). Tohoku Medical Megabank Organization, Tohoku University, https://jmorp.megabank.tohoku.ac.jp/gwas-analyses/TGA000007-ed63d3c3. Accessed 29 Oct 2022
jMorp. TGA000007 > Glucose. Tohoku Medical Megabank Organization, Tohoku University, https://jmorp.megabank.tohoku.ac.jp/gwas-analyses/TGA000007-2c35c1a8. Accessed 29 Oct 2022
BioBank Japan PheWeb. Osaka, Japan: Department of Statistical Genetics, Osaka University Graduate School of Medicine; 2020. HbA1c: HbA1c; https://pheweb.jp/pheno/HbA1c. Accessed 29 Oct 2022
BioBank Japan PheWeb. Osaka, Japan: Department of Statistical Genetics, Osaka University Graduate School of Medicine; 2020. Glucose: Glucose; https://pheweb.jp/pheno/Glucose. Accessed 29 Oct 2022.
Imamura M, Takahashi A, Yamauchi T, et al. Genome-wide association studies in the Japanese population identify seven novel loci for type 2 diabetes. Nat Commun. 2016;7:10531. https://doi.org/10.1038/ncomms10531.
Hamajima N, J-MICC Study Group. The Japan multi-institutional collaborative cohort study (J-MICC Study) to detect gene-environment interactions for cancer. Asian Pac J Cancer Prev. 2007;8:317–23.
Takeuchi K, Naito M, Kawai S, et al. Study profile of the Japan multi-institutional collaborative cohort (J-MICC) study. J Epidemiol. 2021;31(12):660–8. https://doi.org/10.2188/jea.JE20200147.
Kashiwagi A, Kasuga M, Araki E, et al. International clinical harmonization of glycated hemoglobin in Japan: from japan diabetes society to national glycohemoglobin standardization program values. J Diabetes Investig. 2012;3(1):39–40. https://doi.org/10.1111/j.2040-1124.2012.00207.x.
Chang CC, Chow CC, Tellier LC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. https://doi.org/10.1186/s13742-015-0047-8.
Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–9. https://doi.org/10.1038/ng1847.
Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. https://doi.org/10.1038/nature11632.
Auton A, Brooks LD, Durbin RM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. https://doi.org/10.1038/nature15393.
Yamaguchi-Kabata Y, Nakazono K, Takahashi A, et al. Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies. Am J Hum Genet. 2008;83(4):445–56. https://doi.org/10.1016/j.ajhg.2008.08.019.
Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9(2):179–81. https://doi.org/10.1038/nmeth.1785.
Das S, Forer L, Schönherr S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–7. https://doi.org/10.1038/ng.3656.
Das S. DosageConvertor. Updated on 18 January 2019. https://genome.sph.umich.edu/wiki/DosageConvertor. Accessed 1 May 2022.
Gogarten SM, Bhangale T, Conomos MP, et al. GWASTools: an R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics. 2012;28(24):3329–31. https://doi.org/10.1093/bioinformatics/bts610.
Turner SD. qqman: an R package for visualizing GWAS results using Q-Q and Manhattan plots. bioRxiv. 2014. https://doi.org/10.1101/005165.
Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–7. https://doi.org/10.1093/bioinformatics/btq419.
Hozawa A, Tanno K, Nakaya N, et al. Study profile of the tohoku medical megabank community-based cohort study. J Epidemiol. 2021;31(1):65–76. https://doi.org/10.2188/jea.JE20190271.
Sutoh Y, Hachiya T, Suzuki Y, et al. ALDH2 genotype modulates the association between alcohol consumption and AST/ALT ratio among middle-aged Japanese men: a genome-wide G × E interaction analysis. Sci Rep. 2020;10(1):16227. https://doi.org/10.1038/s41598-020-73263-1.
Kawai Y, Mimori T, Kojima K, et al. Japonica array: improved genotype imputation by designing a population-specific SNP array with 1070 Japanese individuals. J Hum Genet. 2015;60(10):581–7. https://doi.org/10.1038/jhg.2015.68.
Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. https://doi.org/10.1371/journal.pgen.1000529.
Yamaguchi-Kabata Y, Yasuda J, Tanabe O, et al. Evaluation of reported pathogenic variants and their frequencies in a Japanese population based on a whole-genome reference panel of 2049 individuals. J Hum Genet. 2018;63(2):213–30. https://doi.org/10.1038/s10038-017-0347-1.
Tadaka S, Katsuoka F, Ueki M, et al. 3.5KJPNv2: an allele frequency panel of 3552 Japanese individuals including the X chromosome. Hum Genome Var. 2019;6:28. https://doi.org/10.1038/s41439-019-0059-5.
Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. https://doi.org/10.1093/bioinformatics/btq340.
