Abstract
Background
We aimed to develop and validate a predictive model for the overall survival (OS) of patients with nasopharyngeal carcinoma (NPC).
Methods
Overall, 519 patients were retrospectively reviewed in this study. In addition, a random forest model was used to identify significant prognostic factors for OS among NPC patients. Then, calibration plot and concordance index (C-index) were utilized to evaluate the predictive accuracy of the nomogram model.
Results
We used a random forest model to select the three most important features, dNLR, HGB and EBV DNA, which were significantly associated with the OS of NPC patients. Furthermore, the C-index of our model for OS were 0.733 (95% CI 0.673 ~ 0.793) and 0.772 (95% CI 0.691 ~ 0.853) in the two cohorts, which was significantly higher than that of the TNM stage, treatment, and EBV DNA. Based on the model risk score, patients were divided into two groups, associated with low-risk and high-risk. Kaplan–Meier curves demonstrated that the two subgroups were significantly associated with OS in the primary cohort, as well as in the validation cohort. The nomogram for OS was established using the risk score, TNM stage and EBV DNA in the two cohorts. The nomogram achieved a higher C-index of 0.783 (95% CI 0.730 ~ 0.836) than that of the risk score model 0.733 (95% CI 0.673 ~ 0.793) in the primary cohort (P = 0.005).
Conclusions
The established risk score model and nomogram resulted in more accurate prognostic prediction for individual patient with NPC.
Similar content being viewed by others
Background
Nasopharyngeal carcinoma (NPC) is an endemic malignancy characterized by its unique geographic distribution [1]. NPC contributes to a large part of the overall cancer burden among prevalent areas, including southern China, southeast Asia and northern Africa [2, 3]. Radiotherapy is the mainstay treatment for non-metastatic NPC. Chemotherapy combined with radiotherapy is recommended for treatment of advanced NPC [4, 5]. However, the current TNM staging system that is utilized for guidance of the different treatment regimens is insufficient, as many varied clinical outcomes of patients at same stages have been reported [6]. Therefore, there is a need for more accurate indicators to predict prognosis to achieve effective clinical treatment.
Recently, there have been several attempts to improve NPC prognostication through the use of blood molecular biomarkers, including circulating Epstein-Barr virus (EBV) DNA, serum lactate dehydrogenase (LDH) [7], globulin (GLOB), hs-CRP [8], and neutrophil/lymphocyte ratio (NLR) [9]. The infection of EBV is virtually 100% associated with NPC in endemic areas. In addition, the plasma EBV DNA has gradually been used in clinical applications and is considered to be the most attractive potential biomarker to complement TNM staging system [10]. The derived neutrophils to leukocytes ratio (dNLR) has been linked to the inflammatory status and clinical outcomes among several cancers, including NPC [11]. This indicates that dNLR likely has prognostic value and also has the advantage of being inexpensive and easy to calculate. Hemoglobin (HGB) levels have been regarded as important determinants of outcome for a number of cancers treated with radiotherapy, especially gynecological tumors and NPC [12]. However, it remains a challenge to screen and incorporate biomarkers into a new staging system for NPC patients.
A random forest model is an effective classifier as it can predict class of input, and select its most important features [13]. Nomograms have currently been proven to be an effective tool to predict prognosis of patients with cancers, including lung cancer [14], rectal cancer [15], and gastric cancer [16]. In this study, we utilized a random forest model to screen factors related to prognosis of NPC, and incorporate them into a new staging system by establishing a nomogram model.
Materials and methods
Patients and clinical characteristic
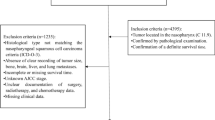
We retrospectively reviewed the patients with first diagnosed NPC at Sun Yat-Sen University Cancer Center (SYSUCC) between January 2009 and December 2011. Patients included criteria: (1) All patients were pathologically diagnosed as NPC in SYSUCC for the first time. (2) Patients were not any malignancies besides NPC. (3) Patients did not undergo anti-tumor therapy, and data was collected prior to anti-tumor therapy. (4) No patients were infected with hepatitis B virus or hepatitis C virus. (5) Complete records in the database of medical information, and follow-up data. All the patients were classified based on the 8th edition of the AJCC TNM staging guidelines.
