Abstract
N6-methyladenosine (m6A) is one of the most common RNA modifications in eukaryotes, mainly in messenger RNA (mRNA). Increasing evidence shows that m6A methylation modification acts an essential role in various physiological and pathological bioprocesses. Noncoding RNAs (ncRNAs), including miRNAs, lncRNAs and circRNAs, are known to participate in regulating cell differentiation, angiogenesis, immune response, inflammatory response and carcinogenesis. m6A regulators, such as METTL3, ALKBH5 and IGF2BP1 have been reported to execute a m6A-dependent modification of ncRNAs involved in carcinogenesis. Meanwhile, ncRNAs can target or modulate m6A regulators to influence cancer development. In this review, we provide an insight into the interplay between m6A modification and ncRNAs in cancer.
Similar content being viewed by others
Introduction
Up to now, more than 100 kinds of RNA modifications have been confirmed [1]. Among them, m6A RNA methylation is one of the most thoroughly studied modifications. m6A RNA modification occurs by methylation of the sixth N atom of adenine (A) in mRNAs or ncRNAs [2]. m6A modification sites tend to be found in the stop codons and 3′-Untranslated region (3′-UTR) of mRNA with a typical consensus sequence RRACH (R = G or A and H = A, C, or U) [3, 4]. Accumulating data show that m6A RNA methylation acts by modulating circadian rhythm, gene expression, cell differentiation, stress response, inflammatory response, and carcinogenesis [5,6,7,8,9,10]. According to the global cancer statistics, there were estimated 18.1 million new cases and 9.6 million deaths in 2018 [11]. Recent studies have shown that m6A modification acts a vital role in the diagnosis, treatment and prognosis of cancer patients as well as in carcinogenesis. It also regulates fly sex, virus genome, meiosis of yeast, tissue differentiation, germination, and collateral generation of Arabidopsis [12,13,14,15].
Noncoding RNAs (ncRNAs) including microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) act pivotal roles in cancer [16,17,18]. m6A modification can affect ncRNA splicing and maturation involved in carcinogenesis (Table 1). In this review, we summarize the latest progress about the interplay between m6A modification and ncRNAs in cancer.
Molecular compositions of m6A RNA methylation
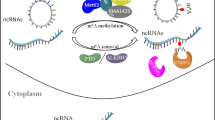
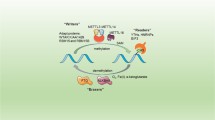
Molecular compositions of m6A RNA methylation include m6A methyltransferase, m6A demethylase, and m6A recognition factors (Fig. 1). m6A methyltransferases, called “writers” contain methyltransferase-like 3 (METTL3) [19], METTL14 [20], Wilms tumor 1-associated protein (WTAP) [2], KIAA1429 [21], METTL16 [22] and RNA-binding motif protein 15/15B (RBM15/15B) [23]. METTL3 regulates the circadian clock of hepatic lipid metabolism and hematopoiesis [24, 25]. METTL3/14 depletion promotes myeloid differentiation and suppresses the progression of acute myeloid leukemia (AML) [26, 27]. METTL16 maintains the levels of methyl donor S-adenosylmethionine (SAM) [28]. WTAP connects METTL3/14 to form a complex, anchored to the nucleus to catalyze m6A methyltransferase [2].
m6A methylation is dynamic and can be reversed by m6A demethylase, also named as m6A “erasers”, containing fat mass and obesity-associated protein (FTO) and alkB homologue 5 (ALKBH5) [29, 30]. FTO shares the motifs with Fe (II)- and 2-oxoglutarate-dependent oxygenase and is related to increased fat mass [31]. FTO harbors an efficient oxidative demethylation activity and reduces the m6A levels of mRNAs [30]. ALKBH5 is responsible for RNA splicing and stability and causes the degradation of abnormal transcripts in spermatocytes and round spermatids [32].
m6A recognition factors, known as “readers,” consist of YT521-B homology (YTH) domain family (YTHDF1/2/3) [33], YTH domain-containing proteins (YTHDC1/2) [12], heterogeneous nuclear ribonucleoprotein (HNRNP) protein families [33], eukaryotic translation initiation factor 3 (eIF3) [23], and insulin-like growth factor-2 mRNA-binding proteins 1/2/3 (IGF2BP1/2/3) [34]. m6A recognition factors act in oligodendrocyte progenitor cells and oligodendrocyte fate [35]. YTHDF1 controls pre-crossing axon guidance in the spinal cord by regulating m6A-modified Robo3.1 [36]. HNRNPA2B1 can initiate the immune response to DNA viruses by regulating interferon-α/β and stimulator of interferon genes (STING)-dependent antiviral signaling [37].
m6A modification of miRNAs in cancer
As is known to us, the dysregulation of miRNAs is involved in various bio-behaviors, such as mouse prenatal development, immune response, inflammatory response and carcinogenesis [38,39,40,41]. METTL3 or HNRNPA2B1 facilitates pri-miRNA processing by recruiting RNA-binding protein DiGeorge syndrome critical region 8 (DGCR8) [42, 43]. METTL3 suppresses osteogenic processes by promoting the maturation of miR-7212-5p and downregulating its target fibroblast growth factor receptor 3 (FGFR3) [44].
