Abstract
Background
A complex cascade of genes, enzymes, and transcription factors regulates AmpC β-lactamase overexpression. We investigated the network of AmpC β-lactamase overexpression in Klebsiella aerogenes and identified the role of AmpG in resistance to β-lactam agents, including cephalosporins and carbapenems.
Methods
A transposon mutant library was created for carbapenem-resistant K. aerogenes YMC2008-M09-943034 (KE-Y1) to screen for candidates with increased susceptibility to carbapenems, which identified the susceptible mutant derivatives KE-Y3 and KE-Y6. All the strains were subjected to highly contiguous de novo assemblies using PacBio sequencing to investigate the loss of resistance due to transposon insertion. Complementation and knock-out experiments using lambda Red-mediated homologous recombinase and CRISPR–Cas9 were performed to confirm the role of gene of interest.
Results
In-depth analysis of KE-Y3 and KE-Y6 revealed the insertion of a transposon at six positions in each strain, at which truncation of the AmpG permease gene was common in both. The disruption of the AmpG permease leads to carbapenem susceptibility, which was further confirmed by complementation. We generated an AmpG permease gene knockout using lambda Red-mediated recombineering in K. aerogenes KE-Y1 and a CRISPR–Cas9-mediated gene knockout in multidrug-resistant Klebsiella pneumoniae-YMC/2013/D to confer carbapenem susceptibility.
Conclusions
These findings suggest that inhibition of the AmpG is a potential strategy to increase the efficacy of β-lactam agents against Klebsiella aerogenes.
Similar content being viewed by others
Background
Gram-negative bacteria are a major threat to hospitalized patients and are associated with high mortality rates [1,2,3,4]. Hospital-acquired infections such as pneumonia, bloodstream infections, urinary tract infections, wound or surgical site infections, and meningitis are of particular concern to the clinicians. In the presence of antibiotic selective pressure, bacteria are capable of acquiring or up-regulating genes that code for antibiotic drug resistance, eventually leading to the emergence and global dissemination of these pathogens [5]. Also, Gram-negative bacteria can have redundant resistance mechanisms, either using single mechanisms against multiple antibiotics or multiple mechanisms against a single antibiotic. The antimicrobial- resistance crisis has escalated for several reasons: the high cost associated with finding novel antibiotic targets and antibiotic discovery, the length of time needed to design the drug and evaluate the efficacy, and the increased frequency of emerging bacterial resistance.
Klebsiella aerogenes (previously known as Enterobacter aerogenes) is a clinically significant bacterium in the family Enterobacteriaceae. K. aerogenes are part of the normal microbiota of the gastrointestinal tract in 40 to 80% of the population [6]. Though not a primary human pathogen, it has been implicated in a variety of healthcare-associated conditions, such as systemic bacteremia and urinary and lower respiratory tract infections, which are intrinsically resistant to ampicillin and narrow-spectrum cephalosporins [7, 8]. K. aerogenes possesses a chromosomal ampC β-lactamase gene that can be induced by antibiotic stress with various β-lactams. Mutations in the AmpC β-lactamase expression pathway can lead to resistance against extended- and broad-spectrum cephalosporins [9, 10].
Carbapenems, one class of β-lactam antibiotics, have historically been successful for treating cephalosporin-resistant K. aerogenes infections [11]. Alarmingly, K. aerogenes have recently emerged worldwide as carbapenem-resistant because of the high frequency of mutations in its ampR and ampD genes [12, 13]. In this study, we examined the mechanism of carbapenem resistance in K. aerogenes using transposon mutagenesis. In addition, we incorporated the CRISPR–Cas9-mediated gene knockout system to delete ampG permease and understand its role in antimicrobial resistance.
