Abstract
Background
Cancers located on the right and left sides of the colon have distinct clinical and molecular characteristics. This study aimed to explore the regulatory mechanisms of location-specific long noncoding RNAs (lncRNAs) as competing endogenous RNAs (ceRNAs) in colon cancer and identify potential prognostic biomarkers.
Method
Differentially expressed lncRNAs (DELs), miRNAs (DEMs), and genes (DEGs) between right- and left-side colon cancers were identified by comparing RNA sequencing profiles. Functional enrichment analysis was performed for the DEGs, and a ceRNA network was constructed. Associations between DELs and patient survival were examined, and a DEL-based signature was constructed to examine the prognostic value of these differences. Clinical colon cancer tissues and Gene Expression Omnibus (GEO) datasets were used to validate the results.
Results
We identified 376 DELs, 35 DEMs, and 805 DEGs between right- and left-side colon cancers. The functional enrichment analysis revealed the functions and pathway involvement of DEGs. A ceRNA network was constructed based on 95 DEL–DEM–DEG interactions. Three DELs (LINC01555, AC015712, and FZD10-AS1) were associated with the overall survival of patients with colon cancer, and a prognostic signature was established based on these three DELs. High risk scores for this signature indicated poor survival, suggesting that the signature has prognostic value for colon cancer. Examination of clinical colon cancer tissues and GEO dataset analysis confirmed the results.
Conclusion
The ceRNA regulatory network suggests roles for location-specific lncRNAs in colon cancer and allowed the development of an lncRNA-based prognostic signature, which could be used to assess prognosis and determine treatment strategies in patients with colon cancer.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC), which most commonly presents as colon cancer, is the third most common cancer worldwide and therefore poses a significant threat to human health [1]. Despite great progress in CRC diagnosis and treatment over the past two decades, the mortality rate of colon cancer remains high, especially for patients with advanced-stage disease [2, 3]. Numerous studies have been conducted to elucidate the carcinogenic mechanisms of CRC, but its exact pathogenesis remains unclear, and a lack of novel efficient prognosis biomarkers affects patient outcome [4, 5].
The colon can be divided into the left and right segments, with the distal transverse colon as the boundary. The left-side colon includes the splenic flexure, descending colon, and sigmoid colon; the right-side colon includes the cecum, ascending colon, liver flexure, and transverse colon [6]. There are significant differences in clinical symptoms and outcomes between left- and right-side colon cancer, which also differ in their molecular characteristics, such as microsatellite instability and gene mutation status [7, 8], as cancers in different sections of the colon are caused by different molecular alterations.
The carcinogenesis of colon cancer is accompanied by changes in many molecules, including long noncoding RNAs (lncRNAs), microRNAs (miRNAs), and mRNAs. The roles of miRNAs and mRNAs in colon cancer pathogenesis have been described in many studies [9, 10], including reports identifying interactions between miRNAs and mRNAs [11, 12] as well as biomarkers to predict patient prognosis [13]. The expression of mRNAs and miRNAs can be regulated by lncRNAs, which play vital roles in the development of colon cancer. Studies have shown that lncRNAs interact with miRNAs and mRNAs, acting as competing endogenous RNAs (ceRNAs) in the regulation of carcinogenesis [14, 15]. However, little is known about the effects of colon cancer location on lncRNA changes. Therefore, in this study, we aimed to investigate location-specific regulatory relationships in colon cancer and identify potential prognostic biomarkers for the disease, which could help to improve treatment efficacy.
Materials and methods
Raw data and annotation
The raw RNA and miRNA sequencing data (level 3) of patients with colon cancer (cohort: COAD) were downloaded from the Broad GDAC Firehose database (http://gdac.broadinstitute.org/; accessed April 3, 2019). Downloading data from this site constitutes agreement to The Cancer Genome Atlas (TCGA) data usage policy. Annotation of lncRNAs and mRNAs was performed using the Ensembl genome browser (Homo_sapiens.GRCh38.96.gtf; http://www.ensembl.org/index.html). The lncRNAs with a given “gene_type” column were selected, including lincRNA, sense intronic, overlap sense, antisense RNA, and processed_transcript. Corresponding clinical data of the patients were also downloaded from the same database. The patients were divided into left- and right-side colon cancer groups based on the cancer location, which included 225 and 315 patients, respectively.
