Abstract
Background
β-Lactam antibiotics have been broadly used for the treatment of Acinetobacter baumannii infections, resulting in development of β-lactam inactivating β-lactamases. Here, we described antibiotic resistance rate, prevalence of β-lactamase-encoding genes, and clonal relationships of A. baumannii strains isolated from children referred to Children’s Medical Center in Tehran, Iran, during 2019–2020.
Methods
A total of 60 non-replicate A. baumannii isolates were recovered from clinical specimens of pediatric patients. Antibiotic susceptibility testing was done by the disc diffusion method. Colistin susceptibility of isolates was performed by the broth microdilution method. β-lactamase-encoding genes were characterized by PCR. The presence of ISAba1 element upstream of the several oxacillinase genes was also checked. Genetic relatedness of isolates was determined by using random amplification of polymorphic DNA (RAPD) typing.
Results
The antimicrobial susceptibility tests showed that 83.3% of A. baumannii isolates were MDR, and 40% XDR. Both MDR and XDR A. baumannii isolates were susceptible to colistin. The frequency of blaOXA-51-like, blaOXA-23-like, blaTEM, blaOXA-24-like, blaPER, blaSHV, blaCTX-M, blaOXA-58-like, and blaIMP was 100, 93.33, 60, 36.67, 28.33, 8.33, 5, 3.33, and 1.67%, respectively. Coexistence of ISAba1/blaOXA-23-like and ISAba1/blaOXA-51-like was observed in 65% and 85% of isolates, respectively. RAPD analysis revealed 4 common types and 2 single types of A. baumannii isolates.
Conclusions
The multiple clones harboring blaOXA-23-like, ISAba1-blaOXA-51-like, and ISAba1-blaOXA-23-like were responsible for the spread of A. baumannii isolates in our clinical wards. Dissemination of the well-established clones is worrisome and would become therapeutic challenges due to the possible transferring genetic elements associated with resistance.
Similar content being viewed by others
Background
Acinetobacter baumannii, a known serious human pathogen, is the most important for rising rates of hospital-acquired infections and rapid development of antibiotic and antimicrobial resistance worldwide [1]. A. baumannii is usually transmitted by medical staff-to-patient contact and exposure to contaminated surfaces; nevertheless, aerosol route from colonized patients has also been reported [2]. The ability of A. baumannii for environmental persistence and desiccation resistance makes it a successful pathogen to spread in healthcare settings [3]. Healthcare-associated pneumonia and bacteremia are the most common clinical manifestations of A. baumannii infections; however, other infection types, including wound infections, urinary tract infections, endocarditis, meningitis, endophthalmitis, and osteomyelitis have also been seen in adults and children [4].
Recently, multidrug-resistant (MDR) A. baumannii has emerged as a significant pathogen in children with high morbidity and mortality and caused serious therapeutic problems due to the rapid acquisition of a large diversity of antibiotic-resistance genes [5]. β-lactam antibiotics (e.g., penicillins, cephalosporins, carbapenems, and monobactams) are empirically prescribed for the treatment of infections caused by A. baumannii. However, several β-lactam hydrolyzing groups such as class A β-lactamases, ESBLs, (e.g., TEM, SHV, CTX-M, PER, VEB, and GES), class B β-lactamases, MBLs, (e.g., IMP, VIM, GIM, and NDM), class C AmpC cephalosporinases, and class D carbapenemases/oxacillinases, OXA types, (e.g., OXA-23-like, OXA-24-like, OXA-51-like, and OXA-58-like) were most frequently described in A baumannii [6, 7]. Carbapenems possess potent antimicrobial activity against MDR A. baumannii, although, new carbapenem-hydrolyzing beta-lactamases have arisen with increasing carbapenem use resulted in a higher risk of treatment failure [8].
Insertion sequences (ISs) cause mutations and genome rearrangements resulting in spread of resistance and enhancing virulence determinants in Acinetobacter species. Previous studies have been showing that ISAba1 element is responsible for the transfer and overexpression of carbapenem resistance genes (e.g., blaOXA-51-like, blaOXA-23-like, and blaOXA-58-like) in A. baumannii [9, 10].
