Abstract
Background
The gastrointestinal microbiome is an important component of the human body and is closely related to human health and disease. This study describes the hotspots of the human gastrointestinal microbiome research and its evolution in the past decade, evaluates the scientific cooperation network, and finally predicts the field’s future development trend using bibliometric analysis and a visualized study.
Methods
We searched the original articles from January 2010 to February 2021 in the Scopus database using the term “gastrointestinal microbiome” and its synonyms. CiteSpace was used to construct country and author co-occurrence map; conduct journal, citation cocitation analysis, and reference co-citation knowledge map; and form a keywords co-occurrence map, a clustering knowledge map, timeline view of keywords, and burst term map.
Result
A total of 4444 documents published from January 2010 to February 2021 were analysed. In approximately the past decade, the number of articles on the human gastrointestinal microbiome has increased rapidly, and the research topics focus on different populations, research methods, and detection methods. All countries and regions in the world, led by the US, are studying the human gastrointestinal microbiome, and many research teams with close cooperation have been formed. The research has been published extensively in microbiology journals and clinical medicine journals, and the highly cited articles mainly describe the relationship between gastrointestinal microorganisms and human health and disease. Regarding the research emphasis, researchers' exploration of the human gastrointestinal microbiome (2011–2013) was at a relatively macro and superficial stage and sought to determine how the gastrointestinal microbiome relates to humans. From 2014 to 2017, increasingly more studies were conducted to determine the interaction between human gastrointestinal flora and various organs and systems. In addition, researchers (2018–2021) focused on the gastrointestinal microbial community and the diversity of certain types of microbes.
Conclusion
Over time, the scope of the research on the clinical uses of the gastrointestinal microbiome gradually increased, and the contents were gradually deepened and developed towards a more precise level. The study of the human gastrointestinal microbiome is an ongoing research hotspot and contributes to human health.
Similar content being viewed by others
Background/introduction
It is currently well appreciated that diverse microbial communities reside within the intestinal tract, on the skin, and on nearly all of the exposed surfaces of the human body [1]. The human gastrointestinal (GI) tract harbours the highest density and complexity of microbial organisms in the body [2], and the gastrointestinal microbiota has a level of complexity comparable to that of an organ system [3]. A key role of the gastrointestinal microbiome in the establishment and maintenance of health, as well as in the pathogenesis of diseases, has been identified over the past two decades [4]. In addition, the relationship between the gastrointestinal microbiome and populations with different ages and genders has been gradually revealed [5, 6]. An increasing number of gastrointestinal microbiome detection methods, such as Polymerase Chain Reaction (PCR) or Fluorescence in situ hybridization (FISH), have also emerged [7, 8]. Importantly, the microbiome (including bacteria, viruses, fungi, etc.) regulates health, and its alterations can contribute to disease [9]. A large number of systematic reviews and meta-analyses have shown that gastrointestinal microorganisms are interrelated with inflammatory bowel disease (IBD), irritable bowel syndrome (IBS), diabetes, hepatitis, and autism in humans [10,11,12,13,14]. For example, IBS can disturb the intestinal microecology, which may continue to aggravate IBS. Conversely, the improvement of the intestinal microecology using probiotics and other means may be conducive to the alleviation of the symptoms of IBS [15,16,17]. However, most research on the gastrointestinal microbiome is still in the stage of animal experiments, and the results of animal studies cannot be directly applied to humans. For instance, bidirectional microbiota-gut-brain communication has mostly been explored in animal models with human research lagging [18].
Characterizing the structure of knowledge, the evolution of research topics, and the emergence of topics have always been an important part of information science (IS) [19]. Bibliometric analysis is an important tool in assessing the research activity and research trends on a particular topic, as well as the most prominent research trends, for future research. A knowledge map, visualizing the connections between complex silos of information, is one way to accurately capture and display disparate pieces of information [20]. Moreover, key researchers, countries, and collaboration networks between leading research groups can be identified [21, 22]. However, previous bibliometric analysis or visualized study of the gastrointestinal microbiome did not exclude animal experiments, and the articles related to the human gastrointestinal microbiome were not analyzed separately. Such research could not describe how well the gastrointestinal microbiome works in humans. Therefore, it is necessary to conduct systematic, intuitive, and scientific bibliometric analysis and visualized study of the growing number of original research articles on the human gastrointestinal microbiome.
This study aims to visualize articles on the human gastrointestinal microbiome in the last ten years by using knowledge maps. We described the research hotspots of the human gastrointestinal microbiome and its evolution in the past decade, evaluated the scientific cooperation network, discussed the relationship between humans and gastrointestinal microbiomes, and predicted the field’s future development trend.
Methods
The literature data used in this study were downloaded from the Scopus database, which is widely accepted among researchers conducting high-quality bibliometric analyses [23,24,25,26]. We used “gastrointestinal microbiome” for topical retrieval and the following search queries in titles, abstracts, and keywords: (gastrointestinal AND microbiome), (gut AND microbiota), (gut AND flora), (intestinal AND microbial AND population), (intestinal AND microecology), (enteric AND microorganism), (gut AND microecology), and (intestinal AND microorganism). In addition, the time was defined as “2010–2021” without any language limitation. The above keywords were chosen from a list of Medical Subject Headings (MeSHs) provided by the National Library of Medicine (NLM)/PubMed. The literature type was defined as “articles”. Studies in the subject areas of veterinary, poultry science, soil biology, dentistry, engineering, material science, animal experiments, in vitro cell culture experiments, and secondary studies were excluded.