Screening of differentially expressed lncRNAs, miRNAs, and mRNAs
The screening of differentially expressed lncRNAs, miRNAs, and mRNAs between left- and right-side colon cancers was performed using the “edgeR” package in R software (version 3.5.1). The “edgeR” package is designed for differential expression analysis of replicate count data, which has been widely applied in the differential expression analysis of high-throughput sequencing data. Those lncRNAs and mRNAs with a null expression value in more than 10% of samples were discarded. Differentially expressed genes (DEGs) and differentially expressed lncRNAs (DELs) were identified at a false discovery rate (FDR) < 0.05. As there was little change in miRNA expression between left- and right-side colon cancers, we included differentially expressed miRNAs (DEMs) with p-values < 0.05.
Functional enrichment analysis
The biological functions (gene ontology, GO) and pathway involvement of DEGs were analyzed using the “clusterProfiler” package [16] implemented in R. In brief, the genes were first converted to their Homo sapiens Entrez gene IDs, then pathway and process enrichment analysis was carried out for each given gene list, including biological processes (BP), cellular components (CC), and molecular functions (MF), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. The p-values were calculated based on the accumulative hypergeometric distribution. GO terms and KEGG pathways with p-values < 0.05 were included.
Identification and construction of the ceRNA network
The ceRNA theory suggests that lncRNAs regulate the expression of miRNAs, and also act as natural miRNA “sponges” to affect mRNA expression, thus forming lncRNA–miRNA–mRNA interactions [14, 15]. In this study, we first predicted the target relationships between DELs and DEMs using the miRcode (http://www.mircode.org/) database and then predicted targeted DEM–DEG relationships using the Targetscan (http://www.targetscan.org/), miRDB (http://www.mirdb.org/), and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) databases. Next, we overlapped the DEMs from the DEL–DEM and DEM–DEG relationships, and ultimately retained DEL, DEM, and DEG intersection relationships. Then, a ceRNA network of DEL–DEM–DEG regulatory relationships was generated and visualized using Cytoscape (version 3.7.1).
Prognostic value of DELs and clinical features in colon cancer
The prognostic value of DELs in patients with colon cancer was assessed using a multivariate Cox regression model by analyzing the survival data. The prognostic value of clinical features (age, gender, T stage, N stage, M stage, and tumor stage) from TCGA on patient survival was determined in the same manner. The significant variables were defined as having p-values lower than 0.05. All statistical analyses were performed in R.
Construction of the DEL-based prognostic signature
To examine the association of DELs with the overall survival (OS) of patients with colon cancer, a multivariate Cox regression analysis was performed to screen DELs significantly associated with the survival of patients. Only DELs with p-values < 0.01 were considered significantly prognostic. A DEL-based prognostic signature was established using the linear combination of the DEL expression levels multiplied by the coefficient (beta value) from multivariate Cox regression analysis. The risk score formula was as follows: risk score = (expression of DEL1 × β1DEL1) + (expression of DEL2 × β2 DEL2) + …(expression of DEL n × βn DELn) [17, 18]. This DEL-based prognostic signature was then employed to divide the patients into high- and low-risk groups using the median risk score as a cutoff. The association between the DEL-based prognostic signature and the survival of patients was analyzed. The nomogram for the prediction of probability of the DEL signature in colon cancer was established using clinical features and risk scores and was visualized using the “rms” package (version 5.1-2) in R.
DEL validation in colon cancer tissues and gene expression omnibus datasets
Tissue samples from 60 cases of colon cancer (30 left-side and 30 right-side) were collected from the Biobank of the Affiliated Tumor Hospital of Guangxi Medical University. This study was approved by the ethics committee of the Affiliated Tumor Hospital of Guangxi Medical University. The patients did not undergo any radiotherapy or chemotherapy before surgery. The RNA of lncRNAs was extracted and the expression of DELs was assessed by real-time quantitative reverse transcription-polymerase chain reaction (qRT-qPCR) assay. Briefly, TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract RNA from the tissues and the PrimeScript RT reagent kit (Takara, Dalian, China) was used to synthesize complementary DNA. The qRT-qPCR assay was performed using a 7500 Fast Real-Time PCR system (Applied Biosystems) using SYBR Green reagent (Applied Biosystems). The primers used for qRT-PCR are listed in Additional file 1: Table S1. The results were analyzed using the 2−∆∆Ct method. The experiments were repeated in triplicate.