We collected information regarding gender, age, family history, smoking, body mass index(BMI), TNM stage, treatment, white blood cells (WBC), neutrophils, lymphocyte, monocyte, platelets, HGB, NLR, neutrophils/(WBC-neutrophils) ratio (dNLR), lymphocyte/monocyte ratio (LMR), platelet/lymphocyte ratio (PLR), systemic immune-inflammation index (SII), total protein (TP), albumin (ALB), GLOB, ALB/GLOB ratio (AGR), C-reactive protein (CRP), CRP/ALB ratio (CAR), apolipoprotein B (APOA), apolipoprotein B (APOB), APOA/APOB ratio (ABR), LDH, high density lipoprotein(HDL), EBV DNA, virus capsid antigen specific immunoglobulin A (VCA-IgA), early antigen immunoglobulin A (EA-IgA), prognostic nutritional index (PNI) and prognostic index (PI). The PNI was calculated based on the following formula: Alb (g/L) + 5 × lymphocyte count × 109/L [17]. The SII was calculated based on this formula: PLR × Neutrophil × 109/L [18]. The PI score was 0 for patients that had CRP levels of 10 mg/L or less, and a WBC count of 11 × 109/L or less. Patients with only one of these abnormalities were given a score of 1, and patients that had an elevation of both CRP and WBC were allocated a score of 2 [19]. The patients’ data were collected prior to any treatment.
Statistical analysis
Data analyses was carried out using SPSS standard version 20.0 (SPSS, Chicago, USA) and R software version 3.6.1 (http://www.R-project.org). The cut-off values were determined using the R package “survival” and “survminer”. The Kaplan–Meier method was utilized to estimate OS of patients in high-risk and low-risk groups. The concordance index (C-index) was utilized to assess discriminative ability and predictive accuracy of established random forest model and nomogram. The C-index was calculated and compared using the “survcomp” package. The area under the curve (AUC) was computed using the “survivalROC” package. Calibration of the nomogram for 1-, 3-, and 5-year OS was executed via comparison of the predicted survival and observed survival. All statistical tests were two-tailed, and a P value less than 0.05 was considered to be statistically significant.
Results
Patients and clinical characteristics
In total, 519 NPC patients were enrolled in this study. All patients were randomly divided into either the primary cohort (n = 363) or the validation cohort (n = 156). The patients’ demographic data and clinical characteristics in both the primary cohort and validation cohort are described in Table 1. In the primary cohort, 209 (57.57%) patients were males and 154 (42.43%) were females. The mean age (SD) of patients in the primary cohort was 46.05 (10.87) years, and the median OS was 51.0 months (interquartile range [IQR]: 42.3–66.7 months). In the validation cohort, 92 (58.97%) patients were males and 64 (41.03%) were females. The mean age (SD) was 46.87 (11.58) years, while the median OS was 50.4 months (IQR: 41.7–66.0 months). In the primary cohort, the 1-, 3-, and 5-year OS rates were 95.0%, 84.0% and 46.8%, respectively. In the validation cohort, the 1-, 3-, and 5-year OS rates were 98.7%, 84.0% and 45.5%, respectively.
Model construction based on clinical characteristics
In the primary cohort, we utilized a random forest model to select the most significant variables from 34 different clinical features. In addition, we used the sliding windows sequential forward feature selection method (SWSFS) to identify important variables by minimizing the ‘out of bag (OOB)’ error rate (Fig. 1A). According to the minimum OOB, three variables including dNLR (HR = 1.14; 95% CI 1.05–1.23; P = 9.14 × 10–4), HGB (HR = 0.98; 95% CI 0.97–0.99; P = 5.24 × 10–3) and EBV DNA (HR = 1.59, 95% CI 1.32–1.93, P = 1.22 × 10–6) were found to be significantly associated with OS among NPC patients (Fig. 1B). Finally, we constructed a risk score model that included dNLR, HGB and EBV DNA. The computational formula of the risk score was as follows: risk score = (0.466 × DNA) + (0.129 × dNLR) − (0.02 × HGB). The heatmap of NPC samples among the two cohorts are shown in Fig. 2, in which red represents upregulated imaging features and blue represents downregulated imaging features. The three feature clusters (C1–C3) were identified within the heatmap using unsupervised hierarchical clustering of 3 imaging features.
