Abstract
Background
The DNA methyltransferase 1 inhibitor, 5-Aza-2′-deoxycytidine (5-Aza-dC) is a potential treatment for breast cancer. However, not all breast tumors will respond similarly to treatment with 5-Aza-dC, and little is known regarding the response of hormone-resistant breast cancers to 5-Aza-dC.
Methods
We demonstrate that 5-Aza-dC-treatment has a stronger effect on an estrogen receptor-negative, Tamoxifen-selected cell line, TMX2-28, than on the estrogen receptor-positive, MCF7, parental cell line. Using data obtained from the HM450 Methylation Bead Chip, pyrosequencing, and RT-qPCR, we identified a panel of genes that are silenced by promoter methylation in TMX2-28 and re-expressed after treatment with 5-Aza-dC.
Results
One of the genes identified, tumor associated calcium signal transducer 2 (TACSTD2), is altered by DNA methylation, and there is evidence that in some cancers decreased expression may result in greater proliferation. Analysis of DNA methylation of TACSTD2 and protein expression of its product, trophoblast antigen protein 2 (TROP2), was extended to a panel of primary (n = 34) and recurrent (n = 34) breast tumors. Stratifying tumors by both recurrence and ER status showed no significant relationship between TROP2 levels and TACSTD2 methylation. Knocking down TACSTD2 expression in MCF7 increased proliferation however; re-expressing TACSTD2 in TMX2-28 did not inhibit proliferation, indicating that TACSTD2 re-expression alone was insufficient to explain the decreased proliferation observed after treatment with 5-Aza-dC.
Conclusions
These results illustrate the complexity of the TROP2 signaling network. However, TROP2 may be a valid therapeutic target for some cancers. Further studies are needed to identify biomarkers that indicate how TROP2 signaling affects tumor growth and whether targeting TROP2 would be beneficial to the patient.
Similar content being viewed by others
Background
DNA methylation, the addition of methyl groups by DNA methyltransferases (DNMTs) to cytosines within CpG dinucleotides, is a key event in tumorigenesis [1, 2]. Although global hypomethylation is characteristic of cancer cells, hypermethylation of CpG islands (CpG-rich regions of the genome approximately 1 kb in length) is also a hallmark of cancer [1, 3]. Hypermethylation of CpG islands in the promoter region of genes frequently leads to transcriptional suppression [4,5,6]. Breast cancer subtypes show varying levels and patterns of aberrant methylation [2, 7]. Estrogen receptor-positive (ERpos) breast tumors tend to have higher levels of methylation than estrogen receptor-negative (ERneg) subtypes [7,8,9,10,11]. Of interest, recurrence following anti-hormonal therapies can result in resistant tumors that are either ERpos or ERneg and the methylation levels in the ERneg recurrent tumors are higher than the ERpos recurrent tumors [12]. Several genes may be silenced by DNA methylation in hormone-resistant breast cancer [13], and breast cancer cell line models have demonstrated a role of DNA methylation in the development of the resistance [14,15,16].
Here we use treatment with the DNMT1 inhibitor, 5-Aza-2′-deoxycytidine or decitabine (5-Aza-dC) of Tamoxifen-sensitive (ERpos, MCF7) and -resistant (ERneg, TMX2-28) cell lines to show that inhibition of methylation affects proliferation only in the cell line model for ERneg, Tamoxifen-resistant breast cancer. The cell-line specific effect of 5-Aza-dC agrees with our previous data demonstrating that the ERneg TMX2-28 line has significantly higher methylation than Tamoxifen-selected lines that retained expression of the estrogen receptor [16]. We identified a panel of genes that were silenced by promoter methylation in the TMX2-28 cell line, but not in the parent MCF7 line. We were particularly interested in a change in the promoter methylation of TACSTD2, which could be reversed by treatment with 5-Aza-dC. The decrease in methylation of TACSTD2 was associated with an increase in TROP2, the protein product for TACSTD2.