Tumor proliferation and tumorigenesis
m6A methylation can modify the maturation of miRNAs involved in cell proliferation and tumorigenesis (Fig. 2). miR-25-3p acts as a pivotal role in pancreatic ductal adenocarcinoma (PDAC). Cigarette smoke condensate (CSC) mediates METTL3 to promote miR-25-3p maturation in PDAC tumorigenesis [45]. METTL3 also enhances the binding of pri-miR-221/222 with DGCR8 involved in the proliferation of bladder cancer [46]. m6A modification affects arsenite-induced carcinogenesis via modifying multiple miRNAs (miR-106b, miR-18a/b, miR-3607, miR-423, miR-30a, miR-320b/d/e) [47].
Tumor invasion and metastasis
METTL14 promotes the maturation of pri-miR-126 and suppresses the invasion and metastasis of hepatocellular carcinoma (HCC) [48]. METTL3 facilitates the maturation of pri-miR-1246 to enhance the metastasis of colorectal cancer (CRC) [49]. METTL3 also accelerates the maturation of miR-143-3p, leading to the formation of METTL3/miR-143-3p/vasohibin-1 axis to favor the metastasis of lung cancers [50].
m6A modification of lncRNAs in cancer
LncRNAs, a subgroup of non-coding RNAs over 200 nucleotides in length can be modified by m6A methylation in cancer (Fig. 3). m6A methylation facilitates lncRNA X-inactive specific transcript (XIST)-mediated transcriptional repression [51,52,53]. YTHDC1 preferentially recognizes the m6A residues of XIST and RBM15/15B and participates in XIST-mediated gene silencing [53]. However, RBM15/m6A-MTase complex is reported to act a minor role in XIST-mediated gene silencing [54]. YTHDF2 recognizes m6A methylation site of lnc-Dpf3 to promote its degradation and enhances the binding of lnc-Dpf3 with hypoxia-inducible factor 1-alpha (HIF-1α), leading to the suppression of the glycolysis and migration of dendritic cells [55]. METTL3 can modify metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) to form the METTL3/MALAT1/miR-145/focal adhesion kinase (FAK) axis, contributing to the aggravation of renal fibrogenesis in obstructive nephropathy [56].
Tumor proliferation and tumorigenesis
ALKBH5 has been found upregulated in glioblastoma and prompts the proliferation of glioblastoma stem-like cells (GSCs). A lncRNA antisense to forkhead box M1 (FOXM1-AS) promotes the interaction of ALKBH5 with forkhead box M1 (FOXM1) nascent transcripts to increase FOXM1 expression and GSCs tumorigenesis [57]. LncRNA Differentiation antagonizing non-protein coding RNA (DANCR) contributes to the tumorigenesis of multiple cancers [58, 59]. IGF2BP2 serves as an m6A reader to modify DANCR and favors the oncogenicity of pancreatic cancer [60]. MALAT1, the first lncRNA to be found associated with lung cancer, possesses a triple helix structure at its 3’end [61,62,63]. METTL16 interacts directly with MALAT1 triple helix and promotes cancer cell proliferation [64].
Tumor invasion and metastasis
Long non-coding RNA 00958 (LINC00958) is upregulated by METTL3 and facilitates HCC cell migration and invasion by sponging miR-3619-5p [65]. METTL3/14 enhance the migration of head and neck squamous cell carcinoma (HNSCC) by upregulating lncRNA activating regulator of DKK1 (LNCAROD) [66]. METTL3-family with sequence similarity 225 member A (FAM225A)-integrin β3 (ITGB3)-FAK/PI3K/Akt axis facilitates the metastasis of nasopharyngeal cancer [67]. METTL3 mediates lncRNA RP11–138 J23.1 (RP11) or MALAT1-miR-1914-3p-Yes associated protein (YAP) axis to enhance the migration and invasion of CRC and non-small cell lung cancer (NSCLC) [68, 69]. METTL14 increases the m6A levels of XIST and suppresses the invasion of CRC [70]. ALKBH5 favors the invasion and metastasis of gastric cancer (GC) by demethylating lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) [71]. YTHDF1 restrains esophageal squamous cell carcinoma (ESCC) by interacting with long intergenic non-protein coding RNA 278 (LINC00278), but ALKBH5 harbors an opposite function [72].
m6A modification of circRNAs in cancer
CircRNAs, a novel subset of ncRNAs generated by back-splicing, play a crucial role in protein translation [73]. METTL3 and YTHDC1 are associated with the metabolism of circular RNA zinc finger protein 609 (circ-ZNF609) and promote its production [74]. Minigenes of ribosomes-circRNAs (Ribo-circRNAs) can facilitate protein translation in drosophila heads and circ-ZNF609 boosts protein translation and myoblasts cell proliferation [75, 76]. m6A methylation has been reported to affect protein translation of cricRNAs [77, 78]. m6A motifs are enriched in circRNAs, and a single m6A site is regarded as a trigger to initiate the translation of circRNAs. m6A regulators METTL3/14, FTO, YTHDF3, and initiation factor eIF4G2 are involved in m6A-driven protein translation [78]. Mammalian cells can recognize the m6A modification on circRNAs to inhibit innate immunity by abrogating immune gene activation and adjuvant activity [79].