Heatmap were generated by unsupervised hierarchical clustering of 3 features (Y axis) across identified NPC patients on X-axis revealed 3 major image feature patterns in the primary cohort (A) and the validation cohort (B). The corresponding treatment and TNM stage that the tumor was derived from are shown above the color bars
Model evaluation
ROCs were utilized to evaluate the accuracy of the established risk score model, TNM stage, treatment, and EBV DNA. In the primary cohort, for the 1-year OS (Fig. 3A), the AUC of TNM stage, treatment, EBV DNA and our established model were 0.748, 0.591, 0.751 and 0.797, respectively. Moreover, our model achieved a higher AUC than the TNM stage, treatment, EBV DNA for both the 3-year OS (Fig. 3B) and 5-year OS (Fig. 3C). In the validation cohort, for the 1-year OS (Fig. 3D), the AUC of TNM stage, treatment, EBV DNA, and our established model were 0.399, 0.588, 0.932 and 0.854, respectively. For 3-year and 5-year OS, the AUC of TNM stage, treatment, EBV DNA, and our established model were 0.728, 0.573, 0.794, 0.821 and 0.725, 0.555, 0.747, 0.791, respectively (Fig. 3E, F). The results of a time-dependent ROC curve for OS in the primary (Fig. 4A) and validation cohort (Fig. 4B) demonstrated the AUCs of TNM stage, treatment, EBV DNA and our established model in more detail.
Moreover, we evaluated C-Index of the established model, TNM stage, treatment and EBV DNA to predict of OS in the primary and validation cohort. In the primary cohort, the established model achieved a higher C-index of 0.733(95% CI 0.673–0.793) than the TNM stage (0.712; 95% CI 0.657 ~ 0.768), treatment (0.542; 95% CI 0.505 ~ 0.580) and EBV DNA (0.691; 95% CI 0.626 ~ 0.756). In the validation cohort, the C-index of our model, TNM stage, treatment and EBV DNA were 0.772 (95% CI 0.691 ~ 0.853), 0.699 (95% CI 0.628 ~ 0.770), 0.551 (95% CI 0.503 ~ 0.600), 0.739 (95% CI 0.652 ~ 0.826), respectively (Table 2).
Performance of the established model in stratifying risk
Based on the computational formula of risk score (0.466 × DNA + 0.129 × dNLR − 0.02 × HGB), NPC patients were subdivided into high risk (risk score ≤ −0.16) and low risk (risk score > −0.16). We used the R package “survival” and “survminer” in order to determine the cut-off values. The optimum cut-off of our model was − 1.46. The results demonstrated that patients with a high-risk score had significantly shorter OS than patients with a low-risk score in the primary cohort (P < 0.001; Fig. 5A) and the validation cohort (P < 0.001; Fig. 5E). In the primary cohort, the 1-, 3-, and 5-year survival probabilities of the high-risk were 90.0%, 71.3% and 37.3%, respectively. On the other hand, for the low-risk patients, the 1-, 3-, and 5-year survival probabilities were 99.0%, 93.0% and 53.5%, respectively. Meanwhile, in the validation cohort, the 1-, 3-, and 5-year survival probabilities of the high-risk and low-risk patients were 97.2%, 70.4%, 35.2% and 98.8%, 95.3%, 54.1%, respectively (Table 3). Moreover, Kaplan–Meier curves demonstrated that high-risk and low-risk subgroups were significantly correlated with OS outcomes in the primary cohort (Fig. 5C, P < 0.001; Fig. 5D, P = 0.011), as well as in the validation cohort (Fig. 5G, P = 0.015, Fig. 5H, P = 0.021) with a respective stage III, and stage IV, with an exception for patients in stage I/II (Fig. 5B, F).