TROP2 (trophoblast antigen protein 2 or tumor-associated calcium signal transducer 2) is a 35.7 kDa transmembrane glycoprotein encoded by the intronless gene TACSTD2 [17, 18]. As reviewed by McDougall [19], TROP2 is involved in development, normal intracellular signaling and epithelial cancers, where it provides a signal for proliferation and invasion. TROP2 is expressed in many normal tissues and increased TROP2 expression has been reported for many, but not all epithelial cell cancers [20]. In breast cancer, membrane localization of TROP2 has been correlated with a poor prognosis while intracellular TROP2 has been associated with increased survival [21]. TROP2 is silenced by promoter methylation in some tumors and cancer cell lines (bile duct, lung and prostate) [22,23,24]. However, TROP2 expression has been associated with both inhibition of proliferation [22, 23] and increased growth and metastasis [20, 25,26,27,28,29,30]. The role of TROP2 in endocrine-resistant breast cancer has not been evaluated previously. Our finding that TROP2 is silenced by promoter methylation in the Tamoxifen-resistant breast cancer cell line prompted us to manipulate TACSTD2 expression in cell lines. While knockdown of TACSTD2 in MCF7 cells did increase proliferation as expected, re-expression of TACSTD2 in the TMX2-28 cell line did not inhibit proliferation, suggesting that its role alone was insufficient to explain the decreased proliferation observed after treatment with 5-Aza-dC in Tamoxifen-resistant cells.
Methods
Cell culture, RNA and DNA isolation
MCF7 and TMX2-28 cells were cultured, with the omission of antibiotics, as previously described [16]. DNA was isolated with the QIAamp DNA Mini Kit (Qiagen Cat. No. 51,304) as per manufacturer’s instructions and protocols described previously [31, 32]. RNA was isolated using TriReagent (Molecular Research Center, Inc. Cat. No. TR118). Purified DNA and RNA were quantified using a NanoDrop 8000 (Thermo Scientific).
Proliferation assay
Cells were seeded in a 96-well plate at 2500 cells/well (for 5-Aza-dC exposure) or 5000 cells/well (for TACSTD2 cloned cell lines) and treated with 2.5 μM 5-Aza-dC (Sigma Aldrich, Cat. No. A3656) or 0.1% DMSO (vehicle control) for 120 h. At 60 h cells were refed with 100 μL of growth media with 5-Aza-dC or DMSO. At the time of the assay, 20 μL of Cell Titer 96® Aqueous One Solution (Promega, Madison, WI) was added to each well containing 100 μL of growth media. The plate was then incubated at 37 °C, 5% CO2 for 1.5 h. Absorbance at 490 nm was read on a VersaMax Tunable Microplate reader (Molecular Devices, Sunnyvale, CA). Cell proliferation was quantified as a percentage of the control for each cell line.
Illumina Human Methylation450 (HM450) BeadChip
DNA samples from control to Aza-treated MCF7 and TMX2-28 cells were sent to the University of Southern California for methylation analysis (HM450 BeadChip; Illumina Cat. No. WG-314-1003). Briefly DNA was quantified using an Alu PCR reaction, bisulfite-treated and quantified by additional PCR reactions prior to running on the array. Then the DNA was enzymatically fragmented at 37 °C for 1 h and precipitated in 100% 2-propanol at 4 °C for 30 min followed by centrifugation at 3000×g at 4 °C for 20 min. After resuspension in hybridization buffer of dried pellets, samples were incubated at 48 °C for 1 h followed by 95 °C for 20 min after which the samples were loaded onto the HM450 BeadChip and incubated at 48 °C for 16–24 h. After hybridization of DNA to the primers on the BeadChip, wash buffers were used to remove non-specific and unhybridized DNA. A single-base extension of the hybridized primers was then conducted using labeled nucleotides and the BeadChip was stained with Cy-3 and Cy-5 fluorescent dyes. The BeadChip was then read using the Illumina iScan Reader. Illumina Genome Studio Methylation Module (v 1.9.0) was then used to analyze the image data to determine the efficiency of the reaction. The ratio of the fluorescent signals of methylated to unmethylated sites (beta values) was used to calculate the methylation of interrogated CpG loci.
Pyrosequencing
The EZ DNA Methylation-Lightning kit (Zymo, Cat. No. D5030) was used to bisulfite treat DNA. PCR primers were designed using the Pyromark Assay Design Software version 1.0 (Qiagen). Bisulfite-treated DNA was then amplified using the EPIK Amplification kit (Bioline, Cat. No. BIO-66025). Gene-specific primers targeting the three CpG sites in the promoter region of TACSTD2 (NM_002353) (GRCh37 HG19 Map position (MAPINFO) Ch1: 59043255, 59043280 and 59043370) analyzed by the BeadChip were designed. Primers for pyrosequencing: FWD GGTTGGGGTTGGGAAAGAA-3′, REV -Biot-5′-ACCCCACCTCCTACTACAAACCTA-3′, SEQ 5′-GGAAAGAAAGAAAAGGGA-3′. The Pyromark vacuum prep tool (Qiagen) was used to isolate single stranded products for pyrosequencing. The Pyromark Q24 system (Qiagen) was used to perform pyrosequencing reactions according to manufacturer’s protocol (Qiagen). Percent methylation at the interrogated CpG sites was determined using the Pyromark Q24 Software.