In addition, the dysregulation of circRNAs is associated with the progression of multiple cancers, such as breast cancer, gastric cancer (GC), gallbladder cancer and cervical cancer [80,81,82,83]. YTHDC1 interacts with circRNA NOP2/Sun RNA methyltransferase 2 (circNSUN2) to facilitate its cytoplasmic export, which leads to colorectal liver metastasis by forming a circNUSN2/IGF2BP2/high mobility group AT-hook 2 (HMGA2) RNA-protein ternary complex in the cytoplasm [84]. m6A modification can be involved in the progression of GC by regulating circRNA poliovirus receptor-related 3 (circPVRL3) [85].
m6A regulators are regulated by ncRNAs in cancer
NcRNAs have the capabilities to affect m6A levels involved in multiple biological processes (Table 2). miRNAs can modulate the binding between METTL3 and its target mRNAs to participate in the reprogramming efficiency of mouse embryonic fibroblasts (MEFs) [86]. miR-149-3p inhibits adipogenesis lineage differentiation and potentiates osteogenic lineage differentiation by targeting FTO [87]. miR-1266 inhibits CRC progression by targeting FTO [88]. miR-145 suppresses the proliferation of HCC by targeting YTHDF2 [89]. Similarly, miR-33a and miR-448 suppress the proliferation of NSCLC by targeting METTL3 and eIF3a [90, 91]. METTL3 is also downregulated by miR-600, which induces the apoptosis of lung cancer [92]. miR-141 suppresses the proliferation of pancreatic cancer by forming the miR-141/IGF2BP2/P13K/Akt axis [93]. Hepatitis B X-interacting protein (HBXIP) inhibits let-7 g expression to upregulate IGF2BP2, thus leading to the formation of a positive feedback loop of HBXIP/let-7 g/IGF2BP2/HBXIP to accelerate cell proliferation in breast cancer [94]. miR-497 partially reverses transforming growth factor beta 1 (TGFβ1)-induced epithelial-mesenchymal transition (EMT) and pulmonary fibroblast proliferation through inhibiting eIF3a in alveolar epithelial cells [95].
lncRNAs also regulate m6A methylation in cancer. LncRNA derived from hepatocytes (lnc-HC) interacts with HNRNPA2B1 to inhibit cholesterol metabolism in hepatocytes [96]. Long intergenic non-protein coding RNA 470 (LINC00470) interacts with METTL3 to suppress the stability of phosphatase and tensin homolog (PTEN) to facilitate GC progression [97]. LncRNA miR503 host gene (miR503HG) also interacts with HNRNPA2B1 to promote its degradation through an ubiquitin-proteasome pathway in HCC [98]. Similarly, long intergenic non-protein coding RNA 1234 (LINC01234) interacts with HNRNPA2B1 to facilitate cell proliferation and inhibit cell apoptosis in NSCLC [99]. Lin-28 homolog B antisense RNA 1 (LIN28B-AS1) interacts with IGF2BP1 to promote the proliferation and metastasis of lung adenocarcinoma (LUAD) [100]. Long intergenic Noncoding RNA for IGF2BP2 Stability (LINRIS) promotes CRC proliferation by stabilizing IGF2BP2 [101]. The antisense RNA of growth arrest special 5 (GAS5-AS1) depends on ALKBH5 to suppresses the growth and metastasis of cervical cancer [102]. Growth arrest special 5 (GAS5) can suppress YAP-mediated YTHDF3 to restrain the proliferative behavior of CRC [103]. Antisense strand of the GATA binding protein 3 gene (GATA3-AS) enhances the interaction between KIAA1429 and GATA binding protein 3 (GATA3) pre-mRNA, leading to the formation of the GATA3-AS/KIAA1429/GATA3 axis in HCC [104].
Clinical application of m6A methylation in cancer
m6A methylation serves as new biomarkers for diagnosis and prognosis in cancer. m6A regulators METTL3, YTHDC2 and HNRNPC are used to predict the prognosis in patients with HNSCC [105]. Upregulated METTL3/FTO or downregulated YTHDF2 and METTL14 can indicate a poor survival in GC, CRC, and HCC [48, 70, 106]. Low expression of METTL14 is associated with tumor differentiation, clinical stage, and microvascular invasion [48]. Low expression of ALKBH5 or FTO predicts an unfavorable marker in lung cancer and HCC [107, 108]. IGF2BP2 is considered as a prognostic marker in pancreatic cancer, esophagogastric junction adenocarcinoma and CRC [60, 109, 110].