The rising rate of drug-resistant A. baumannii in pediatric patients is alarming and limiting the treatment options. The aim of this study was to determine in vitro activity of antibiotics, presence of β-lactamase-encoding genes, and genetic diversity of A. baumannii strains isolated from children referred to Children’s Medical Center in Tehran, Iran, during 2019–2020.
Materials and methods
Clinical specimens, bacterial isolates, and identification
This study was conducted on 60 non-duplicate A. baumannii isolates obtained from pediatric patients, aged from 4 days–14 years old, and hospitalized in different wards of Children’s Medical Center in Tehran, during April 2019 until December 2020. The isolates recovered from various clinical specimens were identified by the routine microbiological tests as A. baumannii. Then isolates were confirmed by PCR amplification of partial RNA polymerase β-subunit (rpoB) gene using a set of forward and reverse primers of 5′-TAYCGYAAAGAYTTGAAAGAAG-3′ and 5′-CMACACCYTTGTTMCCRTGA-3′, as described previously [11]. All identified A. baumannii strains were stored at − 70 °C until further investigations.
Antimicrobial susceptibility test
Antimicrobial susceptibility testing was carried out by the Kirby–Bauer disc diffusion method according to Clinical and Laboratory Standards Institute (CLSI) [12] and breakpoint interpretations for including amikacin (30 µg), gentamicin (10 µg), tobramycin (10 µg), cotrimoxazole (trimethoprim–sulfamethoxazole) (25 µg), tigecycline (15 µg), doxycycline (30 µg), tetracycline (30 µg), minocycline (30 µg) ceftriaxone (30 µg), ceftazidime (30 µg), cefepime (30 µg), cefotaxime (30 µg), piperacillin/tazobactam (100/10 µg), ampicillin-sulbactam (10/10 µg), imipenem (10 µg), meropenem (10 µg), ciprofloxacin (5 µg), levofloxacin (5 µg), (Mast Group Ltd., Bootle, UK). The minimum inhibitory concentrations (MICs) of colistin (Colistin sulfate salt powder, Sigma-Aldrich, St. Louis, MO, USA) were determined using the broth microdilution method according to CLSI guidelines (CLSI, 2020). Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27,853 strains were used as the standard strains.
Detection of β-lactamase-encoding genes
Genomic DNA was extracted by BioFact™ Genomic DNA Prep Kit GD141-100 (BioFact, Daejeon, Korea) according to the manufacturer’s recommendations. The primer sequences, product sizes, and annealing temperatures for blaTEM, blaSHV, blaCTX-M, blaVEB, blaPER, blaGES, blaIMP, blaVIM, blaNDM, ISAba1, blaOXA-51-like, blaOXA-24-like, blaOXA-23-like, blaOXA-58-like, ISAba1/blaOXA-23-like, and ISAba1/blaOXA-51-like gene fragments are presented in Table 1. The presence of ISAba1 insertion upstream of blaOXA-51-like and blaOXA-23-like genes was explored by using the forward primer for ISAba1 and the reverse primers for blaOXA-51-like and blaOXA-23-like. PCR was carried out using the ready-to-use 2X Taq DNA Polymerase Master Mix RED (Ampliqon, Denmark) in a 25 mL total volume reaction containing ~ 30 ng of DNA template and 10 pmol/mL of forward and reverse primers. PCR was performed under conditions of initial denaturation at 94 °C for 5 min, 35 cycles of denaturation at 94 °C for 45 s, annealing ranging from 47 to 58 °C for 45 s, extension at 72 °C for 45 s, and final extension at 72 °C for 5 min. PCR products were separated on agarose gel 1.5% by electrophoresis and visualized under ultraviolet light.