Methods
Bacterial strains, plasmids, antibiotics and oligonucleotides
K. aerogenes YMC2008-M09-943034 (KE-Y1) was collected from a tertiary-care hospital in Korea in 2008, and non-carbapenemase-producing carbapenem-resistant Klebsiella pneumoniae (YMC/2013/D) was collected from a different tertiary-care hospital in Korea in 2013. Bacterial identification was performed using the VITEK 32 GN system (BioMérieux, Marcy l’Etoile, France) and was confirmed using the direct colony method with MALDI-TOF MS (Bruker Daltonics, Bremen, Germany). The MICs of piperacillin, piperacillin-tazobactam, ampicillin, ampicillin-sulbactam, ceftazidime, ceftazidime-clavulanate, cefepime, imipenem, meropenem, ciprofloxacin, and aztreonam were determined by agar-dilution methods and E-test and interpreted according to the Clinical and Laboratory Standards Institute guidelines (2018). Lambda Red donor plasmid pKD46 was obtained from the E. coli Genetic Stock Center at Yale University (New Haven, CT, USA). Plasmid ZpUC-19 was a gift from Dr. Yo Suzuki at the J. Craig Venter Institute, La Jolla, CA, USA. Plasmids pCRISPR (Addgene plasmid # 42875) and pCas9 (Addgene plasmid #42876) were gifts from Luciano Marraffini. Plasmid pKDsgRNA-ack (Addgene plasmid #62654) was a gift from Kristala Prather. Recombinant strains were selected with spectinomycin, chloramphenicol, or kanamycin at a concentration of 50, 34, or 30 mg/l, respectively. Antibiotic concentrations were reduced by half for selecting strains harboring two or more plasmids. Oligonucleotides were synthesized by Macrogen, Inc., Korea.
Transposon mutagenesis and complementation
Conjugation was performed by cross-streaking E. coli SM10 λpir and KE-Y1 on Mueller–Hinton plates. Transconjugants were selected on media containing gentamicin (2 mg/l) and ciprofloxacin (0.1 mg/l). The colonies were further replica-plated on Mueller–Hinton plates containing meropenem to obtain carbapenem-susceptible mutants. The in-silico-determined (see below) novel target functional gene, ampG, was amplified using primers AmpG-F and AmpG-R (Additional file 1: Table S3), cloned into EcoRI- and XbaI-digested pAD123 vector, and transformed into chemically competent transposon-mediated mutants (carbapenem-susceptible K. aerogenes KE-Y3 and K. aerogenes KE-Y6) as described previously [14, 15].
DNA isolation, WGS, and analysis
The DNA of the wild-type and mutant strains were extracted using the Wizard genomic purification kit (Promega, WI, USA) with modifications to the manufacturer's protocol by adding 5 μl RNase solution during cell lysis and incubating the supernatant with DNA at − 20 °C for 1 h after addition of isopropanol. The WGS data were obtained by using the PacBio RS II sequencing system (Pacific Biosciences, Inc., Menlo Park, CA, USA) in a commercial laboratory (DNALink, Korea). To confirm the integrity of PacBio sequencing, all strains were sequenced on a 318 chip using the Ion Torrent PGM system and Ion Sequencing 200 kit (Life Technologies, Carlsbad, CA, USA). Annotations were performed using the RAST annotation pipeline with manual scrutiny [16]. All genomic analyses were performed using Geneious Pro 8.0 [17] (http://www.geneious.com).
Amino acid alignment and AmpG structure modeling
The amino acid sequences of AmpG protein from K. aerogenes KE-Y1 (CP045870), Klebsiella pneumoniae ATCC 13883 (KN046818), Escherichia coli O157:H7 (CP034384) and Pseudomonas aeruginosa PAO1 (CP053028) was aligned using Vector NTI align X. The structure model of AmpG was further established using I-TASSER according to homology modeling method [18]. The TOP 5 models predicted by I-TASSER and each model C-score are − 1.43, − 2.07, − 2.21, − 3.10, − 3.51 respectively. C-score is typically in the range of [− 5, 2], where a C-score of a higher value signifies a model with a higher confidence and vice-versa.