Results
Clinical features of patients with left- and right-side colon cancer
The workflow of this study is shown in Fig. 1. Patients were divided into left- and right-side groups based on their tumor location. As shown in Table 1, there were significant differences in the ages and N, M, and tumor stages between patients with left- and right-side colon cancer (p < 0.05). No significant differences were detected with respect to the patients’ gender and T stages (p > 0.05).
Colon cancer location-specific lncRNAs, miRNAs, and mRNAs
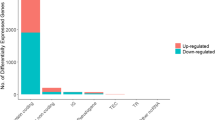
Based on our screening criteria, 376 lncRNAs, 35 miRNAs, and 805 mRNAs were differentially expressed between left- and right-side colon cancers. Of these, 150 and 226 DELs were up- and downregulated, respectively; 16 and 19 DEMs were up- and down-regulated, respectively; and 314 and 491 DEGs were up- and down-regulated, respectively. The top DELs, DEMs, and DEGs are shown in Additional file 1: Figure S1.
Functional enrichment analysis of location-specific DEGs
We analyzed the functional enrichment of the 805 DEGs, and the most significantly enriched biological process, cellular compartment, and molecular function terms were pattern specification (GO: 0007389, score: 5.08), synaptic membrane (GO: 0097060, score: 7.56), and substrate-specific channel activity (GO: 0022838, score: 2.23), respectively. The KEGG pathway enrichment analysis revealed significant enrichment in neuroactive ligand-receptor interaction pathways (hsa04080, score: 6.74; Fig. 2).
Construction of a location-specific ceRNA network for colon cancer
The miRcode database predicted 214 interactions between DELs and DEMs, and the Targetscan, miRDB, and miRTarBase databases predicted 34 interactions between DEMs and DEGs, resulting in 95 DEL–DEM–DEG interactions in the ceRNA network (Fig. 3).
Prognostic value of location-specific DELs in colon cancer
A multivariate Cox regression analysis was performed to examine the association between location-specific DELs and the OS of patients with colon cancer. As shown in Table 2, three DELs (LINC01555, AC015712, and FZD10-AS1) had p-values < 0.001 and were therefore regarded as prognostic DELs and used to establish the prognostic signature. Kaplan–Meier curves of these prognostic DELs are shown in Additional file 1: Figure S2.
Construction of the DEL-based prognostic signature
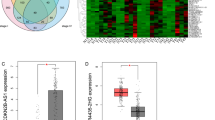
After calculating risk scores, the patients were divided into high and low risk score groups (Fig. 4a), whose survival status and DEL expression levels are shown in Fig. 4b, c. The Kaplan–Meier curve illustrated that patients with high-risk scores had shorter OS than those with low-risk scores (p = 0.014; Fig. 4d).
Association between the risk score and the survival of patients with colon cancer. a Risk score curve of patient survival status and time distributed; b distribution of patient survival status; c heatmap of the expression of the three prognostic DELs; d Kaplan–Meier curve of patients in the low- and high-risk groups
Prognostic value of the DEL-based prognostic signature
We investigated the prognostic values of the DEL-based signature and clinical features on patient survival. As listed in Additional file 1: Table S2, the risk scores showed good predictive value (hazard ratio: 1.25, 95% confidence interval: 1.07–1.36) in the prognosis of patients with colon cancer compared with other clinical features, comparable to that of the tumor stage. The nomogram demonstrated that the DEL-based signature contributed more to the risk than the tumor stage (Fig. 5), indicating the high predictive value of DEL-based prognostic signature.