CiteSpace (Chaomei Chen, China), a freely available software tool for analysis, was used to make visualization maps in this study. Developed by Chaomei Chen in 2004 at Drexel University (USA), CiteSpace is usually used to analyze, detect and visualize trends and patterns in scientific literature [27]. The principle of the software includes coword analysis used to measure the number of occurrences of a group of words (keywords, authors, regions, and citations) in the same group of literature and to perform matrix analysis [28]. In this paper, we use CiteSpace 5.5.R2 to construct country and author co-occurrence map; conduct journal, citation cocitation analysis, and reference co-citation knowledge map; and form a keywords co-occurrence map, a clustering knowledge map, timeline view of keywords, and burst term map.
Results
Distribution of articles by publication years
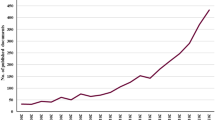
Overall, 4444 documents published from January 2010 to February 2021 were analysed. The number of annual documents during this period showed an exponential growth trend (y = 83.518e0.172x, R2 = 0.4625). The specific numbers of annual documents are shown in Fig. 1.
Scientific cooperation network analysis
In the country co-occurrence knowledge map (Fig. 2), 4444 articles about the human gastrointestinal microbiome were published by research groups in 104 countries or regions. There are 105 nodes and 128 lines, and the centrality is 0.02. The landmark node includes the United States with a count of 1387, China with a count of 756, the United Kingdom with a count of 344, Italy with a count of 311, and Germany (263 texts). The turning points with more connections include Hong Kong, South Africa, the United Kingdom, Switzerland, and Germany. The annual distribution trends in the top five most published countries was shown in Fig. 3. The United States has been leading the way in annual publication.
The country co-occurrence knowledge map of the human gastrointestinal microbiome during 2010–2021. Nodes show in the form of annual rings that the annual ring width represents how many papers the country/region publishes in a given year. The more papers are published, the wider the annual ring is in that year [28]
In the author’s co-occurrence knowledge map (Fig. 4), there are 588 nodes and 1233 lines, and the centrality is 0.007. The landmark nodes include Y Zhang, Y Wang, J Li, J Zhang, and Y Chen. The turning points with more connections include L Wang, L Li, X Yang, Y Chen, and J Wang.
The author co-occurrence knowledge map of the human gastrointestinal microbiome during 2010–2021. The connection between nodes represents the cooperation between authors, and the width of the connection represents the strength of cooperation. The color of the connection represents the author’s first cooperation time. After the network is generated, the author’s cooperation will form several natural clusters. The author cooperation within the cluster is close, and the author cooperation between each class is less [28]
Journal and citation analysis
A total of 1255 journals published the 4444 articles on the human gastrointestinal microbiome. We analyzed the top ten journals that published articles on the human gastrointestinal microbiome (Table 1), reference co-citation knowledge map (Fig. 5) and the details of the top ten articles with the most citations on the human gastrointestinal microbiome in the last ten years (Table 2).
Six of the top 10 journals specialize in microbiology: two are general medicine journals, and the rest are gut and nutrition journals. Most of these journals are of good quality and are European and American journals.
An overview of the 427 top-cited articles among the 1,13,598 articles in the human gastrointestinal microbiome literature and the details of the ten most cited articles on the human gastrointestinal microbiome are summarized in Table 2. The size of a node represents the number of times the corresponding article has been cited in the dataset.
The highly cited articles on the human gastrointestinal microbiome have been published in top journals such as Nature and Science. The topics of these literatures include the relationship between the human gastrointestinal microbiome and diet, diabetes, human health and metabolism, and microbiome analysis methods.
Emerging trends and research focus based on keywords analysis
The articles were imported into CiteSpace, and the keywords were set as nodes. Through a series of software operations, the keyword co-occurrence knowledge map (Fig. 6), clustering knowledge map (Fig. 7), timeline view of keywords (Fig. 8), and keyword burst term map (Fig. 9) were obtained.