The nomogram for the prediction of OS
We established a nomogram for OS, which included risk score, TNM stage and EBV DNA in the two cohorts. In the primary cohort, the nomogram model achieved a C-index of 0.783 (95% CI 0.730 ~ 0.836), which was significantly higher than that of the prognostic model 0.733 (95% CI 0.673–0.793, P = 0.005) (Figs. 6A, 7A). On the other hand, in the validation cohort, the nomogram model achieved a C-index of 0.776 (95% CI 0.709 ~ 0.844), which was much higher than that of the prognostic model of 0.772 (95% CI 0.691 ~ 0.853, P = 0.455) (Figs. 6E, 7B). Calibration curves for probability of survival at 1-, 3-, and 5-years showed optimal agreement between prediction established in the nomogram and actual observation in the primary (Fig. 6 B–D) and validation cohort (Fig. 6F–H). Furthermore, RMS curves demonstrated a larger slope in the primary cohort for nomogram, which indicates superior estimation of survival with nomogram (Fig. 7A, B).
The prognostic model based on risk score, TNM stage and treatment predicting OS in the primary cohort (A) and the validation cohort (E). The calibration curves for predicting patient OS at 1-, 3-, 5-year in the primary cohort (B–D) and the validation cohort (F–H). Total points projected on the bottom scales indicate the probability of 1-, 3-, and 5-year survival
The correlations among the variables in the nomogram model
The relationship among variables of nomogram model were shown in Fig. 8. In this figure, blue indicated positive correlations, while red indicated negative corrections. Moreover, the correlation coefficients were proportional to color intensity and circle size. In the primary cohort, there was a highly significant between EBV DNA and risk score (Fig. 8A). Meanwhile, treatment was moderately correlated to TNM stage. In the meantime, we were able to get consistent results in the validation group (Fig. 8B).
Discussion
The TNM stage is commonly utilized to predict prognosis and guide clinical therapeutic regimen across many cancers. This system for NPC was updated and refined to the 8th edition in 2016 [2]. However, this system has several controversies as it is completely based on anatomical extent of cancer, and neglects the biological heterogeneity of NPC patients. Many other important risk factors need to be considered in the current staging systems.
In the present study, we used a random forest model to investigate prognostic value of many clinical factors and selected the most significant ones. We revealed that EBV DNA, dNLR and HGB levels could be used for prediction of NPC prognosis. The established risk score model, which included EBV DNA, dNLR, and HGB have a higher AUC and C-index than TNM stage, treatment, and EBV DNA model in both the primary and validation cohort. Based on risk score, we stratified NPC patients into two subgroups, including high-risk and low-risk, which were significant in OS outcomes. Moreover, according to results from random forest model analysis, we established nomograms that can help predict OS in NPC patients, which integrated risk score, TNM stage, and treatment. The nomogram model showed better predictive accuracy [C-index: 0.783 (95% CI 0.730 ~ 0.836)] than the risk score model [C-index: 0.733 (95% CI 0.673 ~ 0.793) (P = 0.005)] in the primary cohort. However, there were no differences in the of C-index between nomogram [C-index: 0.776 (95% CI 0.709 ~ 0.844)] and risk score model [C-index: 0.772 (95% CI 0.691 ~ 0.853) (P = 0.455)] in the validation cohort, likely due to our small size of NPC patients in the cohort.
