Abstract
Treatment of infections by Pseudomonas aeruginosa forming biofilms after antimicrobial testing on planktonic bacteria can result in substantial failure. Therefore, we offer a robust and simple experimental platform to test the impact of antimicrobials on biofilms. Antibiotic response patterns varied uniquely within biofilm formation capacity and minimal biofilm eradication concentrations (MBECs) has a significantly better discriminatory power than minimum inhibitory concentrations (MICs) to differentiate the overall efficiency of antibiotics to eradicate biofilm. Our resazurin-based 96-well-plate platform is able to emulate bacterial responses to antibiotics under biofilm conditions in a fast, simple, and cost-effective screening method adaptable to automation, and warrants trials in the clinic.
Similar content being viewed by others
Introduction
The properties of bacteria in biofilms differ from those of planktonic bacteria [1, 2], and bacteria in biofilms have extreme tolerance to immune responses and antimicrobial therapy [3, 4]. Biofilm formation is therefore an obstacle to the treatment of chronic infections with Pseudomonas aeruginosa, most of which are associated with biofilms [1, 2]. Despite the negative impact of biofilms, to our knowledge, no treatment that directly targets bacteria in biofilms has yet been developed [1, 5].
Biofilm recalcitrance to antibiotics is based on a mixture of resistance and tolerance [1, 6]. Clinical treatments with antibiotics are usually determined from minimum inhibitory concentrations (MICs) for planktonic bacteria, and, as a result, patients may suffer from persistent infection over the course of weeks, or even months, often with recurrence of even more aggressive exacerbations [1, 7]. Patients harboring bacteria within biofilms require higher doses of antibiotics and more prolonged courses of treatment than treatment suggested by testing with planktonic bacteria [1, 8].
Patients with chronic infections treated with antibiotic regimens based on biofilm susceptibility-testing have better clinical outcomes than those treated with regimens based on methods measuring susceptibility to planktonic bacteria [5, 9].
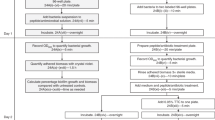
In a previous study, we developed a simplified antibiotic susceptibility assay based on a standardized model to quantify viable cells in biofilms of Acinetobacter baumannii [10, 11]. Our assay is based on the quantitative measurement of metabolically active cells using PrestoBlue, a resazurin (7-hydroxy-3H-phenoxazin-3-one-10-oxide)-based viability indicator. The results clearly demonstrated the significant discriminatory power of the assay (MBEC) to differentiate antibiotic efficacy on biofilms compared with current MIC-based assays [10, 11]. While the new assay has proven to present an effective model of biofilm formation, in this article we describe its reproducibility and applicability for rapid antibiotic susceptibility testing of P. aeruginosa biofilms in a clinical laboratory setting.
Materials and methods
Strains and culture conditions
Clinical isolates used in this study were selected from a P. aeruginosa strain repository in the Department of Microbiology, King Chulalongkorn Memorial Hospital, Bangkok, Thailand. The strains were stored at the repository collection after standard characterization and identification, including 16S rRNA sequencing as described previously [12] (Additional file 1). Clinical strains were isolated during 2016–2017 from chronically-infected patients as part of their standard care. The P. aeruginosa clinical isolates were cultured on Müller-Hinton agar plates at 37 °C. Without preference, we selected 137 unduplicated clinical isolates representing 137 patients and 14 collection sites with relevant chronic infection (including urine, bile, corneal scrapings, nasal swabs, tissue, blood, device related, broncho–alveolar aspirates, ear swabs, eye swabs, conjunctival swabs, wound pus, endotracheal aspirates, and sputum). Strains from patients with multiple sites of infection were excluded, and we only included samples from patients with infection at a single site. All isolates were stored at − 80 °C in tryptic soy broth with 15% glycerol until used in subsequent experiments.
Antibiotics and agents
The biofilm eradication activity of 7 antibiotics was tested against the subset of clinical isolates (n = 137). Gentamicin, amikacin, ciprofloxacin, meropenem, colistin, and ceftazidime were all from Sigma-Aldrich. Susceptibility testing for fosfomycin (Wako Chemicals) was determined by supplementation with 25 μg/mL glucose-6-phosphate. Antibiotic stock solutions were prepared less than 24 h before use. Antibiotics were dissolved in cation-adjusted Müller-Hinton II broth (MHIIB) (Becton Dickinson) medium and sterilized by filtration through a membrane (0.22 μm pores). Serial dilutions of the stocks were prepared in MHIIB immediately before use.