DEL validation in colon cancer tissues and gene expression omnibus datasets
The expression of the prognostic DELs was examined in colon cancer tissues by qRT-PCR. Consistent with the results from TCGA, the expression of LINC01555 and FZD10-AS1 was significantly increased in left-side colon cancer tissues compared with right-side tissues, while the opposite results were observed for AC015712 (Fig. 6a). Associations between the expression of the three DELs and the patients’ clinical features were also analyzed. We found that the expression of LINC01555, AC015712, and FZD10-AS1 was associated with the tumor stage, but not with other clinical features (Table S3). GSE38832 included survival data and lncRNA levels for 122 colon cancer patients. However, only LINC01555 and FZD10-AS1 were included in this dataset, and we, therefore, conducted survival analysis on these two lncRNAs only. Consistent with the results from TCGA, high expression of LINC01555 and FZD10-AS1 was associated with a shorter survival time, compared with low expression (Fig. 6b, c).
Discussion
Many studies have assessed the clinical symptoms, outcomes, and molecular alterations of left- and right-side colon cancers, and many differences in tumor biology and pathophysiology have been identified [8]. These differences alter the treatment and screening approaches used for patients with colon cancer [19, 20]. In this study, we found that many lncRNAs, miRNAs, and mRNAs were differentially expressed between left- and right-side colon cancers, suggesting that these DELs, DEMs, and DEGs were location-specific molecular alterations that might contribute to the differences between left- and right-side colon cancers.
As gene alterations regulate tumor cell biological functions, we examined the functional enrichment of location-specific DEGs using GO and KEGG analysis. The results revealed that many DEGs were enriched in the biological process of pattern specification, which has been shown to participate in the development of cancers, such as lung cancer [21]. Pathway analysis revealed that DEGs were enriched in neuroactive ligand-receptor interaction pathways, which roles have also been documented in several cancers, such as pancreatic cancer and breast cancer [22, 23]. These results indicate that these location-specific DEGs are also involved in the pathogenesis of colon cancer.
lncRNAs perform diverse functions, both by directly regulating mRNA expression and by adsorbing miRNAs to regulate mRNA expression [24,25]. They can interact with miRNAs and mRNAs to form ceRNA networks, which can reveal novel regulatory mechanisms mediated by lncRNAs [26]. Several ceRNA networks have been reported for CRC. For example, Zhang et al. [27] compared differentially expressed lncRNAs and genes between CRC and normal tissues, and found 1143 DELs, 276 DEMs, and 2151 DEGs, which they used to construct a lncRNA–miRNA–mRNA ceRNA network and identify a five-lncRNA prognostic model that was independently prognostic in CRC. Likewise, Yuan et al. [28] constructed a ceRNA network for CRC using DELs, DEMs, and DEGs from TCGA datasets, and found that LINC00400 and LINC00355 were associated with pathological stages of CRC. However, no study had examined DELs between cancers occurring on different sides of the colon yet, and here, we have constructed a location-specific ceRNA network for colon cancer, which has uncovered further potential regulatory mechanisms for colon carcinogenesis.
Aberrant expression of lncRNAs has been implicated in various cancers, and some lncRNAs have been demonstrated to be independent indicators of patient survival [29, 30]. In this study, three DELs were significantly associated with the survival of patients with colon cancer and were used to establish a DEL-based prognostic signature that was significantly associated with patient survival. Furthermore, prognostic value analysis indicated that this DEL signature could faithfully predict the 5-year OS with a prognostic value comparable to that of the tumor stage. Notably, the association of LINC01555 with CRC had been previously reported [31, 32], confirming the credibility of our results. However, the roles of AC015712 and FZD10-AS1 in cancer have not yet been reported. Thus, our results suggest the importance of further investigation of these two DELs in colon cancer. Finally, we verified the expression of the three prognostic DELs in colon cancer tissues, and analyzed their clinical significance, further supporting the credibility of our bioinformatics analysis. There are a few limitations to this study. First, the sample size of the validated cohort was relatively small, and a larger cohort will be needed to confirm these results. Second, although the biological function of LINC01555 had been determined in previous studies, those of AC015712 and FZD10-AS1 remain unknown and will need to be characterized using in vivo and in vitro experiments. Third, the lncRNA–miRNA–mRNA associations identified in the ceRNA network will need to be examined in future studies. Nevertheless, our results provide valuable novel molecular targets that could aid in the screening, prognosis, and treatment of left- and right-side colon cancers.