Two-step reverse transcriptase PCR (RT-qPCR)
RNA was reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosytems) supplemented with the RNase Inhibitor, RNasin (Promega). cDNA was quantified using a NanoDrop 8000 (Thermo Scientific) and diluted to 50 ng/μL. Primers for RT-qPCR were designed to span an exon–exon junction, when possible (TACSTD2 has only one exon) using Primer-BLAST (NIH) (ACTB FWD 5′- GGACTTCGAGCAAGAGATGG -3′, REV 5′- AGCACTGGTTGGCGTACAG -3′; TACSTD2 (in non-transfected cell lines) FWD 5′- AATGTATCCCCTTTCGGTCC -3′, REV 5′- TCCCGGGTTGTCATACAGAT -3′; TACSTD2 (in transfected cell lines) FWD 5′- GCCTTCAACCACTCAGACCT -3′, REV 5′- GAGACTCGCCCTTGATGTCC -3′; CGNL1 FWD 5′- GGCTGAGGAGGAAATCGACA -3′, REV 5′- CTCGGCAGCTTCTTCAGTCTTA -3′). RT-qPCR was conducted using the FastStart Universal SYBR Green Master with Rox Reference Dye (Roche) on the Stratagene MxPro (Mx3005P, Agilent). Relative mRNA expression was quantified using the standard curve method normalized to beta-actin.
Human tissue
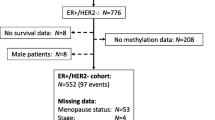
Following Institutional Review Board (IRB) approval from Baystate Medical Center (Springfield, MA), HM450 data were collected for 70 of the previously described 86 samples [12]. The 70 tumors included 18 paired tumors (primary and recurrent) from women with an ERpos primary tumor: 12 paired tumors from women with ERpos primary and ERpos recurrent tumors and 6 paired tumors from women with ERpos primary and ERneg recurrent tumors (patient and tumor characteristics are summarized in Table 1).
Immunohistochemistry (IHC)
Cells were seeded on poly-l-lysine-coated slides (Polysciences, Cat. No. 22247, Warrington, PA) at a density of 2 × 105 cells/mL/slide. Cells were maintained at 37 °C, 5% CO2 in a humidified incubator overnight and fixed 24 h later. First, the slides were rinsed with cold phosphate-buffered saline solution (PBS) twice and then fixed in ice cold 100% methanol for 10 min. The slides were then allowed to dry and stored at − 20 °C until staining. Immunohistochemical staining was performed at Baystate Medical Center. The slides were stained for TROP2 (Human TROP-2 affinity purified polyclonal antibody, R&D Systems, Cat. No. AF650) using the HRP-DAB Cell and Tissue Staining kit (R&D Systems, Cat. No. CTS008).
Formalin-fixed paraffin-embedded (FFPE) tissue blocks were prepared from the breast tumor samples. The blocks were sectioned (5 μm thick) and placed on slides. Using the UltraView Universal DAB Detection Kit on the BenchMark Ultra platform, the slides were stained for ER, PR and HER2. The slides were stained for TROP2. Hematoxylin and eosin (H&E) slides were prepared and used for tumor verification. The antibodies used for estrogen receptor alpha (ER), progesterone receptor (PR) and epidermal growth factor receptor 2 (HER2) were previously optimized: ER (Ventana anti-estrogen receptor SP1 rabbit monoclonal primary antibody), PR (Ventana anti-progesterone receptor 1E2 rabbit monoclonal primary antibody) and HER2 (Ventana PATHWAY anti-hER2/neu antibody 4B5 rabbit monoclonal antibody). Ethanol was used to dehydrate the slides followed by xylene post-staining and addition of coverslips. The Ki67 data were obtained from the pathology records.
Review of pathology
Scoring of slides was conducted by one anatomic pathologist (RJ). Slides were scored for immunoreactivity of 5 antigens; approximate number of positive cells was recorded (%) and intensity of immunoreactivity was reported. For ER and PR, Allred scores were recorded ranging from 0 to 8. Tumors with a score of 3 or greater were considered positive for receptor status and tumors were considered HER2 positive when 30% of the cells contained 3+ membrane staining. Ki67 scores ranged from 0 to 100% as the percentage of positive cells within the area of invasive cells. For TROP2, the scale for percent positive cells was 0: negative, 1: 1–33%, 2: 34–66% and 3: 67–100% and the intensity was reported as a score from 0 (negative) to 3 (strong), this was recorded for both membrane-localized TROP2 and cytoplasmic TROP2. The overall score for membrane or cytoplasmic TROP2 was determined by multiplying the percent positive score by the intensity score (range 0–9). Scoring of TROP2 was completed in one session with a single observer documenting records (SZ). One of the tumor samples was scored twice for TROP2 with similar results.