The Keyword co-occurrence knowledge map of the human gastrointestinal microbiome during 2010–2021. Each node represents a keyword, and the size of the node represents the frequency of their occurrence, and the line between nodes represents the intensity of co-occurrence, and the color of the line corresponds to the time range at the top of the picture. The frequency of keywords and centrality were listed is in the lower-left corner of the graph [28]
The Keyword clustering knowledge map of the human gastrointestinal microbiome during 2010–2021. CiteSpace uses Log-likelihood rate (LLR) to cluster closely related keywords. Different patterns represent a cluster. Tag # was assigned to the cluster, and the smaller the number is, the more keywords are in the cluster. The size of each cluster, the Silhouette value, and the mean publication year of articles in that cluster are shown in the upper right corner [28]
The timeline view of the human gastrointestinal microbiome during 2010–2021. In the timeline view, the keywords on the same horizontal line belong to the right cluster. The colors of lines and keywords in the view correspond to the colors of the time slice at the top [28]
Top 20 Keywords with the Strongest Citation Bursts of the human gastrointestinal microbiome during 2010–2021. *1 The year in which this keyword first appeared. 2 the bursts’ strength of the keyword. 3 The year in which this keyword begins and ends the burst. 4 Red represents the period during which the keyword is burst
The keyword co-occurrence knowledge map (Fig. 6) contains 165 nodes and 270 lines, and the centrality is 0.02. The landmark nodes that are larger include human, intestinal flora, female, male, adult, and controlled study. They represent the most critical components of the human gastrointestinal microbiome field. The turning points with more connections include adults, microbiology, middle-aged, faecal, RNA 16, child, infant, and Bifidobacterium, indicating that they had higher centrality in the domain and were associated with more keywords.
There were 11 clustering patterns in the research field of the human gastrointestinal microbiome, which are shown in the keyword clustering knowledge map (Fig. 7). In addition, the top 20 most typical labels in each cluster are shown in Table 3.
By combining the keyword clustering knowledge map (Fig. 7), timeline view (Fig. 8), and keyword burst map (Fig. 9), we found the evolutionary path of research hotspots. Amoxicillin treatment, vocabulary related to RCTs, the intestines, the gastrointestinal tract, lactobacillus, Escherichia coli, DNA, probiotics host gene regulation, and the metagenome began to attract attention in the early years (2010–2013). The middle stage (2014–2017) focused on microflora, complications, chemistry, newborns, normal humans, on-chronic liver failure, Japanese female patients, autism spectrum disorder, gastric cancer, and HIV-infected patients. In addition, in recent years (2018–2021), researchers have been interested in metagenomics, microbial communities, microbial diversity, inflammation, and other aspects.
Discussion
The annual number of articles on the human gastrointestinal microbiome shows exponential growth (Fig. 1), indicating that this field is a research hotspot, and its popularity will continue to increase. This is consistent with previous studies [23, 39]. Researchers should give continuous attention to trends in related fields to uncover more connections between humans and the gastrointestinal microbiome. All countries and regions in the world have studied the human gastrointestinal microbiome (Fig. 2). The United States has the most publications, which may be related to the Human Microbiome Project (HMP) programme launched by the NIH in 2007 and the Gut Microbiota Brain AXIS programme in 2013 [40, 41]. The second most published country is China, which may be related to the importance attached to the study of the human microbiome mentioned by the National Natural Science Foundation of China, the 14th Five-Year National Key Research and Development Plan of the Ministry of Science and Technology and the Outline of the 2035 Vision Goals [42,43,44]. Although China is the second largest publishing country, there are no Chinese journals in the top 10 journals, which indicates that China can strengthen its construction of periodicals in this field. The largest collaborator is J Zhang's team from Shanghai Jiao Tong Univ, Sch Life Sci & Biotechnol in China, which focuses on probiotics and intestinal microorganisms. It is suggested that researchers from all countries continue to maintain close cooperation and share the latest research results on the human gastrointestinal microbiome.
The top 10 journals (Table 1) may be given priority when researchers publish and read articles on the human gastrointestinal microbiome because they have published a large number of studies on the human gastrointestinal flora. The reference co-citation knowledge map (Fig. 5) is clearly divided into three clusters according to time (from 2010 to 2013, from 2014 to 2018 and from 2019 to 2021), which indicates that the themes of each research stage are different. Researchers can read highly cited papers (those with large circle areas in Fig. 5) to find research hotspots at that stage. Moreover, beginning researchers can read the highly cited literature (Table 2) to help them understand the important findings in the field.
Compared with previous bibliometrics and visualized studies on the gastrointestinal microbiome that did not exclude animal studies, these studies on the human gastrointestinal microbiome mainly focus on the following aspects [23, 39]. As shown in the keyword co-occurrence knowledge map (Fig. 6), the larger landmark nodes can be divided into three categories: population, research methods, and detection methods. A large number of studies have been conducted on the relationship between age (newborn [45], infant [46], child [47], teenager [48], adult [49], middle aged [50] and aged [51]), gender (male [52] and female [53]) and population groups and gastrointestinal microorganisms. This may be related to researchers finding that the gastrointestinal floras of different populations are significantly different, which requires classification to further study the topic. Additionally, various research methods have been used to study this field. Experimental studies often include the effects of probiotics [54], faecal microbiota transplantation [55], Chinese medicine [56], and antibiotic therapy [57, 58] on the human gastrointestinal microbiome. Observational studies often include interactions between the gastrointestinal microbiome and various human diseases such as obesity [59], diabetes [60], and irritable bowel syndrome [61]. In addition, there are a variety of molecular biology technologies that have been used for gastrointestinal microbiome research. These technologies mainly include the following methods: bacterial culture technology based on molecular biotechnology, polymerase chain reaction (PCR), fluorescent in situ hybridization (FISH) [8], gene chips [62], and metagenome sequencing [63], and it is more popular to establish a gene bank of the gastrointestinal microbiome [64]. Each technology has advantages and disadvantages, and researchers can select the technologies suitable for their purposes.