Random amplification of polymorphic DNA (RAPD)-PCR fingerprinting
The genetics relatedness of blaOXA-24-like-positive strains was assessed using RAPD-PCR analysis with a random oligonucleotide 5′-CCGCAGCCAA-3′ (TAG Copenhagen, Denmark). Amplification of random sequences was done by a conventional PCR as follows: initial denaturation at 94 °C for 10 min, followed by 30 cycles consisting of denaturation at 94 °C for 1 min, annealing at 43 °C for 1 min, extension at 72 °C for 3 min, and final extension at 72 °C for 10 min. The RAPD patterns was compared by GelCompar® II v.4.1 software (Applied Maths BVBA, Sint–Martens–Latem, Belgium) and clustered with unweighted pair group method (UPGMA).
Statistical analysis
Data analysis was performed using the Minitab® statistical software package 17 (MINITAB, Inc., State College, Pennsylvania, USA). The P value ≤ 0.05 was considered statistically significant for the analysis.
Results
Clinical isolates and demographic characteristics of pediatric patients
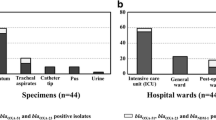
A total of 60 non-replicate A. baumannii clinical isolates were collected from the Children’s Medical Center in a period of April 2019 to December 2020. 43 out of 60 (71.6%) and 17 out of 60 (28.3%) isolates of A. baumannii were belonged to male and female pediatric patients, respectively with an age range of 4 days to 14 years. The majority of isolates were obtained from patients admitted to the neonatal and pediatric intensive care units (NICU, PICU) (65%), followed by the other wards (35%). Distribution of collected isolates from the specimen sources was follows: CSF (6.66%), BAL (23.33%), tracheal tube (18.33%), blood (30%), urine (3.33%), central venous line (CVL) (3.33%), respiratory secretions (10%), drain discharge (3.33%), and dialysis fluid (1.7%). All isolates were molecularly confirmed as A. baumannii using both inherent rpoB and blaOXA-51-like genes. Demographics and clinical features of pediatric patients are accessible in detail in a descriptive Additional file 1: Table S1.
Antimicrobial susceptibility profile
Antimicrobial susceptibility testing was displayed that the highest resistance rate was for imipenem and meropenem (53, 88.3%), followed by cefotaxime, cefepime, ceftriaxone, levofloxacin (52, 86.67%), ceftazidime, amikacin, cotrimoxazole, ciprofloxacin (51, 85%), piperacillin-tazobactam (50, 83.3%), gentamicin (49, 81.67%), tetracycline (44, 73.3%), minocycline (34, 56.6%), doxycycline (32, 53.3%), ampicillin–sulbactam (29, 48.3%), and tigecycline (3, 5%). Fifty-two out of 60 isolates were resistant to both imipenem and meropenem, whereas 6 out of 60 isolates were sensitive to them. However, only one isolate was found to be sensitive to imipenem, and one was susceptible to meropenem.
All isolates were susceptible to colistin with MICs ranging from 0.125 to 2 µg/mL. The MIC50 and MIC90 of colistin were 0.5 and 2 µg/mL, respectively. Fifty-one out of 60 (83.3%) A. baumannii isolates were categorized as the MDR, and 24 out of 60 (40%) were extensively drug-resistant (XDR).
Antimicrobial susceptibility testing for the isolates is summarized in Fig. 1.