Testing susceptibility to antibiotics
The planktonic MIC were established using standard techniques according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria [13] and Clinical and Laboratory Standards Institute (CLSI) guidelines [14]. Escherichia coli ATCC 25922, and P. aeruginosa ATCC 27853 were used as quality control strains. Minimal biofilm eradication concentrations (MBEC) were established using our previously develop fluorometric-based assay to calculate the number of viable cells within the biofilm as described previously [10]. In brief, MBECs were determined by adding the serially diluted antibiotics to mature biofilms and incubating at 37 °C for 24 h before staining with PrestoBlue. Before adding the antibiotics, any nonadherent cells were removed from the mature biofilms by 3 gentle washes with MHIIB. Cell viability was calculated using the following formula: cell viability (%) = ((mean signal of corresponding well − mean signal of negative control well)/(mean signal of positive control well − mean signal of negative control well)) × 100. Two cut-off values (50% and 75% nonviable cells) were used to determine the MBEC. All experiments were performed in triplicate and repeated 3 times. As a comparison we also used the 96‐well Calgary Biofilm Device (CBD) (Innovotech, Calgary, Canada) as described previously to determine MBEC [15].
Biofilm formation quantification and classification
Two methods were used to quantify [16] and classify [17] the biofilm structure by Crystal Violet staining followed by confocal laser scanning microscopy using live or dead bacterial staining as described previously [18]. Mean absorbances and their standard deviations (SDs) were calculated for all tested strains and negative controls, determined in triplicate and repeated 3 times. The clinical isolates were classified as described previously [17].
Statistical analyses
Continuous variables are summarized using means and SDs, and categorical variables as counts and percentages. Levels of P. aeruginosa drug susceptibility are represented in 2 ways: a continuous measure of concentration; and an ordinal categorical form representing biofilm formation (negative, weak, moderate, or strong); both of these outcomes were measured repeatedly over time for each isolate. Linear mixed modeling was used to compare concentrations between test types (MIC vs. MBEC) over time. We then examined which test types (MIC vs. MBEC) were more successful in allowing concentration to be used to distinguish between biofilm formations (negative, weak, moderate, or strong) using ordinal logistic mixed effects regression. Finally, we examined whether concentration could be used to predict biofilm formation using multinomial logistic regression. All analysis was conducted using the R statistical package [19], linear mixed modeling was performed using the R library, lme4 [20], and ordinal logistic mixed effect modeling using the R library, ordinal [21], and multinomial logistic regression using the R library, nnet [22]. P < 0.05 was considered significant for all inferential analysis.
Results
Association between antibiotic resistance and biofilm formation
The planktonic antibiotic resistance profile of each isolate revealed that resistance to meropenem was most common, followed in order by ceftazidime, ciprofloxacin, and fosfomycin (Fig. 1). Most strains showed high susceptibility to colistin, amikacin, and gentamicin. In total, 127 (92%) isolates were positive for biofilm formation, and 56 (46%) isolates formed a stronger biofilm. No significant difference was found in terms of biovolume between the fluorometric assay and the Calgary Biofilm Device (P = 1.0092; Additional file 2). The composition of the biofilm formation categories with respect to resistance profile showed that antibiotic resistant isolates form stronger biofilms than sensitive isolates (P < 0.001; Fig. 1). Strong and moderate biofilms showed similar levels of enhancement in all 3 antibiotic assessment groups.
Correlation between biofilm formation and susceptibility test type
To determine whether biofilm formation is correlated with susceptibility test type, we compared the biofilm forming capacities between strains with 3 types of tests for each of the 7 antibiotics. We found that an overall MBEC susceptibility test significantly modifies the relationship between biofilm formation and antibiotic concentration (P < 0.001; Fig. 2). Strong and moderate biofilms likely exhibit similar trends for all of the antibiotics tested. The trend is very pronounced for amikacin and fosfomycin (MBEC-75 > MBEC-50 > MIC). Variation of the strong and moderate biofilm in MBEC-75 is much more pronounced for amikacin, meropenem, and ceftazidime than other antibiotics, particularly colistin, where variation was comparatively low. MIC tests did not show any differences in association with weak, moderate, or strong biofilms.