CRISPR–Cas9 mediated gene knockout
K. pneumoniae-YMC/2013/D (carbapenemase-non-producing, carbapenem-resistant), was included for CRISPR–Cas9 mediated AmpG permease gene knockout studies. CRISPR/Cas9 system with lambda Red recombineering was used to knockout the chromosomal ampG permease gene as described previously [14]. First, pCas9 was transformed into electrocompetent YMC/2013/D, and clones were selected on chloramphenicol-LB agar plates. pKD46 was digested with XmnI to replace the ampicillin-resistance gene blaamp with the spectinomycin resistance gene blaspec to generate pKD46-spec. The spectinomycin gene was amplified from pKDsgRNA-ack using primers Spec-XmnI-F and Spec-XmnI-R. pCas9 containing YMC/2013/D was made electrocompetent again, and pKD46-spec-transformed cells were selected on spectinomycin LB plates. Plasmid pCRISPR::ampG was constructed by annealing oligonucleotides (ampG.cRNA.S and ampG.cRNA.AS) and ligating the product to BsaI-digested pCRISPR as described in the Marraffini pCRISPR protocol [19]. A 60-nt ssDNA oligonucleotide (ampG::STOP.lead or ampG:STOP.lag) encoding two consecutive stop codons in the ampG open reading frames and pCRISPR::ampG were mixed with 40 µl electrocompetent YMC/2013/D (pKD46-spec, pCas9). Transformants were selected using kanamycin and chloramphenicol.
Allelic replacement mutagenesis using lambda Red recombineering
The ampG deletion mutant strain was constructed by gene replacement via double crossover recombination as described previously with a few modifications [20]. Briefly, three different fragments (including the upstream and downstream fragments of ampG) and the zeocin resistance cassette fragment were amplified using three different primer pairs i.e. Up_ampG_F/R, Zeo_F/R and Down_ampG_F/R (Table S3). The reverse primer of the upstream fragment and forward primer of the downstream fragment of ampG at their 5' ends included a 15–20-nt extension homologous to the primers used to amplify the zeocin marker gene. Nested overlap-extension PCR was performed using an equal concentration of the three fragments to generate a linear DNA template containing the zeocin marker gene flanked by both the upstream and downstream homologous regions. Plasmid pKD-sgRNA-ack was transformed into electrocompetent KE-Y1, and clones were screened on LB plates containing spectinomycin. Strain KE-Y1 containing pKD-sgRNA was grown in LB broth until reaching OD600 = 0.2, and lambda Red recombinase was expressed by adding 0.2% arabinose and with an additional 1.5-h incubation at 30 °C. Cells were harvested and made electrocompetent, and the linear DNA was electroporated. The sample was plated on low-salt LB agar plates containing 50 μg/ml zeocin and incubated at 37 °C. The ampG gene knockout was confirmed using accuPower PCR Premix (Bioneer, Daejeon, Korea) with different primer sets including Up_ampG_F & Down_ampG_R; Up_ampG_F & ZeoR; ZeoF & Down_ampG_R; ZeoF & ZeoR. The reaction mixture was prepared according to manufacturer instructions with 0.2 μM primer concentration..
Results
β-lactam susceptibility induction by mutagenesis
K. aerogenes KE-Y1 was mated with E. coli SM10 λpir containing the transposon donor vector pBTK30 encoding the aacC1 gentamicin 3'-acetyltransferase for clone selection. More than 100,000 colonies were screened to yield two transconjugants of K. aerogenes KE-Y1 susceptible to carbapenem, K. aerogenes KE-Y3 and K. aerogenes KE-Y6. The minimum inhibitory concentrations (MIC) of these strains are shown in Table 1. Transposon insertion led to a 16-fold decrease in ertapenem MIC (from 8 to 0.5 μg/ml) and a 32-fold decrease in the meropenem MIC (from 8 to 0.25 μg/ml). Both mutant strains were also susceptible and showed decreased MICs to piperacillin-tazobactam (32-fold), cefotaxime (64-fold), ceftazidime (64-fold), and aztreonam (64-fold).
Sequencing and comparative analysis
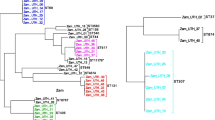
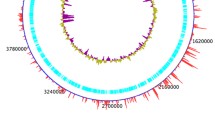
PacBio sequencing yielded circular genomes with 5,296,061, 5,272,156, and 5,266,224 base pairs with 54.8%, 54.9%, and 54.9% GC content for KE-Y1, KE-Y3, and KE-Y6, respectively (Fig. 1). Average Nucleotide Identity (ANI) obtained with MUMmer and BLAST indicated more than 99.8% similarity among all three strains (Additional file 1: Table S1). Whole-genome alignment using Mauve [21] indicated the homology between the three strains with one locally collinear block reversed (Fig. 2).