In vitro antimicrobial susceptibility profile of pediatric A. baumannii isolates. AS Ampicillin–Sulbactam, CAZ Ceftazidime, IMP Imipenem, AN Amikacin, SXT Trimethoprim–Sulfamethoxazole, PT Piperacillin–Tazobactam, CTX Cefotaxime, GM Gentamicin, CPM Cefepime, CIP Ciprofloxacin, MN Minocycline, TOB Tobramycin, LEV Levofloxacin, DXT Doxycycline, CRO Ceftriaxone, TE Tetracycline, MRP Meropenem, TGC Tigecycline, CL Colistin
Characterization of β-lactamase resistance genes profile
Molecular analysis of β-lactamase resistance genes in A. baumannii isolates showed that 93.33% of the clinical isolates (n = 56) were positive for the blaOXA-23-like gene, followed by blaTEM 60% (n = 36), blaOXA−24−like 36.67% (n = 22), blaPER 28.33% (n = 17), blaSHV 8.33% (n = 5), blaCTX-M 5% (n = 3), blaOXA-58-like 3.33% (n = 2), and blaIMP 1.67% (n = 1). In contrast, we could not detect any blaVIM, blaGES, blaVEB, and blaNDM genes. Although, the blaOXA−51−like gene was identified in all A. baumannii isolates, we could not conclude the antibiotic resistance associated with its presence alone (P value > 0.05). The ISAba1 element was found in 95% of the isolates. However, ISAba1 was detected in the upstream of blaOXA-23-like and blaOXA-51-like genes in 65% and 85% of isolates, respectively. Distribution of β-lactamase-encoding genes and ISAba1 upstream of blaOXA genes in pediatric isolates of A. baumannii is presented in Fig. 2 and Additional file 1: Table S1.
RAPD genotyping of A. baumannii
The A. baumannii isolates harboring blaOXA-24-like were designated for further epidemiological analysis by RAPD-PCR. Dendrogram analysis by GelCompar software and UPGMA clustering method by the arithmetic mean calculation and the Dice coefficient (Dice similarity index) with an optimization of 0.5% and a tolerance of 1.5% were used for the similarity with a cut-off value of > 85% and considered the same RAPD type. According to the similarity index, we found 4 common-type (A–D) and 2 single-type (E and F). The major RAPD type A has included 12 strains, followed by RAPD type B with 3, RAPD type C with 2, and RAPD type D with 2 strains. Dendrogram of pediatric A. baumannii isolates based on RAPD-PCR analysis is shown in Fig. 3.
Discussion
The incidence of MDR A. baumannii in pediatric patients is increasing worldwide as far as it has led to therapeutic challenges and has become an ongoing serious public health concern [13]. In this study, we described a high antibiotic resistance rate, rising MDR and XDR strains, and widespread prevalence of β-lactamase-encoding genes in A. baumannii isolated from pediatric patients.
Carbapenems, as the last-line group of β-lactam antibiotics, have been used to treat infections associated with MDR A. baumannii for a long time; however, carbapenem resistant A. baumannii (CRAB) has recently emerged and limited therapeutic options [14, 15]. We found that most isolates were resistant to carbapenem (imipenem and meropenem 88.3% for each, and both imipenem and meropenem 86.6%). A high rate of CRAB in pediatric has been explored in several similar studies [16, 17]. It has been reported that the drug resistance in CRAB is developed by several mechanisms, including production of hydrolyzing enzymes, horizontally transferring resistance determinants, changes in outer membrane permeability, and upregulation of the efflux systems. The major mechanism of carbapenem resistance in A. baumannii is related to the production of carbapenem-hydrolyzing β-lactamases, i.e., carbapenem-hydrolyzing class D β-lactamases (CHDLs), class B metallo-β-lactamases (MBLs), and class A carbapenemases [18]. Distribution of the CHDLs, MBLs, and ESBLs genes has been reported with different prevalence in recent published papers [19,20,21]. We could not find A. baumannii isolates harboring blaVIM, blaGES, blaVEB, and blaNDM genes. In contrast, a high prevalence rate of blaNDM gene has been observed in CRAB isolates from Egyptian patients [22]. The blaOXA-23-like was more prevalent in our study, which is in agreement with previous studies [17, 23]. Moreover, we also detected the blaOXA-24-like and blaOXA-58-like genes, but in lower frequencies, corresponding with similar reports [20, 24]. Although the catalytic activity of OXA-type carbapenemases is far less than MBLs, their expression can be influenced by upstream existence of insertion sequences (ISs) elements such as ISAba1 resulting in increased resistance to carbapenems [25]. Most isolates carried ISAba1; however, 85% of isolates harbored ISAba1-blaOXA−51−like and 65% were also positive for ISAba1/blaOXA-23-like. We found a statistically significant relationship between the presence of ISAba1 upstream of blaOXA-23-like and blaOXA-51-like and carbapenem resistance (P value < 0.05). Over 90% of isolates (54 out of 60) were resistant to carbapenems (imipenem and meropenem) with the presence of ISAba1 upstream of blaOXA-23-like and blaOXA-51-like; though, four isolates despite the existence of ISAba1 upstream of blaOXA-23-like and blaOXA-51-like were susceptible to carbapenems; while it might be expected that the co-existence of the determinants should confer resistance to the carbapenems, as described by Turton et al. [25]. Another study has suggested that ISAba1-blaOXA-51-like alone is insufficient to confer resistance to carbapenems [26]. Nevertheless, it has not been completely clarified that the cause of the contradiction in carbapenem susceptibility levels in isolates harboring the ISAba1-blaOXA-51-like gene [27].