m6A methylation also participates in drug resistance and cancer treatment. METTL3 stabilizes YAP and Rho GTPase activating protein 5 (ARHGAP5) to induce cisplatin resistance in NSCLC and in GC [69, 111]. HNRNPA2B1 is overexpressed in tamoxifen-resistant breast cancer and reduces 4-hydroxytamoxifen sensitivity [112]. In addition to METTL3 and METTL14, FTO and YTHDF2 are overexpressed in AML [26, 27, 113, 114]. A recent study shows that FTO inhibitor (FB23) and its derivative (FB23–2) promote myeloid differentiation and apoptosis in AML by targeting FTO [115]. m6A methylation is also involved in estimating tumor microenvironment and TME infiltration characterization so as to provide insights into an effective immunotherapy for cancer [116]. YTHDF2 is correlated with inflammation infiltration, vascular reconstruction and distant metastasis and predicts a poor prognosis in HCC [117].
In summary, the role of m6A modification in clinical application has been widely validated. As for the core members of m6A methylation, METTL3/14 exert their functions in many biological processes. METTL3/14 can be regarded as the most important and promising m6A regulator and arouse our attention about their modifications on ncRNAs and the clinical application in cancer diagnosis.
Conclusions and perspectives
Accumulating studies have been focused on how m6A methylation modifies the stability, splicing and translation of ncRNAs or ncRNAs regulate m6A regulators in cancer. The interaction between m6A methylation and ncRNAs can impact the different life activities including cancer cell proliferation, invasion and metastasis. As for the clinical application of m6A methylation, they can be regarded as the potential targets for cancer diagnosis, prognosis and treatment. The latest findings show that lncRNA long intergenic non-protein coding RNA 266–1 (LINC00266–1) interacts with IGF2BP1 by encoding a 71-amino acid peptide, named RNA-binding regulatory peptide, thereby promoting tumorigenesis [118]. However, the specific binding sites between m6A methylation and ncRNAs need be further investigated.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its additional files.
Abbreviations
- 3′-UTR:
-
3′ -untranslated region
- ALKBH5:
-
alkB homologue 5
- AML:
-
Acute myeloid leukemia
- ARHGAP5:
-
Rho GTPase activating protein 5
- circRNAs:
-
circular RNAs
- circNSUN2:
-
circular RNA NOP2/Sun RNA methyltransferase 2
- circPVRL3:
-
circular RNA poliovirus receptor-related 3
- circ-ZNF609:
-
circular RNA zinc finger protein 609
- CRC:
-
Colorectal cancer
- CSC:
-
Cigarette smoke condensate
- DANCR:
-
Differentiation antagonizing non-protein coding RNA
- DGCR8:
-
DiGeorge syndrome critical region 8
- eIF3:
-
eukaryotic translation initiation factor 3
- EMT:
-
Epithelial-mesenchymal transition
- ESCC:
-
Esophageal squamous cell carcinoma
- FAK:
-
Focal adhesion kinase
- FGFR3:
-
Fibroblast growth factor receptor 3
- FAM225A:
-
Family with sequence similarity 225 member A
- FTO:
-
Fat mass and obesity-associated protein
- FOXM1:
-
Forkhead box M1
- FOXM1-AS:
-
antisense to forkhead box M1
- GAS5:
-
Growth arrest special 5
- GAS5-AS1:
-
the antisense RNA of GAS5
- GATA3:
-
GATA binding protein 3
- GATA3-AS:
-
Antisense strand of the GATA binding protein 3 gene
- GC:
-
Gastric cancer
- GSCs:
-
Glioblastoma stem-like cells
- HBXIP:
-
Hepatitis B X-interacting protein
- HCC:
-
Hepatocellular carcinoma
- HIF-1α:
-
Hypoxia-inducible factor 1-alpha
- HMGA2:
-
High mobility group AT-hook 2
- HNRNP:
-
Heterogeneous nuclear ribonucleo protein
- HNSCC:
-
Head and neck squamous cell carcinoma
- IGF2BP1/2/3:
-
Insulin-like growth factor-2 mRNA-binding proteins 1/2/3
- ITGB3:
-
Integrin β3
- LINC00266–1:
-
Long intergenic non-protein coding RNA 266–1
- LINC00278:
-
Long intergenic non-protein coding RNA 278
- LINC00470:
-
Long intergenic non-protein coding RNA 470
- LINC00958:
-
Long non-coding RNA 00958
- LINC01234:
-
Long intergenic non-protein coding RNA 1234
- LIN28B-AS1:
-
Lin-28 homolog B antisense RNA 1
- LINRIS:
-
Long intergenic Noncoding RNA for IGF2BP2 Stability
- LNCAROD:
-
lncRNA activating regulator of DKK1
- lnc-HC:
-
LncRNA derived from hepatocytes
- lncRNAs:
-
Long non-coding RNAs
- LUAD:
-
Lung adenocarcinoma
- m6A:
-
N6-methyladenosine
- m6A-seq:
-
N6-methyladenosine-sequensing
- MALAT1:
-
Metastasis-associated lung adenocarcinoma transcript-1
- MEFs:
-
Mouse embryonic fibroblasts
- METTL3/14/16:
-
Methyltransferase-like 3/14/16
- miR503HG:
-
miR503 host gene
- miRNAs:
-
Micro RNAs
- mRNA:
-
Messenger RNA
- ncRNAs:
-
Noncoding RNAs
- NEAT1:
-
Nuclear paraspeckle assembly transcript 1
- NSCLC:
-
Non-small cell lung cancer
- PDAC:
-
Pancreatic ductal adenocarcinoma
- PTEN:
-
Phosphatase and tensin homolog
- RBM15/15B:
-
RNA-binding motif protein 15/15B
- Ribo-circRNAs:
-
Ribosomes-circRNAs
- RP11:
-
RP11–138 J23.1
- SAM:
-
S-adenosylmethionine
- STING:
-
Stimulator of interferon genes
- TGFβ1:
-
Transforming growth factor beta 1
- WTAP:
-
Wilms tumor 1-associated protein
- XIST:
-
X-inactive specific transcript
- YAP:
-
Yes associated protein
- YTH:
-
YT521-B homology
- YTHDC1/2:
-
YTH domain-containing proteins 1/2
- YTHDF1/2/3:
-
YTH domain family 1/2/3
References
Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–7.