Conclusion
In this study, we have identified and validated location-specific lncRNAs, miRNAs, and mRNAs in left- and right-side colon cancers, developed a location-specific ceRNA regulatory network to identify novel molecular mechanisms involved in colon cancer carcinogenesis, and established a location-specific DEL-based prognostic signature, providing potentially valuable biomarkers for patients with different types of colon cancer.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Maida M, Macaluso FS, Ianiro G, et al. Screening of colorectal cancer: present and future. Expert Rev Anticancer Ther. 2017;17(12):1131–46.
Issa IA, Noureddine M. Colorectal cancer screening: an updated review of the available options. World J Gastroenterol. 2017;23(28):5086–96.
Muller MF, Ibrahim AE, Arends MJ. Molecular pathological classification of colorectal cancer. Virchows Arch. 2016;469(2):125–34.
Mohammadi A, Mansoori B, Baradaran B. The role of microRNAs in colorectal cancer. Biomed Pharmacother. 2016;84:705–13.
Mik M, Berut M, Dziki L, Trzcinski R, Dziki A. Right- and left-sided colon cancer—clinical and pathological differences of the disease entity in one organ. Arch Med Sci. 2017;13(1):157–62.
Cannon E, Buechler S. Colon cancer tumor location defined by gene expression may disagree with anatomic tumor location. Clin Colorectal Cancer. 2019;18(2):149–58.
Kim K, Castro EJT, Shim H, Advincula JVG, Kim YW. Differences regarding the molecular features and gut microbiota between right and left colon cancer. Ann Coloproctol. 2018;34(6):280–5.
Tang XJ, Wang W, Hann SS. Interactions among lncRNAs, miRNAs and mRNA in colorectal cancer. Biochimie. 2019;163:58–72.
Tam C, Wong JH, Tsui SKW, Zuo T, Chan TF, Ng TB. LncRNAs with miRNAs in regulation of gastric, liver, and colorectal cancers: updates in recent years. Appl Microbiol Biotechnol. 2019;103(12):4649–77.
Nasri Nasrabadi P, Zareian S, Nayeri Z, et al. A detailed image of rutin underlying intracellular signaling pathways in human SW480 colorectal cancer cells based on miRNAs-lncRNAs-mRNAs-TFs interactions. J Cell Physiol. 2019;234(9):15570–80.
Zhu Y, Bian Y, Zhang Q, et al. Construction and analysis of dysregulated lncRNA-associated ceRNA network in colorectal cancer. J Cell Biochem. 2019;120(6):9250–63.
Chen Y, Yu X, Xu Y, Shen H. Identification of dysregulated lncRNAs profiling and metastasis-associated lncRNAs in colorectal cancer by genome-wide analysis. Cancer Med. 2017;6(10):2321–30.
He M, Lin Y, Xu Y. Identification of prognostic biomarkers in colorectal cancer using a long non-coding RNA-mediated competitive endogenous RNA network. Oncol Lett. 2019;17(3):2687–94.
Liu J, Li H, Zheng B, Sun L, Yuan Y, Xing C. Competitive endogenous RNA (ceRNA) regulation Network of lncRNA-miRNA-mRNA in colorectal carcinogenesis. Dig Dis Sci. 2019;64(7):1868–77.
Yu G, Wang LG, Han Y, He QY. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–7.
Hu CE, Du PZ, Zhang HD, Huang GJ. Long noncoding RNA CRNDE promotes proliferation of gastric cancer cells by targeting miR-145. Cell Physiol Biochem. 2017;42(1):13–21.
Yan X, Liu XP, Guo ZX, Liu TZ, Li S. Identification of hub genes associated with progression and prognosis in patients with bladder cancer. Front Genet. 2019;10:408.
Ghidini M, Petrelli F, Tomasello G. Right versus left colon cancer: resectable and metastatic disease. Curr Treat Options Oncol. 2018;19(6):31.
Stintzing S, Tejpar S, Gibbs P, Thiebach L, Lenz HJ. Understanding the role of primary tumour localisation in colorectal cancer treatment and outcomes. Eur J Cancer. 2017;84:69–80.
Kalari S, Jung M, Kernstine KH, Takahashi T, Pfeifer GP. The DNA methylation landscape of small cell lung cancer suggests a differentiation defect of neuroendocrine cells. Oncogene. 2013;32(30):3559–68.