Numerous reports have indicated that colistin (polymyxin E) and tigecycline, the only active antibiotics against A. baumannii, have become the last resort of treatment for MDR A. baumannii [28]. Now, resistance to colistin is relatively low; however, colistin-resistant strains have been introduced from different regions of the world [29]. A recent study has reported inhibitory potential effects of methylene blue for improving colistin susceptibility in the treatment of colistin-resistant strains of A. baumannii [30]. We investigated the in vitro activity of colistin, indicating that all A. baumannii isolates were susceptible to colistin according to the MIC50 and MIC90 values of < 2 µg/ml. Significantly, both MDR and XDR A. baumannii were susceptible to colistin which are in concordance with previous studies [13, 31].
RAPD is a simple and rapid technique and possesses high discriminative power for source tracking of bacteria, and it has been used in epidemiological studies of Acinetobacter species to identify circulating clones in clinical settings [32, 33]. The 21 blaOXA-24-like-positive isolates were distributed into 4 RAPD types belonging to four clusters (A, B, C, D) and two additional RAPD types E and F. Twelve strains were categorized in cluster A. All strains belonging to cluster A were MDR. All strains of cluster A were positive for ISAba1 and blaOXA-23-like, and the genetic profile of the strains was also varied. However, antibiotic resistance profiles were different between the isolates within the same group. Several strains with the same genetic profile and common origin were observed in clusters B and D. All strains of clusters E and F were susceptible to all antibiotic groups. Significantly, strains were collected from different infection sites were genetically categorized in the same cluster, suggesting the same origin and upsurging threat of multiple infection abilities for these genotypes. We successfully applied RAPD for distinguishing the genetic diversity of A. baumannii clones circulating in clinical setting.
Epidemiological studies on the dissemination of drug-resistant clones and their resistance gene profile are prerequisites to control infection and prevent policies in clinical settings.
There were a number of limitations to the current study; (a) the present literature was limited to single-center experiences, (b) our population size was small, (c) there was not enough data about pediatric patients’ records. Further researches are required to detect the expression level of resistance genes by real-time PCR assay. Other methods such as next-generation sequencing (NGS) can also be considered for well understanding the molecular mechanisms of resistance.
Conclusions
The distribution of resistance determinants in A. baumannii isolates is a great concern for our pediatric center and would become therapeutic challenges soon.
The high prevalence of the MDR clones circulating in different wards is a problematic issue for infection control, surveillance programs, and health care strategies, especially in the pediatric population. It is recommended that the development of novel therapeutic strategies or reassessment of old drugs is scheduled to make insight for pediatricians to address and find alternative antibiotics with high efficiency. In addition, longitudinal studies are needed for continuous monitoring and tracking dispersion dynamics of successful clones associated with the persistence of drug-resistant phenotypes.
Availability of data and materials
All data generated or analyzed during this study were included in this article.