Ping X-L, Sun B-F, Wang L, Xiao W, Yang X, Wang W-J, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–89.
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6.
Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–46.
Fustin J-M, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806.
Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18:31–42.
Edens BM, Vissers C, Su J, Arumugam S, Xu Z, Shi H, et al. FMRP Modulates Neural Differentiation through m6A-Dependent mRNA Nuclear Export. Cell Rep. 2019;28:845–854.e5.
Engel M, Eggert C, Kaplick PM, Eder M, Röh S, Tietze L, et al. The Role of m6A/m-RNA Methylation in Stress Response Regulation. Neuron. 2018;99:389 403.e9.
Yu R, Li Q, Feng Z, Cai L, Xu Q. m6A Reader YTHDF2 Regulates LPS-Induced Inflammatory Response. Int J Mol Sci. 2019;20:1323.
Chen X-Y, Zhang J, Zhu J-S. The role of m6A RNA methylation in human cancer. Mol Cancer. 2019;18:103.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301–4.
Fleming AM, Nguyen NLB, Burrows CJ. Colocalization of m6A and G-Quadruplex-Forming Sequences in Viral RNA (HIV, Zika, Hepatitis B, and SV40) Suggests Topological Control of Adenosine N6-Methylation. ACS Cent Sci. 2019;5:218–28.
Shah JC, Clancy MJ. IME4, a gene that mediates MAT and nutritional control of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1078–86.
Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20:1278–88.
Cesarini V, Silvestris DA, Tassinari V, Tomaselli S, Alon S, Eisenberg E, et al. ADAR2/miR-589-3p axis controls glioblastoma cell migration/invasion. Nucleic Acids Res. 2018;46:2045–59.
Luo H, Zhu G, Xu J, Lai Q, Yan B, Guo Y, et al. HOTTIP lncRNA promotes hematopoietic stem cell self-renewal leading to AML-like disease in mice. Cancer Cell. 2019;36:645–59.
Li J, Huang C, Zou Y, Ye J, Yu J, Gui Y. CircTLK1 promotes the proliferation and metastasis of renal cell carcinoma by sponging miR-136-5p. Mol Cancer. 2020;19:103.
Schumann U, Shafik A, Preiss T. METTL3 Gains R/W Access to the Epitranscriptome. Mol Cell. 2016;62:323–4.
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–5.
Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014;8:284–96.
Koh CWQ, Goh YT, Goh WSS. Atlas of quantitative single-base-resolution N6-methyl-adenine methylomes. Nat Commun. 2019;10:5636.
Meyer KD, Jaffrey SR. Rethinking m6A Readers, Writers, and Erasers. Annu Rev Cell Dev Biol. 2017;33:319–42.
Zhong X, Yu J, Frazier K, Weng X, Li Y, Cham CM, et al. Circadian Clock Regulation of Hepatic Lipid Metabolism by Modulation of m6A mRNA Methylation. Cell Rep. 2018;25:1816–1828.e4.
Cheng Y, Luo H, Izzo F, Pickering BF, Nguyen D, Myers R, et al. m6A RNA Methylation Maintains Hematopoietic Stem Cell Identity and Symmetric Commitment. Cell Rep. 2019;28:1703–1716.e6.
Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m6A Modification. Cell Stem Cell. 2018;22:191–205.e9.
Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–76.
Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, et al. The U6 snRNA m6A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell. 2017;169:824–835.e14.
Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29.
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–7.
Gerken T, Girard CA, Tung Y-CL, Webby CJ, Saudek V, Hewitson KS, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–72.
Tang C, Klukovich R, Peng H, Wang Z, Yu T, Zhang Y, et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3’-UTR mRNAs in male germ cells. Proc Natl Acad Sci U S A. 2018;115:E325–33.