As for research emphases, researchers’ exploration of the human gastrointestinal microbiome from 2010 to 2013 was at a relatively macro and superficial stage. Researchers have sought to determine how the gastrointestinal microbiome relates to humans. In 2011, one study combined 22 newly sequenced faecal metagenomes of individuals from four countries with previously published data sets to identify three robust clusters (referred to as enterotypes), which attracted intense attention at the time [32]. But since then, the discussion of enterotype has become less and less popular. Every year, a small number of studies look at the relationship between human enterotype and diet [33, 65], feces [66], human population [67], obesity [68], etc. Some researchers have suggested that grouping the microbiota of individual subjects into enterotypes, based on the dominance of certain genera may have oversimplified a complex situation [69]. Researchers have also explored the link between human gastrointestinal flora and certain diseases (such as type 2 diabetes, autism, obesity, irritable bowel syndrome, etc.) at this stage [70,71,72] and the relationship between Lactobacillus and human gastrointestinal flora [73,74,75]. In addition, a series of randomized controlled trials on the human gastrointestinal flora began to emerge at this stage [76,77,78,79,80]. The effect of antibiotics on human gastrointestinal flora was also a research hotspot during this period [81, 82].
From 2014 to 2017, increasingly more studies were conducted to determine the interaction between human gastrointestinal flora and various organs and systems (such as liver cirrhosis, Parkinson’s disease, rheumatoid arthritis, etc.) [83,84,85]. For instance, Francesco’s research suggested that the gut mycobiota contributed to the alteration of the intestinal microbial community structure in ASDs, which made it possible to develop new potential intervention strategies aimed at the relief of gastrointestinal symptoms in ASDs [86]. Dillon’s study suggested that an important relationship existed between altered mucosal bacterial communities and intestinal inflammation during chronic HIV-1 infection [87]. Moreover, the relationship between diet and human gastrointestinal flora is a research hotspot during this period [30, 88, 89].
From 2018 to 2021, researchers shifted their focus from certain types of gastrointestinal bacteria to the gastrointestinal microbial community [90] and microbial diversity [90]. Studies [91, 92] on the effects of the gastrointestinal microbiome and its metabolites on inflammation [93] and immunology [94] and their application as biomarkers [95] at the molecular level have also gradually become hotspots.
It is worth noting that researchers are often inspired by animal experiments to explore the mechanisms of diseases caused by the gastrointestinal microbiome and modify the gastrointestinal microbiome to treat disease; then, they judge the feasibility and safety of treatment methods [96]. However, human research has lagged behind animal models, and applying the results of animal experiments to humans requires more rigorous experiments and theories [18, 97]. At present, theories such as microbiota-gut-brain communication [98], gut-lung axis [99, 100], and enterohepatic circulation [101] can explain parts of the relationship between the gastrointestinal microbiome and various human organs and systems, but it is not yet completely clear. With the development of new technologies, such as omics and sequencing, the detection of the gastrointestinal microbiome has become more accurate. Big data also makes it possible to conduct comprehensive artificial intelligence research on multicentre, multidisease, and human gastrointestinal microbiome databases. Future research needs to be based on previous research results, combined with emerging technologies, and explore the relationship between gastrointestinal flora and humans at the molecular mechanism level to improve health.
There are some limitations to this study. In this study, only articles in the Scopus database were retrieved. Although using the Scopus database to conduct high-quality bibliometric analyses is widely accepted by researchers, it is still possible that some studies related to the human gastrointestinal microbiome have not been included, which may change the results of the study. This study examined only the last ten years of research on the human gastrointestinal microbiome, which may miss the development process of the human gastrointestinal microbiome from the start.
Conclusion
In this study, 4444 original studies from January 2010 to February 2021 related to the human gastrointestinal microbiome were downloaded from the Scopus database and analyzed using CiteSpace to generate knowledge maps. The number of articles on the human gastrointestinal microbiome has increased rapidly in the past decade, and the scientific cooperation network showed that cooperation between different countries and institutions has been sufficient. The research topics focus on different populations, research methods, and detection methods. In addition, the research scope has gradually increased over time, and the research content has been gradually deeper and moving towards precision medicine. In short, the study of the human gastrointestinal microbiome is an ongoing research hotspot and contributes to human health.
Availability of data and materials
Not applicable.
References
Brown JM, Hazen SL. Microbial modulation of cardiovascular disease. Nat Rev Microbiol. 2018;16:171–81.
Li M, Gao J, Tang Y, Liu M, Wu S, Qu K, Long X, Li H, Liu M, Liu Y, et al. Traditional herbal medicine-derived sulforaphene LFS-01 reverses colitis in mice by selectively altering the gut microbiota and promoting intestinal gamma–delta T cells. Front Pharmacol. 2017;8:959.
Liu H, Wang HH. Impact of microbiota transplant on resistome of gut microbiota in gnotobiotic piglets and human subjects. Front Microbiol. 2020;11:932.
Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17:223–37.
Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, Mele MC. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 2019;7:14.