DNA purification (human tissues)
DNA was purified from formalin-fixed paraffin embedded (FFPE) blocks as previously described [12] using the BiOstic FFPE tissue DNA isolation kit (Mo Bio, Carlsbad, CA). Briefly, the number of sections needed to purify a minimum of 500 ng of DNA was determined using measurements of the H&E stained slides. Tumor sections were prepared for DNA extraction by aligning the marked tumor H&E slide underneath the unstained slide. A sterile needle was used to remove tissue from the slide.
Plasmids and DNA transfection
The following vectors were received from the Alberti Lab: pSUPER (empty, control vector), hTrop-2siRNA#19 (TACSTD2 knockdown construct), pΔEYFP-N1 (empty, control vector) and hTrop2 in pΔEYFP-N1 (TACSTD2 overexpression construct). MCF7 cells were transfected with 5 μg of either pSUPER (empty, control vector) or hTrop2-siRNA#19 (knockdown construct) plus 200 ng of pΔEYFP-N1 (G418 selection) [20]. TMX2-28 cells were transfected with 2 μg of pΔEYFP-N1 (empty, control vector) or pΔEYFP-N1-hTrop2 with SuperFect (Qiagen No. 301305) per manufacturer’s instructions. Cells were maintained in medium (described above) supplemented with 300 μg/mL of G418 disulfate salt (Sigma Aldrich, Cat No. A1720) prior to seeding for colony isolation. Cells were seeded in a 6-well plate at a density of 1000–2000 cells/well and cultured in selective media until colonies were a sufficient size to harvest (approximately 300+ cells). Colonies were isolated using small cloning cylinders (BellCo Glass, Inc., Vineland, NJ), generating the control-vector transfected cell lines (MCF7-Control and TMX2-28-Control), knockdown (MCF7-TACSTD2-Kd) and overexpression cell lines (TMX2-28-TACSTD2).
Data analysis
The methylation data obtained from the HM450 BeadChip was analyzed using Genome Studio Methylation Module (v.1.9.0). Detection p-values of < 0.01 were used to select statistically significant CpG site data. Methylation data were exported from Genome Studio and averages were calculated in Excel. Fold change values for each CpG site were calculated by division of average beta values. Additional file 1: Table S1, Additional file 2: Table S2, Additional file 3: Table S3 provide the filtering parameters used for specific analyses. Statistical analyses were conducted using the StatPlus application (v. 5.8.2.0) as follows: RT-qPCR data were assessed using un-paired t-tests, CpG methylation of tumor samples was assessed using Two-way ANOVAs and IHC data were assessed using paired t-tests.
Results
5-Aza-dC treatment differentially alters the behavior and DNA methylation of the Tamoxifen-resistant, TMX2-28, compared to the parental cell line, MCF7
Using a MTS assay, a colorimetric assay that indicates metabolic activity, treatment with 5-Aza-dC for 120 h was shown to decrease proliferation of TMX2-28 by approximately 39% compared to the DMSO-control-treated TMX2-28. In contrast, treatment of MCF7 with 5-Aza-dC did not alter proliferation (Fig. 1).