Shi H, Wei J, He C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol Cell. 2019;74:640–50.
Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95.
Wu R, Li A, Sun B, Sun J-G, Zhang J, Zhang T, et al. A novel m6A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019;29:23–41.
Zhuang M, Li X, Zhu J, Zhang J, Niu F, Liang F, et al. The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res. 2019;47:4765–77.
Wang L, Wen M, Cao X. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science. 2019;365. https://doi.org/10.1126/science.aav0758.
Rahmanian S, Murad R, Breschi A, Zeng W, Mackiewicz M, Williams B, et al. Dynamics of microRNA expression during mouse prenatal development. Genome Res. 2019;29:1900–9.
Wu C-J, Cho S, Huang H-Y, Lu C-H, Russ J, Cruz LO, et al. MiR-23~27~24-mediated control of humoral immunity reveals a TOX-driven regulatory circuit in follicular helper T cell differentiation. Sci Adv. 2019;5:eaaw1715.
Cho YK, Son Y, Kim S-N, Song H-D, Kim M, Park J-H, et al. MicroRNA-10a-5p regulates macrophage polarization and promotes therapeutic adipose tissue remodeling. Mol Metab. 2019;29:86–98.
Cen B, Lang JD, Du Y, Wei J, Xiong Y, Bradley N, et al. Prostaglandin E2 induces MIR675-5p to promote colorectal tumor metastasis via modulation of p53 expression. Gastroenterology. 2019;158:971–984.e10.
Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–5.
Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015;162:1299–308.
Mi B, Xiong Y, Yan C, Chen L, Xue H, Panayi AC, et al. Methyltransferase-like 3-mediated N6-methyladenosine modification of miR-7212-5p drives osteoblast differentiation and fracture healing. J Cell Mol Med. 2020;24:6385–96.
Zhang J, Bai R, Li M, Ye H, Wu C, Wang C, et al. Excessive miR-25-3p maturation via N6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019;10:1858.
Han J, Wang J-Z, Yang X, Yu H, Zhou R, Lu H-C, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18:110.
Gu S, Sun D, Dai H, Zhang Z. N6-methyladenosine mediates the cellular proliferation and apoptosis via microRNAs in arsenite-transformed cells. Toxicol Lett. 2018;292:1–11.
Ma J-Z, Yang F, Zhou C-C, Liu F, Yuan J-H, Wang F, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6 -methyladenosine-dependent primary MicroRNA processing. Hepatol Baltim Md. 2017;65:529–43.
Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res CR. 2019;38:393.
Wang H, Deng Q, Lv Z, Ling Y, Hou X, Chen Z, et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer. 2019;18:181.
Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, et al. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–16.
Monfort A, Di Minin G, Postlmayr A, Freimann R, Arieti F, Thore S, et al. Identification of Spen as a Crucial Factor for Xist Function through Forward Genetic Screening in Haploid Embryonic Stem Cells. Cell Rep. 2015;12:554–61.
Patil DP, Chen C-K, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–73.
Nesterova TB, Wei G, Coker H, Pintacuda G, Bowness JS, Zhang T, et al. Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat Commun. 2019;10:3129.
Liu J, Zhang X, Chen K, Cheng Y, Liu S, Xia M, et al. CCR7 Chemokine Receptor-Inducible lnc-Dpf3 Restrains Dendritic Cell Migration by Inhibiting HIF-1α-Mediated Glycolysis. Immunity. 2019;50:600–615.e15.
Liu P, Zhang B, Chen Z, He Y, Du Y, Liu Y, et al. m6A-induced lncRNA MALAT1 aggravates renal fibrogenesis in obstructive nephropathy through the miR-145/FAK pathway. Aging. 2020;12:5280–99.
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell. 2017;31:591–606.e6.
Zhang K-J, Tan X-L, Guo L. The long non-coding RNA DANCR regulates the inflammatory phenotype of breast cancer cells and promotes breast cancer progression via EZH2-dependent suppression of SOCS3 transcription. Mol Oncol. 2020;14:309–28.
Lin X, Yang F, Qi X, Li Q, Wang D, Yi T, et al. LncRNA DANCR promotes tumor growth and angiogenesis in ovarian cancer through direct targeting of miR-145. Mol Carcinog. 2019;58:2286–96.
Hu X, Peng W-X, Zhou H, Jiang J, Zhou X, Huang D, et al. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2019;27:1782–94.
Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–41.
Amodio N, Raimondi L, Juli G, Stamato MA, Caracciolo D, Tagliaferri P, et al. MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches. J Hematol Oncol. 2018;11:63.
Brown JA, Bulkley D, Wang J, Valenstein ML, Yario TA, Steitz TA, et al. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat Struct Mol Biol. 2014;21:633–40.
Brown JA, Kinzig CG, DeGregorio SJ, Steitz JA. Methyltransferase-like protein 16 binds the 3’-terminal triple helix of MALAT1 long noncoding RNA. Proc Natl Acad Sci U S A. 2016;113:14013–8.