Circular view of whole-genome alignment of the chromosome of KE-Y1, KE-Y3, and KE-Y6. The first, second and third ring (outermost to inner) represent the BLAST comparisons of KE-Y1, KE-Y3, and KE-Y6 against the wildtype KE-Y1, respectively. The fourth and fifth layers indicate the GC (guanine-cytosine) skew (purple-green) and the GC content (black), respectively. The GC skew indicate the deviation from the average GC content of the three whole-genome sequences. The positions of the genome are marked in the innermost layer. This image was created using BLAST Ring Image Generator
Mauve alignment of KE-Y1 (top), KE-Y3 (middle) and KE-Y6 (bottom) strains. As determined by progressive Mauve alignment, homology (colored blocks) was noted among the three strains. In the KE-Y6 strain, one locally collinear block (~ 36 Mb) was reversed; however, there was no loss of function for any genes
Amino acid alignment and AmpG structure modeling
The amino acid sequence comparison of AmpG KE-Y1 with K. pneumoniae, E. coli and P. aeruginosa showed similarity of 96.33%, 93.08% and 41.58%, respectively. Also, all known activation motif residues (G25, A122, Q124, A181) were conserved (Fig. 3). Based on the homology modeling structure of AmpG KE-Y1, the activation motif residues are located inside the transmembrane. Therefore, it is expected to be similar to the previously known AmpG modeling structure [22].
Amino acid sequence alignment and AmpG KE-Y1 structure modeling. A The Amino acid sequence alignment results of AmpG in 4 species; Klebsiella aerogenes (CP045870), Klebsiella pneumoniae (KN046818), Escherichia coli (CP034384), Pseudomonas aeruginosa (CP053028). Red stars are conserved activation motif residues. B The picture shows the homology modeling structure of AmpG K.aerogenes KE-Y1. The labeled residues are conserved activation motif residues
Open reading frames deactivated by transposon insertions
Whole-genome sequencing (WGS) analysis of KE-Y1, KE-Y3, and KE-Y6 revealed six insertions of gentamicin-encoded transposons. The transposon-mediated inactivated genes are listed in Additional file 1: Table S2. AmpG permease gene insertions were seen in both carbapenem-susceptible transposon insertion strains KE-Y3 and KE-Y6. In strain KE-Y3, the AmpG permease gene was truncated in the middle by the transposon insertion. However, in strain KE-Y6, the transposon inserted immediately upstream of the gene, leading to the loss of the promoter required for AmpG permease gene expression (Fig. 4). The genome position of the genes responsible for induced β-lactamase expression is illustrated in Fig. 5, along with its comparison to that of the wild-type and its transposon mutant derivatives. AmpC-AmpR and AmpD-AmpE are closely linked with each other. The localization of the AmpG, AmpC-AmpR, and AmpD-AmpE genes are similar and identical in all the three strains.
Linear comparison of multiple genomic loci around the AmpG permease between the KE-Y3 vs KE-Y1 (top) and KE-Y6 vs KE-Y1 (bottom). Both images show the transposon insertions in the KE-Y3 and KE-Y6 mutant derivatives. In KE-Y6, AmpG has been truncated in the middle due to the insertion of transposon. However, in strain KE-Y3, transposon insertion is immediately upstream of AmpG, resulting in the loss of the promoter required for its expression
AmpG permease and carbapenem resistance
Complementation of susceptible strains KE-Y3 and KE-Y6 with plasmid pAD123::AmpG reverted the strain's resistance phenotype. We observed a 16-fold (0.5 to 8 μg/ml) increase in ertapenem resistance for both KE-Y3 and KE-Y6, and meropenem resistance was increased eightfold (0.25 to 2 μg/ml) and 16-fold (0.25 to 4 μg/ml) for both strains, respectively. In addition, the complemented strains were resistant to piperacillin-tazobactam (≥ 128 μg/ml), cefotaxime (≥ 128 μg/ml), ceftazidime (≥ 32 μg/ml), and aztreonam (≥ 32 μg/ml). AmpG gene knockout from the wild-type KE-1 using allelic replacement mutagenesis induced susceptibility to most of the antibiotics tested, and the MICs were similar to the transposon mutants KE-Y3 and KE-Y6.