Abbreviations
- A. baumannii :
-
Acinetobacter baumannii
- RAPD:
-
Random amplification of polymorphic DNA
- MDR:
-
Multidrug-resistant
- XDR:
-
Extensively drug-resistant
- bla :
-
Beta-lactamase
- OXA:
-
Oxacillinase
- IS:
-
Insertion sequence
- ESBLs:
-
Extended spectrum beta-lactamases
- TEM:
-
Temoniera, patient’s name
- SHV:
-
Sulfhydryl variable
- CTX-M:
-
Referred to its preferential hydrolytic activity against cefotaxime (CTX as its acronym, -M from Munich)
- PER:
-
Pseudomonas extended resistant
- VEB:
-
Vietnam extended-spectrum β-lactamase
- GES:
-
Guiana extended-spectrum β-lactamase
- IMP:
-
Imipenemase
- VIM:
-
Verona integron-encoded metallo-β-lactamase
- GIM:
-
German imipenemase
- NDM:
-
New Delhi metallo-β-lactamase
- AmpC:
-
Ampicillinase C
- rpoB :
-
RNA polymerase β subunit
- CLSI:
-
Clinical and Laboratory Standards Institute
- MIC:
-
Minimum inhibitory concentration
- ATCC:
-
American Type Culture Collection
- UPGMA:
-
Unweighted pair group method
- ICU:
-
Intensive care unit
- NICU:
-
neonatal intensive care unit
- PICU:
-
Pediatric intensive care unit
- OPD:
-
Outpatient department
- CSF:
-
Cerebrospinal fluid
- BAL:
-
Bronchoalveolar lavage
- CVL:
-
Central venous line
- CRAM:
-
Carbapenem resistant A. baumannii
- CHDLs:
-
Carbapenem-hydrolyzing class D β-lactamases
References
Alsan M, Klompas M. Acinetobacter baumannii: an emerging and important pathogen. J Clin Outcomes Manag. 2010;17:363–9.
Weinberg SE, Villedieu A, Bagdasarian N, Karah N, Teare L, Elamin WF. Control and management of multidrug resistant Acinetobacter baumannii: a review of the evidence and proposal of novel approaches. Infect Prev Pract. 2020;2:100077.
Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2018;16:91–102.
Bergogne-Bérézin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9:148–65.
Logan LK, Gandra S, Trett A, Weinstein RA, Laxminarayan R. Acinetobacter baumannii resistance trends in children in the United States, 1999–2012. J Pediatr Infect Dis Soc. 2019;8:136–42.
Bonomo RA. β-Lactamases: a focus on current challenges. Cold Spring Harb Perspect Med. 2017;7:a025239.
Sawa T, Kooguchi K, Moriyama K. Molecular diversity of extended-spectrum β-lactamases and carbapenemases, and antimicrobial resistance. J Intensive Care. 2020;8:13.
Tooke CL, Hinchliffe P, Bragginton EC, Colenso CK, Hirvonen VHA, Takebayashi Y, et al. β-Lactamases and β-lactamase inhibitors in the 21st century. J Mol Biol. 2019;431:3472–500.
Vijayakumar S, Anandan S, MS DP, Kanthan K, Vijayabaskar S, Kapil A, et al. Insertion sequences and sequence types profile of clinical isolates of carbapenem-resistant A. baumannii collected across India over four year period. J Infect Public Health. 2020;13:1022–8.
Yazdansetad S, Najari E, Ghaemi EA, Javid N, Hashemi A, Ardebili A. Carbapenem-resistant Acinetobacter baumannii isolates carrying blaOXA genes with upstream ISAba1: first report of a novel OXA subclass from Iran. J Glob Antimicrob Resist. 2019;18:95–9.
Gundi VAKB, Dijkshoorn L, Burignat S, Raoult D, La Scola B. Validation of partial rpoB gene sequence analysis for the identification of clinically important and emerging Acinetobacter species. Microbiology. 2009;155:2333–41.
CLSI. Performance standards for antimicrobial susceptibility testing, twenty-fifth informational supplement in M100-S30. Wayne: Clinical and Laboratory Standards Institute; 2020.
Shi J, Sun T, Cui Y, Wang C, Wang F, Zhou Y, et al. Multidrug resistant and extensively drug resistant Acinetobacter baumannii hospital infection associated with high mortality: a retrospective study in the pediatric intensive care unit. BMC Infect Dis. 2020;20:597.