Association between the level of biofilm formation (negative, weak, moderate, or strong) and susceptibility test types to 7 antibiotics for P. aeruginosa clinical isolates. a gentamicin, b amikacin, c ciprofloxacin, d meropenem, e colistin, f fosfomycin, and g ceftazidime. MIC minimum inhibitory concentration of planktonic cells based on conventional susceptibility test, MBEC minimum biofilm eradication concentration based on PrestoBlue cell viability indicator
Relationship between susceptibility test types and antibiotics
A linear mixed model revealed a significant relationship between the type of susceptibility test and antibiotics (Z2LRT = 312.26, 12 df, P < 0.001) showing that the magnitude of differences between tests was modified by antibiotics. Figure 3 shows all antibiotics except meropenem and colistin tended to have the same general pattern (MBEC-75 > MBEC-50 > MIC). Whereas with meropenem, the difference between MIC and MBEC-50 is much more pronounced, and for colistin the difference is much less pronounced. No significant difference was found in terms of MBEC between the fluorometric assay and the Calgary Biofilm Device (P = 0.998; Table 1).
Relationship between susceptibility tests and 7 antibiotics for P. aeruginosa clinical isolates: (1) gentamicin, (2) amikacin, (3) ciprofloxacin, (4) meropenem, (5) colistin, (6) fosfomycin, and (7) ceftazidime. MIC minimum inhibitory concentration of planktonic cells based on conventional susceptibility test, MBEC minimum biofilm eradication concentration based on PrestoBlue cell viability indicator
Association of susceptibility test types, biofilm formation, and antibiotic concentrations
The associations between odd ratios of MIC, MBEC (50 and 75) and concentration attribute of each antibiotic are shown in Table 2. It is important to note that for this analysis we employed standardized concentrations (Z-scores) to avoid higher (raw) values of concentrations making associations appear more trivial. For each antibiotic, the odds ratios from MBEC-50 and 75 tests are a reflection of the higher level of associations than with MIC, except for fosfomycin. Notably, for gentamicin and amikacin the odds ratio of MBEC-50 was higher than MBEC-75, but both displayed a similar level of significance between MBEC-50 and 75. However, for colistin, a similar level of significance was observed for the association between MIC and MBEC-50.
For all the strains tested, the accuracy of biofilm classification was higher for both MBEC-50 and 75 tests compared with a MIC test for each antibiotic. We can see that concentrations using MBEC-50 correctly predicted the biofilm formation in gentamicin, ciprofloxacin, meropenem, and fosfomycin, followed by ceftazidime. MBEC-75 is able to predict biofilm formation for colistin with 58% accuracy, and for amikacin both MBEC-50 and 75 displayed similar levels.
Discussion
Current guidelines or antibiotic therapies, based on planktonic bacteria are often unable to offer a successful path to the treatment of biofilm infections [1, 6]. As biofilm bacteria are inherently more tolerant to antibiotics, it is necessary to determine biofilm specific antibiotic susceptibility to predict therapeutic success.
Therefore, the key advantages of the present assay are: first, that it simplifies the steps of biofilm formation and is able to produce biofilms equivalent to those produced by the Calgary Biofilm Device, making it standard assay system compliant with antibiotic susceptibility testing for biofilm infections. The reproducibility of the results for biofilms formed on each well of the 96-well plate and reproducible biofilm categories (weak, moderate, or strong) attributed to each clinical isolate demonstrate that this fluorometric assay can produce biofilms equivalent to those of each peg of the Calgary Biofilm Device lid. It is therefore possible to provide equivalent and clinically relevant biofilm that can be exposed to multiple antibiotics in a single 96-well plate with viability assessment to provide accurate antibiotic selection in a clinically useful time frame. The assay requires no specific peg lid plates or changing bottom-well plates at each step, making the process much simpler to set up than the Calgary Biofilm Device, and thus eliminates possible contamination and reduces expense. The use of a single 96-well plate for each testing step greatly reduces the time required to determine the antibiotic susceptibilities of biofilms and minimized the workload. The fluorometric-based assay is also amenable to automation because it is based on typical standard 96-well plates.