The infection of EBV is common in NPC in endemic areas. Levels of plasma EBV DNA have been shown to be the most attractive potential biomarker to predict prognosis and provide accurate risk stratification in NPC [20]. Intriguingly, a recent prospective screening study that involved 20,174 participants showed that plasma EBV DNA detection was useful to screen for NPC, as it was associated with 97.1% sensitivity and 98.6% specificity. NPC was found to be detected significantly earlier by EBV DNA, with a significantly higher proportion of stage I or II disease than in a historical cohort (71% vs 20%), and had superior 3-year progression-free survival (97% vs. 70%; hazard ratio, 0.10) [21]. However, EBV DNA alone for prognosis has limitations, as the methodology of EBV DNA measurement is not globally standardized and measurement is not routinely available in many medical institutions. Moreover, there is accumulating evidence indicating that inflammation plays an important role in carcinogenesis and tumor proliferation [22]. Studies have reported that inflammation-based markers can be used as potential prognostic factors for many cancers, including CRP, neutrophils, lymphocytes, dNLR and ALB [23,24,25]. Neutrophils secrete proangiogenic cytokines, which include IL-8, MMP-9, MMP-8, and VEGF, which are known to contribute to tumor angiogenesis and progression [26, 27]. Lymphocytes, especially the CD8+ T cells, which mediated immune response and increased OS of patients with gallbladder cancer [28]. Furthermore, low HGB is a risk factor in cancer patient survival, and HGB level is an important predictor in evaluation and treatment anemia [29, 30]. In our study, we used a random forest model to identify that EBV DNA, dNLR and HGB levels can be used to predict NPC prognosis.
There are several limitations to this study. First, this is a retrospective study, and there may be a selection bias during data collection. Secondly, this is a single-center study with a limited number of NPC patients. Third, this study established models for predicting OS among patients with NPC. However, models of disease-free survival are unknown. Therefore, our next aim is to validate our models on a large-scale with a multi-center study.
Conclusions
This study established a risk score model based on EBV DNA, dNLR and HGB levels. Compared to TNM stage, treatment and EBV DNA models, risk score model achieved a higher AUC. This easy-to-use scoring prognostic model can provide a more precise estimation for clinicians and patients.
Availability of data and materials
The datasets analyzed during the current study are not publicly available due to patient privacy concerns, but are available from the corresponding author on reasonable request.
Abbreviations
- BMI:
-
Body mass index
- TNM:
-
Tumor Node Metastasis stage
- WBC:
-
White blood cell
- HGB:
-
Hemoglobin
- NLR:
-
Neutrophil/lymphocyte ratio
- dNLR:
-
Neutrophil/WBC-neutrophil ratio
- LMR:
-
Lymphocyte/monocyte ratio
- PLR:
-
Platelet/lymphocyte ratio
- SII:
-
Systemic immune-inflammation index
- TP:
-
Total protein
- ALB:
-
Albumin
- GLOB:
-
Globulin
- AGR:
-
ALB/GLOB ratio
- CRP:
-
C-reactive protein
- CAR:
-
C-reactive protein/albumin ratio
- APOA:
-
Apolipoprotein AI
- APOB:
-
Apolipoprotein B
- ABR:
-
APOA/APOB ratio
- LDH:
-
Lactic dehydrogenase
- HDL:
-
High density lipoprotein
- EBV:
-
Epstein-Barr virus
- VCA-IgA:
-
Viral capsid antigen specific immunoglobulin A
- EA-IgA:
-
Early antigen immunoglobulin A
- PNI:
-
Prognostic nutritional index
- PI:
-
Prognostic index
References
Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394(10192):64–80.
Tang LL, Chen YP, Mao YP, Wang ZX, Guo R, Chen L, Tian L, Lin AH, Li L, Sun Y, et al. Validation of the 8th edition of the UICC/AJCC staging system for nasopharyngeal carcinoma from endemic areas in the intensity-modulated radiotherapy era. J Natl Compr Canc Netw. 2017;15(7):913–9.
Wei KR, Zheng RS, Zhang SW, Liang ZH, Li ZM, Chen WQ. Nasopharyngeal carcinoma incidence and mortality in China, 2013. Chin J Cancer. 2017;36(1):90.
Colevas AD, Yom SS, Pfister DG, Spencer S, Adelstein D, Adkins D, Brizel DM, Burtness B, Busse PM, Caudell JJ, et al. NCCN guidelines insights: head and neck cancers, version 1.2018. J Natl Compr Canc Netw. 2018;16(5):479–90.