Tyner C, Barber GP, Casper J, et al. The UCSC genome browser database: 2017 update. Nucleic Acids Res. 2017;45(D1):D626–34. https://doi.org/10.1093/nar/gkw1134.
Aken BL, Achuthan P, Akanni W, et al. Ensembl 2017. Nucleic Acids Res. 2017;45(D1):D635–42. https://doi.org/10.1093/nar/gkw1104.
GTEx Consortium Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–60. https://doi.org/10.1126/science.1262110.
Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. https://doi.org/10.1093/bioinformatics/bth457.
Unoki H, Takahashi A, Kawaguchi T, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008;40(9):1098–102. https://doi.org/10.1038/ng.208.
Hanson RL, Guo T, Muller YL, et al. Strong parent-of-origin effects in the association of KCNQ1 variants with type 2 diabetes in American Indians. Diabetes. 2013;62(8):2984–91. https://doi.org/10.2337/db12-1767.
Müssig K, Staiger H, Machicao F, et al. Association of type 2 diabetes candidate polymorphisms in KCNQ1 with incretin and insulin secretion. Diabetes. 2009;58(7):1715–20. https://doi.org/10.2337/db08-1589.
Dimitriadis G, Mitrou P, Lambadiari V, et al. Insulin effects in muscle and adipose tissue. Diabetes Res Clin Pract. 2011;93(Suppl 1):S52–9. https://doi.org/10.1016/S0168-8227(11)70014-6.
BioBank Japan PheWeb. Osaka, Japan: Department of Statistical Genetics, Osaka University Graduate School of Medicine; 2020. BMI: Body mass index; https://pheweb.jp/pheno/BMI. Accessed 29 Oct 2022.
National Library of Medicine, Bethesda, MD: HGNC. TMC6 transmembrane channel like 6. Updated on 13 May 2022. https://www.ncbi.nlm.nih.gov/gene/11322. Accessed 1 May 2022.
Hachiya T, Furukawa R, Shiwa Y, et al. Genome-wide identification of inter-individually variable DNA methylation sites improves the efficacy of epigenetic association studies. NPJ Genom Med. 2017;2:11. https://doi.org/10.1038/s41525-017-0016-5.
Hugill AJ, Stewart ME, Yon MA, et al. Loss of arylformamidase with reduced thymidine kinase expression leads to impaired glucose tolerance. Biol Open. 2015;4(11):1367–75. https://doi.org/10.1242/bio.013342.
Acknowledgements
The authors thank the staff of the Laboratory for Genotyping Development, Center for Integrative Medical Sciences at RIKEN, and the staff of the BioBank Japan project. We also thank Dr. Nobuyuki Hamajima and Dr. Hideo Tanaka, the previous principal investigators of the J-MICC study, for their continued support of the current study. This work was supported by Grants-in-Aid for Scientific Research for Priority Areas of Cancer [grant number 17015018], Innovative Areas [grant number 221S0001], and JSPS KAKENHI [grant numbers 17390186, 20390184, 24390165, 16H06277, and 19H03902] from the Japanese Ministry of Education, Culture, Sports, Science, and Technology. This study was also supported, in part, by the BioBank Japan Project of the Japan Agency for Medical Research and Development since April 2015 and the Ministry of Education, Culture, Sports, Science and Technology from April 2003 to March 2015.
Author information
Authors and Affiliations
Contributions
Conceptualization, TS and YN; methodology, TS and YN; validation, YS, AS, TH and YO-Y; formal analysis, TS, YN, YS, YO-Y and MN; writing—original draft, TS and YN; writing—review and editing; YN and YS; visualization, YN and MN; supervision, YN; investigation and resources, YN, YS, AS, TH, YO-Y, NT, AK, KM, YK, HI, JO, KT, CS, TK, IW, SS, HN-S, AH, TT, YK, RO, KK, SK-K, TW, ST, CK, IO, YNK, YN, MK, MN, YM, KW and KM. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Takuya Sakashita is an employee of Takara Bio, Inc., Japan. Dr. Hachiya is a board member of Genome Analytics Japan Inc. Dr. Nakatochi reports receiving grants from Boehringer Ingelheim outside the submitted work. All other authors declare that they have no conflicts of interest associated with this study.
Human rights statement and informed consent
All procedures were in accordance with the technical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and later revisions. Informed consent was obtained from all participants before inclusion into the J-MICC study. The main study protocol of the J-MICC study was approved on July 20, 2005 by the Ethics Committee at Nagoya University School of Medicine (approval number 253).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Sakashita, T., Nakamura, Y., Sutoh, Y. et al. Comparison of the loci associated with HbA1c and blood glucose levels identified by a genome-wide association study in the Japanese population. Diabetol Int 14, 188–198 (2023). https://doi.org/10.1007/s13340-023-00618-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13340-023-00618-0