Kim YS, Unno T, Kim BY, Park MS. Sex differences in gut microbiota. World J Mens Health. 2020;38:48–60.
Durk RP, Castillo E, Márquez-Magaña L, Grosicki GJ, Bolter ND, Lee CM, Bagley JR. Gut microbiota composition is related to cardiorespiratory fitness in healthy young adults. Int J Sport Nutr Exerc Metab. 2019;29:249–53.
Conterno L, Martinelli F, Tamburini M, Fava F, Mancini A, Sordo M, Pindo M, Martens S, Masuero D, Vrhovsek U, et al. Measuring the impact of olive pomace enriched biscuits on the gut microbiota and its metabolic activity in mildly hypercholesterolaemic subjects. Eur J Nutr. 2019;58:63–81.
Song M, Chan AT, Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology. 2020;158:322–40.
Pittayanon R, Lau JT, Leontiadis GI, Tse F, Yuan Y, Surette M, Moayyedi P. Differences in gut microbiota in patients with vs without inflammatory bowel diseases: a systematic review. Gastroenterology. 2020;158:930–46.
Pittayanon R, Lau JT, Yuan Y, Leontiadis GI, Tse F, Surette M, Moayyedi P. Gut microbiota in patients with irritable bowel syndrome—a systematic review. Gastroenterology. 2019;157:97–108.
Liu F, Li J, Wu F, Zheng H, Peng Q, Zhou H. Altered composition and function of intestinal microbiota in autism spectrum disorders: a systematic review. Transl Psychiatry. 2019;9:43.
Jamshidi P, Hasanzadeh S, Tahvildari A, Farsi Y, Arbabi M, Mota JF, Sechi LA, Nasiri MJ. Is there any association between gut microbiota and type 1 diabetes? A systematic review. Gut Pathog. 2019;11:49.
Sharpton SR, Maraj B, Harding-Theobald E, Vittinghoff E, Terrault NA. Gut microbiome-targeted therapies in nonalcoholic fatty liver disease: a systematic review, meta-analysis, and meta-regression. Am J Clin Nutr. 2019;110:139–49.
Duan R, Zhu S, Wang B, Duan L. Alterations of gut microbiota in patients with irritable bowel syndrome based on 16S rRNA-targeted sequencing: a systematic review. Clin Transl Gastroenterol. 2019;10:e12.
El-Salhy M, Hatlebakk JG, Hausken T. Diet in irritable bowel syndrome (ibs): interaction with gut microbiota and gut hormones. Nutrients. 2019;11:1824.
Wang Z, Xu C, Liu Y, Wang X, Zhang L, Li M, Zhu S, Xie Z, Wang P, Duan L, Zhu H. Characteristic dysbiosis of gut microbiota of Chinese patients with diarrhea-predominant irritable bowel syndrome by an insight into the pan-microbiome. Chinese Med J. 2019;132:889–904.
Valles-Colomer M, Falony G, Darzi Y, Tigchelaar EF, Wang J, Tito RY, Schiweck C, Kurilshikov A, Joossens M, Wijmenga C, et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat Microbiol. 2019;4:623–32.
Hou J, Yang X, Chen C. Emerging trends and new developments in information science: a document co-citation analysis (2009–2016). Scientometrics. 2018;115:869–92.
Chen C. Searching for intellectual turning points: progressive knowledge domain visualization. Proc Natl Acad Sci USA. 2004;101(Suppl 1):5303–10.
Sun J, Zhou Z, Huang J, Li G. A bibliometric analysis of the impacts of air pollution on children. Int J Environ Res Public Health. 2020;17:1277.
Aleixandre-Tudó JL, Castelló-Cogollos L, Aleixandre JL, Aleixandre-Benavent R. Tendencies and challenges in worldwide scientific research on probiotics. Probiotics Antimicrob Proteins. 2020;12:785–97.
Yue Y, Fan X, Zhang Q, Lu Y, Wu S, Wang S, Yu M, Cui C, Sun Z. Bibliometric analysis of subject trends and knowledge structures of gut microbiota. World J Clin Cases. 2020;8:2817–32.
Pathirana MM, Lassi ZS, Roberts CT, Andraweera PH. Cardiovascular risk factors in offspring exposed to gestational diabetes mellitus in utero: systematic review and meta-analysis. J Dev Orig Health Dis. 2020;11:599–616.
Xiong J, Lipsitz O, Nasri F, Lui LMW, Gill H, Phan L, Chen-Li D, Iacobucci M, Ho R, Majeed A, McIntyre RS. Impact of COVID-19 pandemic on mental health in the general population: a systematic review. J Affect Disord. 2020;277:55–64.
Shields GS, Spahr CM, Slavich GM. Psychosocial interventions and immune system function: a systematic review and meta-analysis of randomized clinical trials. JAMA Psychiatry. 2020;77:1031–43.
Pan X, Yan E, Cui M, Hua W. Examining the usage, citation, and diffusion patterns of bibliometric mapping software: a comparative study of three tools. J Informetr. 2018;12:481–93.
Chaomei C. How to use CiteSpace. 2020. https://leanpub.com/howtousecitespace. Accessed 15 Jan 2021.
Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7.