Second, the assay is a valid way to differentiate antibiofilm effectiveness based on biofilm formation capacity by resembling clinical situations. We observed marked differences between MBEC concentration patterns of each tested antibiotics to weak, moderate, and strong biofilms. Some antibiotics are able to penetrate moderate biofilms readily, but strong biofilms poorly. This is an interesting observation, in that such differences may also reflect more fundamental differences in the biological characteristics of biofilm structure [4, 23], and metabolic or physiological factors [1, 4, 24] of clinical isolates that are not accounted for in MIC testing. Third, a clear difference in antibiotic susceptibility was seen between planktonic populations of each of the isolates tested and the biofilm populations of the same isolate. Each of the isolates had a unique biofilm susceptibility to the each of antibiotics tested. The biofilms of clinical isolates of P. aeruginosa proved to be very difficult to eradicate, with only colistin being effective at achievable drug concentrations, with the other aminoglycoside tested (gentamicin), and ciprofloxacin showing just some activity against weak biofilms. Indeed, the higher levels of significant odds ratios with biofilm formation classification accuracy of MBEC-50 and 75 tests suggest that they have better discriminatory power than an MIC test. The accuracy of biofilm formation classification reflects that, to overcome the 50% cell death in a biofilm is crucial for the efficacy of particular antibiotics. The manner in which biofilm-induced tolerance and intrinsic resistance become integrated to promote biofilm-specific antibiotic resistance was shown. These data could be interpreted to indicate that the MIC is predictive of antibiotic efficacy against planktonic bacterial cells, but not for those living within biofilms. This is consistent with what is often seen as symptomatic relief by eliminating the planktonic population [1, 6, 25]. However, because the biofilm is not eliminated by antibiotic treatment, reinfection occurs once the antibiotic is removed [1, 6, 25].
The present work is limited by the following considerations. In the presently described assay, the effect of antibiotics on biofilm were determined, although appropriate standard reference values required to clear infections in vivo remain unclear. Combining the present quantitative screening of bacterial biofilm-specific antibiotic resistance with clinical trials of antibiotics would clarify the clinical applicability of the assay.
In conclusion, the presently described quantitative screening assay of bacterial biofilm-specific antibiotic resistance assay is a versatile, easy to manage, and robust method that should help to improve treatment of infections that are threats in the clinic.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its Additional information.
Abbreviations
- ATCC:
-
American Type Culture Collection
- CLSI:
-
Clinical and Laboratory Standards Institute
- EUCAST:
-
European Committee on Antimicrobial Susceptibility Testing
- MHIIB:
-
Müller-Hinton II broth
- MBEC:
-
Minimum biofilm eradication concentration
- MIC:
-
Minimum inhibitory concentration
- MBEC-75:
-
75% Nonviable cells
- MBEC-50:
-
50% Nonviable cells
- OD:
-
Optical density
- SD:
-
Standard deviations
References
Lebeaux D, Ghigo JM, Beloin C. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev. 2014;78:510–43.
Grant SS, Hung DT. Persistent bacterial infections, antibiotic tolerance, and the oxidative stress response. Virulence. 2013;4:273–83.
Gellatly SL, Hancock RE. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathogens Dis. 2013;67:159–73.
Rybtke M, Hultqvist LD, Givskov M, Tolker-Nielsen T. Pseudomonas aeruginosa biofilm infections: community structure, antimicrobial tolerance and immune response. J Mol Biol. 2015;427:3628–45.
Malone M, Goeres DM, Gosbell I, Vickery K, Jensen S, Stoodley P. Approaches to biofilm-associated infections: the need for standardized and relevant biofilm methods for clinical applications. Expert Rev Anti Infect Therapy. 2017;15:147–56.
Pragman AA, Berger JP, Williams BJ. Understanding persistent bacterial lung infections: clinical implications informed by the biology of the microbiota and biofilms. Clin Pulm Med. 2016;23:57–66.
Barsoumian AE, Mende K, Sanchez CJ Jr, Beckius ML, Wenke JC, Murray CK, et al. Clinical infectious outcomes associated with biofilm-related bacterial infections: a retrospective chart review. BMC Infect Dis. 2015;15:223. https://doi.org/10.1186/s12879-015-0972-2.
Høiby N, Bjarnsholt T, Moser C, Bassi GL, Coenye T, Donelli G, et al. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect. 2015;21(1):S1–25.
Waters V, Ratjen F. Standard versus biofilm antimicrobial susceptibility testing to guide antibiotic therapy in cystic fibrosis. Cochrane Database Syst Rev. 2017;5(10):CD009528.
Wannigama DL, Hurst C, Pearson L, Saethang T, Singkham-in U, Luk-in S, et al. Simple fluorometric-based assay of antibiotic effectiveness for Acinetobacter baumannii biofilms. Sci Rep. 2019;9:6300. https://doi.org/10.1038/s41598-019-42353-0.