CRISPR–Cas9 mediated AmpG permease knockout
The clinical isolate K. pneumoniae-YMC/2013/D was included for CRISPR–Cas9-mediated gene knockout studies to evaluate the role of AmpG permease in another Gram-negative multidrug resistant pathogen. The CRISPR/Cas9 knockout system, coupled with lambda Red recombineering, has been used to overcome cell death caused by double-stranded DNA breaks in Enterobacteriaceae. An AmpG knockout in K. pneumoniae-YMC/2013/D induced susceptibility to carbapenems. MIC data indicated a ≥ fourfold (≥ 0.32 to 8 μg/ml) and twofold (8 to 4 μg/ml) decrease in resistance to imipenem and meropenem, respectively, compared to the wild-type isolate.
Discussion
Our results demonstrate that carbapenemases do not mediate carbapenem resistance in K. aerogenes. Thus, control and management of carbapenem resistance should not be focused solely on the use of carbapenemases. Carbapenem resistance can also be due to overexpression of AmpC, efflux pumps, and porin loss (or a combination of these). To the best of our knowledge, this is the first report of K. aerogenes and the role of AmpG permease on carbapenem resistance following induction of ampC during antibiotic-induced stress. This stress may lead to many other mutations in the bacterial genome that cause resistance to a wide variety of non-carbapenem antibiotics such as cefotaxime, ceftazidime and cefepime [10].
AmpC is a chromosomally encoded group I, class C cephalosporinase produced by K. aerogenes at basal levels. The presence of β-lactams, such as cefoxitin and imipenem, highly induce AmpC expression [23], which involves a complex network of regulatory genes closely linked with peptidoglycan recycling [24]. During antibiotic treatment, the balance of peptidoglycan synthesis is compromised, releasing GlcNAc-anhydro-MurNAc-oligopeptides into the periplasm. Resulting murapeptides are transported into the cytosol by an AmpG transporter encoded by ampG [25]. However, AmpG has no influence on ampC induction, nor does it show a gene dosage effect [26]. Upon entry into the cytosol, the GlcNAc sugar residue is removed by β-N-acetylglucosaminidase (NagZ) to generate 1,6-anhydromuropeptide, which is processed by N-acetyl-anhydromuramyl-L-alanine amidase (AmpD) during the non-induced state [27, 28]. However, growth in the presence of β-lactams leads to increased breakdown of peptidoglycan or mutations in the ampD gene and may eventually lead to an increased intracellular concentration of murapeptides [29]. Another gene, ampE, located near ampD; AmpD modulates the response exerted on β-lactamase expression by AmpE [30].
In the carbapenem-resistant KE-Y1 wild-type strain, this increased concentration of intracellular murapeptides might have induced the AmpC production by interacting with the LysR-type transcriptional regulator AmpR, making the strain resistant to carbapenems [31]. The ampR gene is located immediately upstream of ampC and encodes a DNA-binding protein that activates ampC [32, 33].
Consistent with the above observations and data obtained in this study, the ampG knockout implicates this protein as a potential pharmaceutical target for controlling ampC hyper-expression. Studies indicate that cells lacking AmpD or AmpG lose 40% of the peptidoglycan layer per generation [31]. Both carbapenem-susceptible transposon insertion mutants KE-Y3 and KE-Y6 contained an ampD gene; therefore, the susceptibility to the carbapenem was due to the loss of functional ampG. Similar studies previously suggested the role of AmpG in the antimicrobial resistance of P. aeruginosa and K. cloacae [26, 34].