David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63.
Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60.
Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende D, Fernandes G, Tap J, Bruls T, Batto J, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80.
Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8.
Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto J, Kennedy S, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–6.
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65.
Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–21.
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3.
Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–4.
Huang X, Fan X, Ying J, Chen S. Emerging trends and research foci in gastrointestinal microbiome. J Transl Med. 2019;17:67.
Integrative HMP (iHMP) Research network consortium. The integrative human microbiome project. Nature. 2019;569:641–8.
Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–10.
The Central People’s Government of the People’s Republic of China: The Fourteenth Five-Year Plan for the National Economic and Social Development of the People’s Republic of China and the Outline of the Long-range Goals to 2035. http://www.gov.cn. Accessed 12 May 2021.
National Natural science Foundation of China: National Natural Science Foundation Project Guide for 2021. http://www.nsfc.gov.cn/. Accessed 10 May 2021.
Guidelines for the application of the key special project of biomacromolecules and microbiome in National Policy Fund Advisory Network: 2021 under the 14th Five-Year National Key Research and Development Program. http://chinazx.org.cn. Accessed 12 May 2021.
Zanella A, Silveira RC, Roesch LFW, Corso AL, Dobbler PT, Mai V, Procianoy RS. Influence of own mother’s milk and different proportions of formula on intestinal microbiota of very preterm newborns. PLoS One. 2019;14:e217296.
Gasparrini AJ, Wang B, Sun X, Kennedy EA, Hernandez-Leyva A, Ndao IM, Tarr PI, Warner BB, Dantas G. Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nat Microbiol. 2019;4:2285–97.
Xi W, Gao X, Zhao H, Luo X, Li J, Tan X, Wang L, Zhao J, Wang J, Yang G, et al. Depicting the composition of gut microbiota in children with tic disorders: an exploratory study. J Child Psychol Psychiatry. 2021. https://doi.org/10.1111/jcpp.13409.
Jobira B, Frank DN, Pyle L, Silveira LJ, Kelsey MM, Garcia-Reyes Y, Robertson CE, Ir D, Nadeau KJ, Cree-Green M. Obese adolescents with PCOS have altered biodiversity and relative abundance in gastrointestinal microbiota. J Clin Endocrinol Metab. 2020;105:e2134–44.
Charoenngam N, Shirvani A, Kalajian TA, Song A, Holick MF. The effect of various doses of oral vitamin D(3) supplementation on gut microbiota in healthy adults: a randomized, double-blinded dose-response study. Anticancer Res. 2020;40:551–6.
Boehme M, van de Wouw M, Bastiaanssen TFS, Olavarría-Ramírez L, Lyons K, Fouhy F, Golubeva AV, Moloney GM, Minuto C, Sandhu KV, et al. Mid-life microbiota crises: middle age is associated with pervasive neuroimmune alterations that are reversed by targeting the gut microbiome. Mol Psychiatry. 2020;25:2567–83.
Ghosh TS, Rampelli S, Jeffery IB, Santoro A, Neto M, Capri M, Giampieri E, Jennings A, Candela M, Turroni S, et al. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: the NU-AGE 1-year dietary intervention across five European countries. Gut. 2020;69:1218–28.
Zeb F, Wu X, Chen L, Fatima S, Haq I, Chen A, Majeed F, Feng Q, Li M. Effect of time-restricted feeding on metabolic risk and circadian rhythm associated with gut microbiome in healthy males. Br J Nutr. 2020;123:1216–26.
Al Assal K, Prifti E, Belda E, Sala P, Clément K, Dao M, Doré J, Levenez F, Taddei CR, Fonseca DC, et al. Gut microbiota profile of obese diabetic women submitted to roux-en-Y gastric bypass and its association with food intake and postoperative diabetes remission. Nutrients. 2020;12:278.
Kakiuchi T, Mizoe A, Yamamoto K, Imamura I, Hashiguchi K, Kawakubo H, Yamaguchi D, Fujioka Y, Nakayama A, Okuda M, Matsuo M. Effect of probiotics during vonoprazan-containing triple therapy on gut microbiota in Helicobacter pylori infection: a randomized controlled trial. Helicobacter. 2020;25:e12690.
Craven L, Rahman A, Nair Parvathy S, Beaton M, Silverman J, Qumosani K, Hramiak I, Hegele R, Joy T, Meddings J, et al. Allogenic fecal microbiota transplantation in patients with nonalcoholic fatty liver disease improves abnormal small intestinal permeability: a randomized control trial. Am J Gastroenterol. 2020;115:1055–65.
Cui H, Zhang L, Luo Y, Yuan K, Huang Z, Guo Y. A purified anthraquinone-glycoside preparation from rhubarb ameliorates type 2 diabetes mellitus by modulating the gut microbiota and reducing inflammation. Front Microbiol. 2019;10:1423.
Liou J, Chen C, Chang C, Fang Y, Bair M, Chen P, Chang C, Hsu Y, Chen M, Chen C, et al. Long-term changes of gut microbiota, antibiotic resistance, and metabolic parameters after Helicobacter pylori eradication: a multicentre, open-label, randomised trial. Lancet Infect Dis. 2019;19:1109–20.