Wierzbicka M, Napierala J. Updated National Comprehensive Cancer Network guidelines for treatment of head and neck cancers 2010–2017. Otolaryngol Pol. 2017;71(6):1–6.
Liu MZ, Tang LL, Zong JF, Huang Y, Sun Y, Mao YP, Liu LZ, Lin AH, Ma J. Evaluation of sixth edition of AJCC staging system for nasopharyngeal carcinoma and proposed improvement. Int J Radiat Oncol Biol Phys. 2008;70(4):1115–23.
Huang L, Sim AYL, Wu Y, Liang Z, Li K, Du Y, Ong EHW, Tan HQ, Wee JTS, Xie Y, et al. Lactate dehydrogenase kinetics predict chemotherapy response in recurrent metastatic nasopharyngeal carcinoma. Ther Adv Med Oncol. 2020;12:1758835920970050.
Li QJ, Mao YP, Guo R, Huang CL, Fang XL, Ma J, Tang LL, Chen L. A nomogram based on serum biomarkers and clinical characteristics to predict survival in patients with non-metastatic nasopharyngeal carcinoma. Front Oncol. 2020;10:594363.
Feng Y, Zhang N, Wang S, Zou W, He Y, Ma JA, Liu P, Liu X, Hu C, Hou T. Systemic inflammation response index is a predictor of poor survival in locally advanced nasopharyngeal carcinoma: a propensity score matching study. Front Oncol. 2020;10:575417.
Tang LQ, Li CF, Li J, Chen WH, Chen QY, Yuan LX, Lai XP, He Y, Xu YX, Hu DP, et al. Establishment and validation of prognostic nomograms for endemic nasopharyngeal carcinoma. J Natl Cancer Inst. 2016;108(1):djv291.
Zhao L, Chen H, Hu B, Zhang H, Lin Q. Prognostic significance of Ki67 expression and the derived neutrophil-lymphocyte ratio in nasopharyngeal carcinoma. Cancer Manag Res. 2018;10:1919–26.
Guo SS, Tang LQ, Chen QY, Zhang L, Liu LT, Huang PY, Cao KJ, Guo L, Mo HY, Guo X, et al. Is hemoglobin level in patients with nasopharyngeal carcinoma still a significant prognostic factor in the era of intensity-modulated radiotherapy technology? PLoS ONE. 2015;10(8):e0136033.
He B, Ji T, Zhang H, Zhu Y, Shu R, Zhao W, Wang K. MRI-based radiomics signature for tumor grading of rectal carcinoma using random forest model. J Cell Physiol. 2019;234(11):20501–9.
Huang Z, Xing S, Zhu Y, Qu Y, Jiang L, Sheng J, Wang Q, Xu S, Xue N. Establishment and validation of nomogram model integrated with inflammation-based factors for the prognosis of advanced non-small cell lung cancer. Technol Cancer Res Treat. 2020;19:1533033820971605.
Li C, Qin X, Yang Z, Guo W, Huang R, Wang H, Wang H. A nomogram to predict the incidence of permanent stoma in elderly patients with rectal cancer. Ann Transl Med. 2021;9(4):342.
Ding Y, Chen Y, Wu M, Li L, Huang Y, Wang H, Wang H, Yu X, Xu N, Teng L. Identification of genes associated with gastric cancer survival and construction of a nomogram to improve risk stratification for patients with gastric cancer. Oncol Lett. 2020;20(1):215–25.
He X, Xue N, Liu X, Tang X, Peng S, Qu Y, Jiang L, Xu Q, Liu W, Chen S. A novel clinical model for predicting malignancy of solitary pulmonary nodules: a multicenter study in Chinese population. Cancer Cell Int. 2021;21(1):115.
Chen JH, Zhai ET, Yuan YJ, Wu KM, Xu JB, Peng JJ, Chen CQ, He YL, Cai SR. Systemic immune-inflammation index for predicting prognosis of colorectal cancer. World J Gastroenterol. 2017;23(34):6261–72.