Lowe M, Ehlers MM, Ismail F, Peirano G, Becker PJ, Pitout JDD, et al. Acinetobacter baumannii: epidemiological and beta-lactamase data from two tertiary academic hospitals in Tshwane, South Africa. Front Microbiol. 2018;9:1280.
Lukovic B, Gajic I, Dimkic I, Kekic D, Zornic S, Pozder T, et al. The first nationwide multicenter study of Acinetobacter baumannii recovered in Serbia: emergence of OXA-72, OXA-23 and NDM-1-producing isolates. Antimicrob Resist Infect Control. 2020;9:101.
Katragkou A, Kotsiou M, Antachopoulos C, Benos A, Sofianou D, Tamiolaki M, et al. Acquisition of imipenem-resistant Acinetobacter baumannii in a pediatric intensive care unit: a case-control study. Intensive Care Med. 2006;32:1384–91.
Chen Y, Ai L, Guo P, Huang H, Wu Z, Liang X, et al. Molecular characterization of multidrug resistant strains of Acinetobacter baumannii isolated from pediatric intensive care unit in a Chinese tertiary hospital. BMC Infect Dis. 2018;18:614.
Kyriakidis I, Vasileiou E, Pana ZD, Tragiannidis A. Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens. 2021;10:373.
Tavares LCB, de Vasconcellos FM, de Sousa WV, Rocchetti TT, Mondelli AL, Ferreira AM, et al. Emergence and persistence of high-risk clones among MDR and XDR A. baumannii at a Brazilian Teaching Hospital. Front Microbiol. 2019;9:2898.
Al-Agamy MH, Khalaf NG, Tawfick MM, Shibl AM, Kholy A, El. Molecular characterization of carbapenem-insensitive Acinetobacter baumannii in Egypt. Int J Infect Dis. 2014;22:49–54.
Ranjbar R, Farahani A. Study of genetic diversity, biofilm formation, and detection of Carbapenemase, MBL, ESBL, and tetracycline resistance genes in multidrug-resistant Acinetobacter baumannii isolated from burn wound infections in Iran. Antimicrob Resist Infect Control. 2019;8:172.
Wasfi R, Rasslan F, Hassan SS, Ashour HM, Abd El-Rahman OA. Co-existence of carbapenemase-encoding genes in Acinetobacter baumannii from cancer patients. Infect Dis Ther. 2021;10:291–305.
Zowawi HM, Sartor AL, Sidjabat HE, Balkhy HH, Walsh TR, Al Johani SM, et al. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii Isolates in the gulf cooperation council states: dominance of OXA-23-type producers. J Clin Microbiol. 2015;53:896–903.
Thirapanmethee K, Srisiri-a-nun T, Houngsaitong J, Montakantikul P, Khuntayaporn P, Chomnawang M. Prevalence of OXA-Type β-lactamase genes among carbapenem-resistant Acinetobacter baumannii clinical isolates in Thailand. Antibiotics. 2020;9:864.
Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, et al. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett. 2006;258:72–7.
Pagano M, Martins AF, Machado ABMP, Barin J, Barth AL. Carbapenem-susceptible Acinetobacter baumannii carrying the ISAba1 upstream blaOXA–51–like gene in Porto Alegre, southern Brazil. Epidemiol Infect. 2013;141:330–3.
Chen T-L, Lee Y-T, Kuo S-C, Hsueh P-R, Chang F-Y, Siu L-K, et al. Emergence and distribution of plasmids bearing the blaOXA–51–Like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in Taiwan. Antimicrob Agents Chemother. 2010;54:4575–81.
Vázquez-López R, Solano-Gálvez SG, Juárez Vignon-Whaley JJ, Abello Vaamonde JA, Padró Alonzo LA, Rivera Reséndiz A, et al. Acinetobacter baumannii resistance: a real challenge for clinicians. Antibiotics. 2020;9:205.
Talebi Bezmin Abadi A, Rizvanov AA, Haertlé T, Blatt NL. World Health Organization report: current crisis of antibiotic resistance. Bionanoscience. 2019;9:778–88.