Fodor AA, Pimentel M, Chey WD, Lembo A, Golden PL, Israel RJ, Carroll IM. Rifaximin is associated with modest, transient decreases in multiple taxa in the gut microbiota of patients with diarrhoea-predominant irritable bowel syndrome. Gut Microbes. 2019;10:22–33.
de la Cuesta-Zuluaga J, Mueller NT, Álvarez-Quintero R, Velásquez-Mejía EP, Sierra JA, Corrales-Agudelo V, Carmona JA, Abad JM, Escobar JS. Higher fecal short-chain fatty acid levels are associated with gut microbiome dysbiosis, obesity, hypertension and cardiometabolic disease risk factors. Nutrients. 2018;11:51.
Harbison JE, Roth-Schulze AJ, Giles LC, Tran CD, Ngui KM, Penno MA, Thomson RL, Wentworth JM, Colman PG, Craig ME, et al. Gut microbiome dysbiosis and increased intestinal permeability in children with islet autoimmunity and type 1 diabetes: a prospective cohort study. Pediatr Diabetes. 2019;20:574–83.
Berentsen B, Nagaraja BH, Teige EP, Lied GA, Lundervold AJ, Lundervold K, Steinsvik EK, Hillestad ER, Valeur J, Brønstad I, et al. Study protocol of the Bergen brain-gut-microbiota-axis study: a prospective case-report characterization and dietary intervention study to evaluate the effects of microbiota alterations on cognition and anatomical and functional brain connectivity in patients with irritable bowel syndrome. Medicine. 2020;99:e21950.
Beaumont M, Portune KJ, Steuer N, Lan A, Cerrudo V, Audebert M, Dumont F, Mancano G, Khodorova N, Andriamihaja M, et al. Quantity and source of dietary protein influence metabolite production by gut microbiota and rectal mucosa gene expression: a randomized, parallel, double-blind trial in overweight humans. Am J Clin Nutr. 2017;106:1005–19.
Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, Vatanen T, Hall AB, Mallick H, McIver LJ, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4:293–305.
Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, Lawley TD, Finn RD. A new genomic blueprint of the human gut microbiota. Nature. 2019;568:499–504.
Roager HM, Licht TR, Poulsen SK, Larsen TM, Bahl MI. Microbial enterotypes, inferred by the prevotella-to-bacteroides ratio, remained stable during a 6-month randomized controlled diet intervention with the new nordic diet. Appl Environ Microb. 2014;80:1142–9.
Vandeputte D, Falony G, Vieira-Silva S, Tito RY, Joossens M, Raes J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2016;65:57–62.
Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. Plos Comput Biol. 2013;9:e1002863.
Christensen L, Roager HM, Astrup A, Hjorth MF. Microbial enterotypes in personalized nutrition and obesity management. Am J Clin Nutr. 2018;108:645–51.
Jeffery IB, Claesson MJ, O’Toole PW, Shanahan F. Categorization of the gut microbiota: enterotypes or gradients? Nat Rev Microbiol. 2012;10:591–2.
Larsen N, Vogensen FK, van den Berg FWJ, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sørensen SJ, Hansen LH, Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. Plos One. 2010;5:e9085.
Adams JB, Johansen LJ, Powell LD, Quig D, Rubin RA. Gastrointestinal flora and gastrointestinal status in children with autism–comparisons to typical children and correlation with autism severity. Bmc Gastroenterol. 2011;11:22.
Million M, Maraninchi M, Henry M, Armougom F, Richet H, Carrieri P, Valero R, Raccah D, Vialettes B, Raoult D. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int J Obes. 2012;36:817–25.
Fallani M, Young D, Scott J, Norin E, Amarri S, Adam R, Aguilera M, Khanna S, Gil A, Edwards CA, Doré J. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastr Nutr. 2010;51:77–84.
Karlsson CLJ, Onnerfält J, Xu J, Molin G, Ahrné S, Thorngren-Jerneck K. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity. 2012;20:2257–61.
Bervoets L, Van Hoorenbeeck K, Kortleven I, Van Noten C, Hens N, Vael C, Goossens H, Desager KN, Vankerckhoven V. Differences in gut microbiota composition between obese and lean children: a cross-sectional study. Gut Pathog. 2013;5:10.
Zimmermann MB, Chassard C, Rohner F, N’goran EK, Nindjin C, Dostal A, Utzinger J, Ghattas H, Lacroix C, Hurrell RF. The effects of iron fortification on the gut microbiota in African children: a randomized controlled trial in Cote d’Ivoire. Am J Clin Nutr. 2010;92:1406–15.
Pärtty A, Luoto R, Kalliomäki M, Salminen S, Isolauri E. Effects of early prebiotic and probiotic supplementation on development of gut microbiota and fussing and crying in preterm infants: a randomized, double-blind, placebo-controlled trial. J Pediatr. 2013;163:1272–7.
Michail S, Kenche H. Gut microbiota is not modified by randomized, double-blind, placebo-controlled trial of VSL#3 in diarrhea-predominant irritable bowel syndrome. Probiotics Antimicrob Proteins. 2011;3:1–7.