Zuo X, Chen Z, Gao W, Zhang Y, Wang J, Wang J, et al. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. 2020;13:5.
Ban Y, Tan P, Cai J, Li J, Hu M, Zhou Y, et al. LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma. Mol Oncol. 2020;14:1282–96.
Zheng Z-Q, Li Z-X, Zhou G-Q, Lin L, Zhang L-L, Lv J-W, et al. Long Noncoding RNA FAM225A Promotes Nasopharyngeal Carcinoma Tumorigenesis and Metastasis by Acting as ceRNA to Sponge miR-590-3p/miR-1275 and Upregulate ITGB3. Cancer Res. 2019;79:4612–26.
Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F, et al. m6A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer. 2019;18:87.
Jin D, Guo J, Wu Y, Du J, Yang L, Wang X, et al. m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12:135.
Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19:46.
Zhang J, Guo S, Piao H-Y, Wang Y, Wu Y, Meng X-Y, et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. 2019;75:379–89.
Wu S, Zhang L, Deng J, Guo B, Li F, Wang Y, et al. A novel micropeptide encoded by Y-Linked LINC00278 links cigarette smoking and AR signaling in male esophageal squamous cell carcinoma. Cancer Res. 2020;80:2790–803.
Li X, Yang L, Chen L-L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell. 2018;71:428–42.
Di Timoteo G, Dattilo D, Centrón-Broco A, Colantoni A, Guarnacci M, Rossi F, et al. Modulation of circRNA Metabolism by m6A Modification. Cell Rep. 2020;31:107641.
Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell. 2017;66:9–21.e7.
Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66:22–37.e9.
Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N, et al. Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep. 2017;20:2262–76.
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–41.
Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol Cell. 2019;76:96–109.e9.
Du WW, Yang W, Li X, Awan FM, Yang Z, Fang L, et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. 2018;37:5829–42.
Ding L, Zhao Y, Dang S, Wang Y, Li X, Yu X, et al. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol Cancer. 2019;18:45.
Wang S, Zhang Y, Cai Q, Ma M, Jin LY, Weng M, et al. Circular RNA FOXP1 promotes tumor progression and Warburg effect in gallbladder cancer by regulating PKLR expression. Mol Cancer. 2019;18:145.
Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, Ni C, et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat Commun. 2019;10:2300.
Chen R-X, Chen X, Xia L-P, Zhang J-X, Pan Z-Z, Ma X-D, et al. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695.
Sun H-D, Xu Z-P, Sun Z-Q, Zhu B, Wang Q, Zhou J, et al. Down-regulation of circPVRL3 promotes the proliferation and migration of gastric cancer cells. Sci Rep. 2018;8:10111.
Chen T, Hao Y-J, Zhang Y, Li M-M, Wang M, Han W, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289–301.
Li Y, Yang F, Gao M, Gong R, Jin M, Liu T, et al. miR-149-3p Regulates the Switch between Adipogenic and Osteogenic Differentiation of BMSCs by Targeting FTO. Mol Ther Nucleic Acids. 2019;17:590–600.
Shen X-P, Ling X, Lu H, Zhou C-X, Zhang J-K, Yu Q. Low expression of microRNA-1266 promotes colorectal cancer progression via targeting FTO. Eur Rev Med Pharmacol Sci. 2018;22:8220–6.
Yang Z, Li J, Feng G, Gao S, Wang Y, Zhang S, et al. MicroRNA-145 Modulates N6-Methyladenosine Levels by Targeting the 3’-Untranslated mRNA Region of the N6-Methyladenosine Binding YTH Domain Family 2 Protein. J Biol Chem. 2017;292:3614–23.
Du M, Zhang Y, Mao Y, Mou J, Zhao J, Xue Q, et al. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem Biophys Res Commun. 2017;482:582–9.
Fang C, Chen Y-X, Wu N-Y, Yin J-Y, Li X-P, Huang H-S, et al. MiR-488 inhibits proliferation and cisplatin sensibility in non-small-cell lung cancer (NSCLC) cells by activating the eIF3a-mediated NER signaling pathway. Sci Rep. 2017;7:40384.
Wei W, Huo B, Shi X. miR-600 inhibits lung cancer via downregulating the expression of METTL3. Cancer Manag Res. 2019;11:1177–87.
Xu X, Yu Y, Zong K, Lv P, Gu Y. Up-regulation of IGF2BP2 by multiple mechanisms in pancreatic cancer promotes cancer proliferation by activating the PI3K/Akt signaling pathway. J Exp Clin Cancer Res CR. 2019;38:497.
Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–9.
Guo R, Lv Y, Ouyang Y, Liu S, Li D. The Role of miR-497/EIF3A Axis in TGFβ1-Induced Epithelial-Mesenchymal Transition and Extracellular Matrix in Rat Alveolar Epithelial Cells and Pulmonary Fibroblasts. J Cell Biochem. 2017;118:3401–8.