Wannigama DL, Hurst C, Pearson L, Saethang T, Singkham-in U, Luk-in S, et al. A simple antibiotic susceptibility assay for Pseudomonas aeruginosa and Acinetobacter baumannii biofilm could lead to effective treatment selection for chronic lung infections. Chest. 2019;155:77A.
Spilker T, Coenye T, Vandamme P, LiPuma JJ. PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol. 2004;42(5):2074–9.
EUCAST. Clinical Breakpoints. European Committee for Antimicrobial Susceptibility Testing 2016; version 6.
CLSI. Performance standards for antimicrobial susceptibility testing—Twenty-ninth informational supplement. Clinical and Laboratory Standards Institute. 2019;M100(S29).
Macià MD, Rojo-Molinero E, Oliver A. Antimicrobial susceptibility testing in biofilm-growing bacteria. Clin Microbiol Infect. 2014;20:981–90.
O’Toole GA. Microtiter dish biofilm formation assay. J Vis Exp. 2011. https://doi.org/10.3791/2437.
Stepanović S, Vuković D, Hola V, Di Bonaventura G, Djukić S, Ćirković I, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. 2007;115:891–9.
Musken M, Di Fiore S, Romling U, Haussler S. A 96-well-plate-based optical method for the quantitative and qualitative evaluation of Pseudomonas aeruginosa biofilm formation and its application to susceptibility testing. Nat Protoc. 2010;5:1460–9.
RCore Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2016.
Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:1–48.
Christensen RHB. ordinal—Regression models for ordinal data. R package, version 20156-28; http://www.cranr-projectorg/package=ordinal/. 2015.
Ripley BD, Venables WN. Modern applied statistics with S. 4th ed. New York: Springer; 2002.
Taylor PK, Yeung ATY, Hancock REW. Antibiotic resistance in Pseudomonas aeruginosa biofilms: towards the development of novel anti-biofilm therapies. J Biotechnol. 2014;191:121–30.
Balcazar JL, Subirats J, Borrego CM. The role of biofilms as environmental reservoirs of antibiotic resistance. Front Microbiol. 2015;6:1216. https://doi.org/10.3389/fmicb.2015.01216.
Romling U, Balsalobre C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J Intern Med. 2012;272:541–61.
Acknowledgements
We thank the staff of the bacteriology division, Department of Microbiology at King Chulalongkorn Memorial Hospital, for providing the P. aeruginosa clinical isolates.
Funding
This work was supported by a grant from the 90th Year Anniversary Ratchadapiseksompotch Endowment Fund from the Faculty of Medicine and Graduate School, Chulalongkorn University, Bangkok, Thailand (batch No. 39 (2/61)). Dhammika Leshan Wannigama was supported under the 100th Anniversary Chulalongkorn University Fund for Doctoral Scholarships, The University of Western Australia for Cross Institutional Exchange Doctoral Fellowship and Graduate School, Chulalongkorn University, for Overseas Research Experience Scholarship. The sponsor(s) had no role in study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Author information
Authors and Affiliations
Contributions
DLW: Conceptualization, investigation, data curation, formal analysis, writing—original draft. CH, PH, LP: conceptualization, data curation, formal analysis, supervision, writing—review and editing. TS: Data curation, formal analysis, methodology, validation, writing—review and editing. US-in, SL-in: Data curation, investigation. RJS: Conceptualization, formal analysis, supervision, writing—review and editing. TC: conceptualization, funding acquisition, supervision, writing—review and editing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study protocol was approved by the Institutional Review Board (IRB) of the Faculty of medicine, Chulalongkorn University, Bangkok, Thailand (certificate of Approval No. 745/2017, IRB No. 414/60).
Competing interests
All authors have read the journal’s authorship agreement and policy on disclosure of potential conflicts of interest and none of the authors declare any potential competing interests.
Informed consent
For this retrospective study of anonymous clinical isolates, the requirement for informed consent from patients was waived by the IRB.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
Biofilm formation and Testing susceptibility to antibiotics.
Additional file 2: Figure S1.
Distribution of clinical isolates (n = 137) of P. aeruginosa biovolume within the fluorometric-based assay and Calgary Biofilm Device.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wannigama, D.L., Hurst, C., Hongsing, P. et al. A rapid and simple method for routine determination of antibiotic sensitivity to biofilm populations of Pseudomonas aeruginosa. Ann Clin Microbiol Antimicrob 19, 8 (2020). https://doi.org/10.1186/s12941-020-00350-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12941-020-00350-6