Kasymjanova G, MacDonald N, Agulnik JS, Cohen V, Pepe C, Kreisman H, Sharma R, Small D. The predictive value of pre-treatment inflammatory markers in advanced non-small-cell lung cancer. Curr Oncol. 2010;17(4):52–8.
Li W, Chen J, Liang B, Li Z, Li J, Yuan X, Wu S, Zeng F, Peng X, Li Y, et al. Long-term monitoring of dynamic changes in plasma EBV DNA for improved prognosis prediction of nasopharyngeal carcinoma. Cancer Med. 2021;10(3):883–94.
Chan KCA, Woo JKS, King A, Zee BCY, Lam WKJ, Chan SL, Chu SWI, Mak C, Tse IOL, Leung SYM, et al. Analysis of plasma Epstein-Barr virus DNA to screen for nasopharyngeal cancer. N Engl J Med. 2017;377(6):513–22.
McMillan DC. Cancer and systemic inflammation: stage the tumour and stage the host. Br J Cancer. 2013;109(3):529.
Proctor MJ, McMillan DC, Morrison DS, Fletcher CD, Horgan PG, Clarke SJ. A derived neutrophil to lymphocyte ratio predicts survival in patients with cancer. Br J Cancer. 2012;107(4):695–9.
Partl R, Lukasiak K, Thurner EM, Renner W, Stranzl-Lawatsch H, Langsenlehner T. The elevated pre-treatment C-reactive protein predicts poor prognosis in patients with locally advanced rectal cancer treated with neo-adjuvant radiochemotherapy. Diagnostics. 2020;10(10):780.
Guo S, He X, Chen Q, Yang G, Yao K, Dong P, Ye Y, Chen D, Zhang Z, Qin Z, et al. The C-reactive protein/albumin ratio, a validated prognostic score, predicts outcome of surgical renal cell carcinoma patients. BMC Cancer. 2017;17(1):171.
Kusumanto YH, Dam WA, Hospers GA, Meijer C, Mulder NH. Platelets and granulocytes, in particular the neutrophils, form important compartments for circulating vascular endothelial growth factor. Angiogenesis. 2003;6(4):283–7.
Weitzman SA, Gordon LI. Inflammation and cancer: role of phagocyte-generated oxidants in carcinogenesis. Blood. 1990;76(4):655–63.
Fluxa P, Rojas-Sepulveda D, Gleisner MA, Tittarelli A, Villegas P, Tapia L, Rivera MT, Lopez MN, Catan F, Uribe M, et al. High CD8(+) and absence of Foxp3(+) T lymphocytes infiltration in gallbladder tumors correlate with prolonged patients survival. BMC Cancer. 2018;18(1):243.
Rodgers GM 3rd, Becker PS, Blinder M, Cella D, Chanan-Khan A, Cleeland C, Coccia PF, Djulbegovic B, Gilreath JA, Kraut EH, et al. Cancer and chemotherapy-induced anemia. J Natl Compr Canc Netw. 2012;10(5):628–53.
Wan S, Lai Y, Myers RE, Li B, Palazzo JP, Burkart AL, Chen G, Xing J, Yang H. Post-diagnosis hemoglobin change associates with overall survival of multiple malignancies—results from a 14-year hospital-based cohort of lung, breast, colorectal, and liver cancers. BMC Cancer. 2013;13:340.
Acknowledgements
Not applicable.
Funding
This study was supported by Science and Technology Department of Henan Province (Grant No. LHGJ20190637).
Author information
Authors and Affiliations
Contributions
QXX, YBL, and MMJ designed this study. NX, and GPO collected clinical data and wrote the manuscript. WGM, LNJ, and JHS performed data analysis. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Clinical Research Ethics Committee of Sun Yat-sen University Cancer Center.
Consent for publication
Not applicable.
Competing interests
The authors have declared no conflicts of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Xue, N., Ou, G., Ma, W. et al. Development and validation of a risk prediction score for patients with nasopharyngeal carcinoma. Cancer Cell Int 21, 452 (2021). https://doi.org/10.1186/s12935-021-02158-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-021-02158-6