Conclusions
To our knowledge, this is the first report illustrating the role of AmpG in carbapenem resistance in K. aerogenes. We used knockout studies with transposon mutagenesis in K. aerogenes KE-1 and the CRISPR–Cas9 gene knockout system to inactivate AmpG permease in a multidrug-resistant clinical isolate of K. pneumoniae. Additionally, using gene complementation, we reversed carbapenem resistance to validate our findings. Future studies should explore additional AmpG protein inhibitors as therapeutic drugs for controlling antibiotic resistance. Also, the other inactivated genes in our study due to mutagenesis should be further analyzed to detect any additional change in the phenotype. The transposon mutagenesis approach could be used to understand novel resistance mechanisms in other classes of bacteria to potentially identify other antibacterial targets.
Availability of data and materials
The WGS data of KE-Y1, KE-Y2, and KE-Y3 generated by the PacBio RS II sequencing system are under the GenBank accession numbers CP045870, CP045869 and CP045868, respectively.
References
Mouloudi E, Protonotariou E, Zagorianou A, Iosifidis E, Karapanagiotou A, Giasnetsova T, et al. Bloodstream infections caused by Metallo-β-Lactamase/Klebsiella pneumoniae Carbapenemase-Producing K pneumoniae among intensive care unit patients in greece: risk factors for infection and impact of type of resistance on outcomes. Infect Contr Hosp Epidemiol. 2010;31(12):1250–6.
Perl TM, Dvorak L, Hwang T, Wenzel RP. Long-term survival and function after suspected gram-negative sepsis. JAMA. 1995;274(4):338–45.
Mehrad B, Clark NM, Zhanel GG, Lynch JP 3rd. Antimicrobial resistance in hospital-acquired gram-negative bacterial infections. Chest. 2015;147(5):1413–21.
Golan Y. Empiric therapy for hospital-acquired, Gram-negative complicated intra-abdominal infection and complicated urinary tract infections: a systematic literature review of current and emerging treatment options. BMC Infect Dis. 2015;15(1):1–7.
Donskey CJ. Antibiotic regimens and intestinal colonization with antibiotic-resistant gram-negative bacilli. Clin Infect Dis. 2006;43(Suppl 2):S62–9.
Todar K. Todar's Online Textbook of Bacteriology 2004. Available from: http://textbookofbacteriology.net/normalflora_3.html.
Diene SM, Merhej V, Henry M, El Filali A, Roux V, Robert C, et al. The rhizome of the multidrug-resistant Enterobacter aerogenes genome reveals how new “killer bugs” are created because of a sympatric lifestyle. Mol Biol Evol. 2013;30(2):369–83.
Davin-Regli A, Pages JM. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front Microbiol. 2015;6:392.
Arpin C, Coze C, Rogues AM, Gachie JP, Bebear C, Quentin C. Epidemiological study of an outbreak due to multidrug-resistant Enterobacter aerogenes in a medical intensive care unit. J Clin Microbiol. 1996;34(9):2163–9.
Jacoby GA. AmpC beta-lactamases. Clin Microbiol Rev. 2009;22(1):161–82.
Siedner MJ, Galar A, Guzman-Suarez BB, Kubiak DW, Baghdady N, Ferraro MJ, et al. Cefepime vs other antibacterial agents for the treatment of Enterobacter species bacteremia. Clin Infect Dis. 2014;58(11):1554–63.
Kuga A, Okamoto R, Inoue M. ampR gene mutations that greatly increase class C beta-lactamase activity in Enterobacter cloacae. Antimicrob Agents Chemother. 2000;44(3):561–7.
Schmidtke AJ, Hanson ND. Model system to evaluate the effect of ampD mutations on AmpC-mediated beta-lactam resistance. Antimicrob Agents Chemother. 2006;50(6):2030–7.
Pyne ME, Moo-Young M, Chung DA, Chou CP. Coupling the CRISPR/Cas9 system with lambda red recombineering enables simplified chromosomal gene replacement in Escherichia coli. Appl Environ Microbiol. 2015;81(15):5103–14.
Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166(4):557–80.
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–9.
Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–38.
Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31(3):233–9.