Gazel D, Tatman Otkun M, Akçalı A. In vitro activity of methylene blue and eosin methylene blue agar on colistin-resistant A. baumannii: an experimental study. J Med Microbiol. 2019;68:1607–13.
Li KL, Abad CLR. The clinical profile and outcomes of adult patients given intravenous colistin for multidrug-resistant gram negative infections in a Philippine tertiary hospital. Int J Infect Dis. 2020;93:9–14.
D’Arezzo S, Capone A, Petrosillo N, Visca P. Epidemic multidrug-resistant Acinetobacter baumannii related to European clonal types I and II in Rome (Italy). Clin Microbiol Infect. 2009;15:347–57.
El-Kazzaz W, Metwally L, Yahia R, Al-Harbi N, El-Taher A, Hetta HF, Antibiogram. Prevalence of OXA carbapenemase encoding genes, and RAPD-genotyping of multidrug-resistant Acinetobacter baumannii incriminated in hidden community-acquired infections. Antibiotics. 2020;9:603.
Ghalavand Z, Heidary Rouchi A, Bahraminasab H, Ravanasa E, Mirsamadi ES, Nodeh Farahani N, et al. Molecular testing of Klebsiella pneumoniae contaminating tissue allografts recovered from deceased donors. Cell Tissue Bank. 2018;19:391–8.
Eckert C, Gautier V, Arlet G. DNA sequence analysis of the genetic environment of various blaCTX–M genes. J Antimicrob Chemother. 2006;57:14–23.
Naas T, Poirel L, Karim A, Nordmann P. Molecular characterization of In50, a class 1 integron encoding the gene for the extended-spectrum β-lactamase VEB-1 in Pseudomonas aeruginosa. FEMS Microbiol Lett. 1999;176:411–9.
Fallah F, Noori M, Hashemi A, Goudarzi H, Karimi A, Erfanimanesh S, et al. Prevalence of blaNDM, blaPER, blaVEB, blaIMP, and blaVIM genes among Acinetobacter baumannii isolated from two hospitals of Tehran, Iran. Scientifica (Cairo). 2014;2014:1–6.
Salehi B, Ghalavand Z, Mohammadzadeh M, Maleki DT, Kodori M, Kadkhoda H. Clonal relatedness and resistance characteristics of OXA-24 and ‐58 producing carbapenem‐resistant Acinetobacter baumannii isolates in Tehran, Iran. J Appl Microbiol. 2019;127:1421–9.
Goudarzi H, Mirsamadi ES, Ghalavand Z, Hakemi Vala M, Mirjalali H, Hashemi A. Rapid detection and molecular survey of blaVIM, blaIMP and blaNDM genes among clinical isolates of Acinetobacter baumannii using new multiplex real-time PCR and melting curve analysis. BMC Microbiol. 2019;19:122.
Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27:351–3.
Acknowledgements
This article has been extracted from the Ph.D thesis written by Mrs. Neda Yousefi Nojookambari in school of medicine, Shahid Beheshti University of Medical Sciences (Registration No. 183). The authors would like to thank department staff of microbiology, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
ZG: conception of the research idea, study design, and the drafting of the manuscript; NY, MS, and RD: laboratory works; GE and MV: analysis and data interpretation; BN: collection of the strains SY: write up and language edition; All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was authorized by the ethical committee of Shahid Beheshti University of Medical Sciences with reference number IR.SBMU.MSP.REC.1399.262.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Clinical and demographics data of pediatricpatients, drug resistance profile, and distribution pattern of β-lactamase genes in A. baumannii isolates.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yousefi Nojookambari, N., Sadredinamin, M., Dehbanipour, R. et al. Prevalence of β-lactamase-encoding genes and molecular typing of Acinetobacter baumannii isolates carrying carbapenemase OXA-24 in children. Ann Clin Microbiol Antimicrob 20, 75 (2021). https://doi.org/10.1186/s12941-021-00480-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12941-021-00480-5