Lan X, Yan J, Ren J, Zhong B, Li J, Li Y, et al. A novel long noncoding RNA Lnc-HC binds hnRNPA2B1 to regulate expressions of Cyp7a1 and Abca1 in hepatocytic cholesterol metabolism. Hepatol Baltim Md. 2016;64:58–72.
Yan J, Huang X, Zhang X, Chen Z, Ye C, Xiang W, et al. LncRNA LINC00470 promotes the degradation of PTEN mRNA to facilitate malignant behavior in gastric cancer cells. Biochem Biophys Res Commun. 2019;521:887–93.
Wang H, Liang L, Dong Q, Huan L, He J, Li B, et al. Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-κB pathway in hepatocellular carcinoma. Theranostics. 2018;8:2814–29.
Chen Z, Chen X, Lei T, Gu Y, Gu J, Huang J, et al. Integrative Analysis of NSCLC Identifies LINC01234 as an Oncogenic lncRNA that Interacts with HNRNPA2B1 and Regulates miR-106b Biogenesis. Mol Ther J Am Soc Gene Ther. 2020;28:1479–93.
Wang C, Gu Y, Zhang E, Zhang K, Qin N, Dai J, et al. A cancer-testis non-coding RNA LIN28B-AS1 activates driver gene LIN28B by interacting with IGF2BP1 in lung adenocarcinoma. Oncogene. 2019;38:1611–24.
Wang Y, Lu J-H, Wu Q-N, Jin Y, Wang D-S, Chen Y-X, et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer. 2019;18:174.
Wang X, Zhang J, Wang Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am J Transl Res. 2019;11:4909–21.
Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol Cancer. 2019;18:143.
Lan T, Li H, Zhang D, Xu L, Liu H, Hao X, et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol Cancer. 2019;18:186.
Zhao X, Cui L. Development and validation of a m6A RNA methylation regulators-based signature for predicting the prognosis of head and neck squamous cell carcinoma. Am J Cancer Res. 2019;9:2156–69.
Liu T, Yang S, Sui J, Xu S-Y, Cheng Y-P, Shen B, et al. Dysregulated N6-methyladenosine methylation writer METTL3 contributes to the proliferation and migration of gastric cancer. J Cell Physiol. 2020;235:548–62.
Jin D, Guo J, Wu Y, Yang L, Wang X, Du J, et al. m6A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC. Mol Cancer. 2020;19:40.
Liu X, Liu J, Xiao W, Zeng Q, Bo H, Zhu Y, et al. SIRT1 regulates N6 -methyladenosine RNA modification in hepatocarcinogenesis by inducing RANBP2-dependent FTO SUMOylation. Hepatol Baltim Md. 2020. https://doi.org/10.1002/hep.31222.
Wu X-L, Lu R-Y, Wang L-K, Wang Y-Y, Dai Y-J, Wang C-Y, et al. Long noncoding RNA HOTAIR silencing inhibits invasion and proliferation of human colon cancer LoVo cells via regulating IGF2BP2. J Cell Biochem. 2018;120:1221–31.
Tang W, Chen S, Liu J, Liu C, Wang Y, Kang M. Investigation of IGF1, IGF2BP2, and IGFBP3 variants with lymph node status and esophagogastric junction adenocarcinoma risk. J Cell Biochem. 2019;120:5510–8.
Zhu L, Zhu Y, Han S, Chen M, Song P, Dai D, et al. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis. 2019;10:383.
Klinge CM, Piell KM, Tooley CS, Rouchka EC. HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells. Sci Rep. 2019;9:9430.
Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N 6 -Methyladenosine RNA Demethylase. Cancer Cell. 2017;31:127–41.
Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, et al. Targeting the RNA m6A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell. 2019;25:137–148.e6.
Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell. 2019;35:677–691.e10.
Zhang B, Wu Q, Li B, Wang D, Wang L, Zhou YL. m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer. 2020;19:53.
Hou J, Zhang H, Liu J, Zhao Z, Wang J, Lu Z, et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol Cancer. 2019;18:163.
Zhu S, Wang J-Z, Chen D, He Y-T, Meng N, Chen M, et al. An oncopeptide regulates m6A recognition by the m6A reader IGF2BP1 and tumorigenesis. Nat Commun. 2020;11:1685.
Acknowledgements
None.
Funding
Our work was supported by the grants from National Natural Science Foundation of China (No. 81873143) and Double-Hundred Talent Plan of Shanghai Jiao Tong University School of Medicine (No. 20191831).
Author information
Authors and Affiliations
Contributions
JZ and JSZ designed this study and YCY drafted the manuscript. YCY and XYC collected the data and conducted the picture processing. JZ revised the paper and all authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
None.
Consent for publication
Consent for publication has been obtained from the authors.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yi, YC., Chen, XY., Zhang, J. et al. Novel insights into the interplay between m6A modification and noncoding RNAs in cancer. Mol Cancer 19, 121 (2020). https://doi.org/10.1186/s12943-020-01233-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12943-020-01233-2