Aranda J, Poza M, Pardo BG, Rumbo S, Rumbo C, Parreira JR, et al. A rapid and simple method for constructing stable mutants of Acinetobacter baumannii. BMC Microbiol. 2010;10:279.
Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5(6):e11147.
Li P, Ying J, Yang G, Li A, Wang J, Lu J, et al. Structure-function analysis of the transmembrane protein AmpG from Pseudomonas aeruginosa. PLoS ONE. 2016;11(12):e0168060.
Livermore DM. beta-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8(4):557–84.
Normark S. beta-Lactamase induction in gram-negative bacteria is intimately linked to peptidoglycan recycling. Microb Drug Resist. 1995;1(2):111–4.
Chahboune A, Decaffmeyer M, Brasseur R, Joris B. Membrane topology of the Escherichia coli AmpG permease required for recycling of cell wall anhydromuropeptides and AmpC beta-lactamase induction. Antimicrob Agents Chemother. 2005;49(3):1145–9.
Korfmann G, Sanders CC. ampG is essential for high-level expression of AmpC beta-lactamase in Enterobacter cloacae. Antimicrob Agents Chemother. 1989;33(11):1946–51.
Holtje JV, Kopp U, Ursinus A, Wiedemann B. The negative regulator of beta-lactamase induction AmpD is a N-acetyl-anhydromuramyl-L-alanine amidase. FEMS Microbiol Lett. 1994;122(1–2):159–64.
Lee M, Zhang W, Hesek D, Noll BC, Boggess B, Mobashery S. Bacterial AmpD at the crossroads of peptidoglycan recycling and manifestation of antibiotic resistance. J Am Chem Soc. 2009;131(25):8742–3.
Lindquist S, Galleni M, Lindberg F, Normark S. Signalling proteins in enterobacterial AmpC beta-lactamase regulation. Mol Microbiol. 1989;3(8):1091–102.
Kong KF, Schneper L, Mathee K. Beta-lactam antibiotics: from antibiosis to resistance and bacteriology. APMIS. 2010;118(1):1–36.
Park JT. Identification of a dedicated recycling pathway for anhydro-N-acetylmuramic acid and N-acetylglucosamine derived from Escherichia coli cell wall murein. J Bacteriol. 2001;183(13):3842–7.
Honore N, Nicolas MH, Cole ST. Inducible cephalosporinase production in clinical isolates of Enterobacter cloacae is controlled by a regulatory gene that has been deleted from Escherichia coli. EMBO J. 1986;5(13):3709–14.
Nicolas MH, Honore N, Jarlier V, Philippon A, Cole ST. Molecular genetic analysis of cephalosporinase production and its role in beta-lactam resistance in clinical isolates of Enterobacter cloacae. Antimicrob Agents Chemother. 1987;31(2):295–9.
Zamorano L, Reeve TM, Juan C, Moya B, Cabot G, Vocadlo DJ, et al. AmpG inactivation restores susceptibility of pan-beta-lactam-resistant Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother. 2011;55(5):1990–6.
Funding
This work was supported by the BioNano Health-Guard Research Center funded by the Ministry of Science, ICT & Future Planning (MSIP) of Korea as a Global Frontier Project (H-GUARD_2014M3A6B2060509); the Research Program funded by the Korea Centers for Disease Control and Prevention(2019-ER5403-00); a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (Grant Number: HI14C1324). This work was also supported by the Brain Korea 21 PLUS Project for Medical Science, Yonsei University.
Author information
Authors and Affiliations
Contributions
DY and RD designed the study. RD, NP, NLP, NTV and HL performed the experiments. RD, NLP and DY analyzed and interpreted the data and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
None.
Consent for publication
None.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Average nucleotide analysis using BLAST and MUMmer. Table S2. List of genes interrupted due to transposon insertion in the mutagenized strains. Table S3. Oligonucleotides used in this study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
D’Souza, R., Nguyen, L.P., Pinto, N.A. et al. Role of AmpG in the resistance to β-lactam agents, including cephalosporins and carbapenems: candidate for a novel antimicrobial target. Ann Clin Microbiol Antimicrob 20, 45 (2021). https://doi.org/10.1186/s12941-021-00446-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12941-021-00446-7