Leong KSW, Jayasinghe TN, Wilson BC, Derraik JGB, Albert BB, Chiavaroli V, Svirskis DM, Beck KL, Conlon CA, Jiang Y, et al. Effects of fecal microbiome transfer in adolescents with obesity: the gut bugs randomized controlled trial. JAMA Netw Open. 2020;3:e2030415.
Søndergaard B, Olsson J, Ohlson K, Svensson U, Bytzer P, Ekesbo R. Effects of probiotic fermented milk on symptoms and intestinal flora in patients with irritable bowel syndrome: a randomized, placebo-controlled trial. Scand J Gastroenterol. 2011;46:663–72.
Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4554–61.
Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, Murphy B, Ross RP, Fitzgerald GF, Stanton C, Cotter PD. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother. 2012;56:5811–20.
Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64.
Scheperjans F, Aho V, Pereira PAB, Koskinen K, Paulin L, Pekkonen E, Haapaniemi E, Kaakkola S, Eerola-Rautio J, Pohja M, et al. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov Disord. 2015;30:350–8.
Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21:895–905.
Strati F, Cavalieri D, Albanese D, De Felice C, Donati C, Hayek J, Jousson O, Leoncini S, Renzi D, Calabrò A, De Filippo C. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome. 2017;5:24.
Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, Gianella S, Siewe B, Smith DM, Landay AL, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7:983–94.
Suez J, Korem T, Zeevi D, Zilberman-Schapira G, Thaiss CA, Maza O, Israeli D, Zmora N, Gilad S, Weinberger A, et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014;514:181–6.
Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. A Dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167:1339–53.
Venturelli OS, Carr AC, Fisher G, Hsu RH, Lau R, Bowen BP, Hromada S, Northen T, Arkin AP. Deciphering microbial interactions in synthetic human gut microbiome communities. Mol Syst Biol. 2018;14:e8157.
Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vich Vila A, Võsa U, Mujagic Z, Masclee AAM, Jonkers DMAE, Oosting M, et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. 2019;51:600–5.
Strandwitz P, Kim KH, Terekhova D, Liu JK, Sharma A, Levering J, McDonald D, Dietrich D, Ramadhar TR, Lekbua A, et al. GABA-modulating bacteria of the human gut microbiota. Nat Microbiol. 2019;4:396–403.
Ponziani FR, Bhoori S, Castelli C, Putignani L, Rivoltini L, Del Chierico F, Sanguinetti M, Morelli D, Paroni Sterbini F, Petito V, et al. Hepatocellular carcinoma is associated with gut microbiota profile and inflammation in nonalcoholic fatty liver disease. Hepatology. 2019;69:107–20.
Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 2018;33:570–80.
Shen Y, Xu J, Li Z, Huang Y, Yuan Y, Wang J, Zhang M, Hu S, Liang Y. Analysis of gut microbiota diversity and auxiliary diagnosis as a biomarker in patients with schizophrenia: a cross-sectional study. Schizophr Res. 2018;197:470–7.
Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103.
Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, Kotler E, Zur M, Regev-Lehavi D, Brik RB, et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. 2018;174:1388–405.
Cryan JF, O’Riordan KJ, Cowan CSM, Sandhu KV, Bastiaanssen TFS, Boehme M, Codagnone MG, Cussotto S, Fulling C, Golubeva AV, et al. The Microbiota–gut–brain axis. Physiol Rev. 2019;99:1877–2013.
Bingula R, Filaire M, Radosevic-Robin N, Berthon J, Bernalier-Donadille A, Vasson M, Thivat E, Kwiatkowski F, Filaire E. Characterisation of gut, lung, and upper airways microbiota in patients with non-small cell lung carcinoma: study protocol for case-control observational trial. Medicine. 2018;97:e13676.
Bingula R, Filaire M, Radosevic-Robin N, Bey M, Berthon J, Bernalier-Donadille A, Vasson M, Filaire E. Desired turbulence? Gut–lung axis, immunity, and lung cancer. J Oncol. 2017;2017:5035371.
Le Roy T, Lécuyer E, Chassaing B, Rhimi M, Lhomme M, Boudebbouze S, Ichou F, Haro Barceló J, Huby T, Guerin M, et al. The intestinal microbiota regulates host cholesterol homeostasis. Bmc Biol. 2019;17:94.
Acknowledgements
Authors would like to thank West China School of Nursing, Sichuan University/West China Hospital, Sichuan University and Sichuan University Library for supporting the work.
Funding
This work was supported by the Key research and development project of Science & Technology Department of Sichuan Province (No. 2021YFS0022). The funder had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Guarantor of the article: LK. LK conceived the study and performed critical revision of manuscript. YX designed the study, performed statistical analyses and drafted the manuscript. CC designed the study and wrote the manuscript. CX performed the article retrieval, data interpretation and provided supervision. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors agree publication.
Competing interests
We declare that all authors have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yuan, X., Chang, C., Chen, X. et al. Emerging trends and focus of human gastrointestinal microbiome research from 2010–2021: a visualized study. J Transl Med 19, 327 (2021). https://doi.org/10.1186/s12967-021-03009-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-021-03009-8