Wei P, Tang H, Li D. Insights into pancreatic cancer etiology from pathway analysis of genome-wide association study data. PLoS ONE. 2012;7(10):e46887.
Liu H, Ye H. Screening of the prognostic targets for breast cancer based co-expression modules analysis. Mol Med Rep. 2017;16(4):4038–44.
Zhang Y, Zhang D, Lv J, Wang S, Zhang Q. LncRNA SNHG15 acts as an oncogene in prostate cancer by regulating miR-338-3p/FKBP1A axis. Gene. 2019;705:44–50.
Yang Y, Wang F, Huang H, Zhang Y, Xie H, Men T. lncRNA SLCO4A1-AS1 promotes growth and invasion of bladder cancer through sponging miR-335-5p to upregulate OCT4. Onco Targets Ther. 2019;12:1351–8.
Xie JJ, Guo QY, Jin JY, Jin D. SP1-mediated overexpression of lncRNA LINC01234 as a ceRNA facilitates non-small-cell lung cancer progression via regulating OTUB1. J Cell Physiol. 2019;234(12):22845–56.
Zhang H, Wang Z, Wu J, Ma R, Feng J. Long noncoding RNAs predict the survival of patients with colorectal cancer as revealed by constructing an endogenous RNA network using bioinformation analysis. Cancer Med. 2019;8(3):863–73.
Yuan W, Li X, Liu L, et al. Comprehensive analysis of lncRNA-associated ceRNA network in colorectal cancer. Biochem Biophys Res Commun. 2019;508(2):374–9.
Chen XD, Zhu MX, Wang SJ. Expression of long non-coding RNA MAGI2AS3 in human gliomas and its prognostic significance. Eur Rev Med Pharmacol Sci. 2019;23(8):3455–60.
Zhang Y, Li Y, Han L, Zhang P, Sun S. SBF2-AS1: an oncogenic lncRNA in small-cell lung cancer. J Cell Biochem. 2019;120(9):15422–8.
Wang X, Chen X, Zhou H, et al. The long noncoding RNA, LINC01555, promotes invasion and metastasis of colorectal cancer by activating the neuropeptide. Neuromedin U Med Sci Monit. 2019;25:4014–24.
Zeng JH, Liang L, He RQ, et al. Comprehensive investigation of a novel differentially expressed lncRNA expression profile signature to assess the survival of patients with colorectal adenocarcinoma. Oncotarget. 2017;8(10):16811–28.
Acknowledgements
We would like to thank the native English-speaking scientists of Editage Company for editing our manuscript.
Funding
This study was partially supported by research funding from the National Natural Science Foundation (No. 81860417), Natural Science Foundation of Guangxi (No. 2018JJA140136), Basic Competence Promotion Project for Young and Middle-aged Teachers in Guangxi (No. 2020KY0348), Guangxi Medicine Science Project (Z20200334), and Guangxi University Students Innovation and Entrepreneurship Project (WLXSZX19039; 201910598012; 202010598086; 201910598241). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Study concept and design: LKZ and HBL; Collection and assembly of data: LKZ and TYP; Performed the experiment: TYB, LKZ and LJL; Data analysis and interpretation: HBL, LKZ, XMZ and TYP; Revised manuscript: YYX, DK; Manuscript writing and review: All authors. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file
1: Figure S1: Differential RNA expression in left- and right-side colon cancers. (A) Differentially expressed (A) lncRNAs, (B) miRNAs, and (C) genes between left- and right-side colon cancers. Figure S2: Prognostic DEL expression and survival in patients with colon cancer. (A) Kaplan-Meier curves for high and low (A) LINC01555, (B) AC015712, and (C) FZD10-AS1 expression in patients with colon cancer. Table S1 Primer sequences for qRT-PCR. Table S2 Multivariate Cox regression of risk score and clinical features in colon cancer patients. Table S3 Association of LINC01555, AC015712, FZD10-AS1 with clinical features.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, Kz., Yin, Yx., Tang, Yp. et al. Construction of a long noncoding RNA-based competing endogenous RNA network and prognostic signatures of left- and right-side colon cancer. Cancer Cell Int 21, 211 (2021). https://doi.org/10.1186/s12935-021-01901-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-021-01901-3