5-Aza-dC treatment decreases proliferation in the Tamoxifen-resistant cell line, TMX2-28. Proliferation, quantified by MTS assay, in control and 120-h 5-Aza-dC-treated cells. Data represented as percentage of control, for the parental line, MCF7, and the ERneg Tamoxifen-resistant cell line, TMX2-28 (n = 8, *p < 0.005)
To determine the role of promoter methylation in the differential proliferation of MCF7 and TMX2-28 in response to treatment with 5-Aza-dC, we examined global DNA methylation using the HM450 BeadChip, which interrogates over 450,000 CpGs. We had previously shown that the Tamoxifen-resistant ERneg cell line is significantly hypermethylated throughout the genome as compared to the parental MCF7 [16]. Here we confirm the increased methylation and demonstrate for the first time that the Tamoxifen-resistant cell line is more sensitive to demethylation by treatment with the DNMT1-inhibitor, 5-Aza-dC. Using the filtering parameters provided in Additional file 1: Table S1 there are 37,501 significantly hypermethylated CpGs in TMX2-28 as compared to MCF7, while there are less than half that number of hypomethylated CpGs (14,956). These differentially methylated CpGs are shown in Fig. 2a as dots above and below the outer red lines. Comparison of 96-h 5-Aza-dC-treated cell cultures with control cell cultures shows a greater effect of the DNMT1-inhibitor in TMX2-28 (Fig. 2c) than in MCF7 (Fig. 2b). Using the filtering parameters provided in Additional file 2: Table S2, the 5-Aza-dC-treated TMX2-28 have 6637 CpGs that are significantly less methylated than the TMX2-28 control, and only 59 CpGs that are hypermethylated as compared to the control. In contrast, the 5-Aza-dC-treated MCF7 has only 1050 hypomethylated and 32 hypermethylated CpGs as compared to the MCF7 control. Accordingly, the correlation of the beta values between the treated and control cell cultures is lower for TMX2-28 (r2 = 0.94) than for MCF7 (r2 = 0.98).
Tamoxifen-resistant cell line is more sensitive than MCF7 to demethylation by 5-Aza-dC treatment. Scatter plots indicating genome-wide methylation differences between the Tamoxifen-resistant cell line, TMX2-28, and the parental line, MCF7 (a), between 96-h 5-Aza-dC-treated MCF7 and MCF7-Control (b), and between 96-h 5-Aza-dC-treated TMX2-28 and TMX2-28-Control (c). Each black dot on the plot represents a CpG site analyzed on the Human Methylation 450 BeadChip. Center red line represents equal average beta values in the two samples and outer red and green triangles indicate the number of CpGs with increased or decreased methylation relative to the cell line on the x-axis, respectively, based on a 1.8-fold change in average beta values
Identification of genes with increased promoter methylation in the Tamoxifen-resistant cell line that are demethylated by 5-Aza-dC
Our goal is to identify genes in Tamoxifen-resistant breast cancer that may provide novel targets for therapy. We approached this by determining the set of CpGs in the promoter region that are both methylated in the Tamoxifen-selected cell line and demethylated by short-term treatment with 5-Aza-dC, as these CpGs may regulate the expression of genes responsible for the decrease in proliferation observed in the TMX2-28 after treatment with 5-Aza-dC. Using the filtering parameters provided in Additional file 3: Table S3, only 707 of the 37,501 CpG sites with increased methylation in TMX2-28 are significantly decreased after treatment with 5-Aza-dC (Additional file 4: Table S4). To determine the extent to which these CpGs are likely to be involved in regulating gene expression, we determined their distribution in the genome. Of the 707 CpG sites, 251 (35%) are located in the promoter (TSS200 or TSS1500 region), 129 (18%) are in the 5′UTR/1st Exon, 223 (32%) are in the body, 13 (2%) are in the 3′UTR, and 91 (13%) are intergenic (neighborhood locations are shown in Additional file 5: Figure S1 and an overview of the selection process in Additional file 6: Figure S2). The 251 CpG sites in the promoter region that are hypermethylated in TMX2-28 and have decreased methylation after 5-Aza-dC treatment are located in 219 genes, of which 27 genes contain two or more of the CpGs (Table 2 and highlighted in Additional file 4: Table S4). We predicted that these target genes are likely re-expressed after 5-Aza-dC treatment and involved in regulating cell behavior. Using RT-qPCR and mRNA expression arrays we examined the expression of 11 of the 27 genes and determined that 6 of these genes were downregulated in TMX2-28 compared to MCF7. Two of these 6 genes (TACSTD2 and CGNL1) were significantly re-expressed in TMX2-28 after 5-Aza-dC treatment (Table 2).
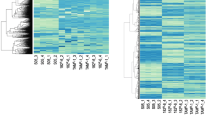
We selected one gene, TACSTD2, for detailed study. As shown in Fig. 3a, three CpG sites in the promoter of TACSTD2 were identified as significantly hypermethylated in TMX2-28 as compared to MCF7 and show decreased methylation after treatment with 5-Aza-dC. The mean average beta values for these three CpGs in MCF7-Control, TMX2-28-Control and TMX2-28-5-Aza-dC-treated cell lines are 0.04, 0.71 and 0.32, respectively. Mean methylation of these three CpGs in TMX2-28 decreased by 45% after treatment with 5-Aza-dC but remained significantly higher than MCF7. Next, we confirmed the results from the HM450 BeadChip using pyrosequencing. Interrogation of six CpG sites in the promoter region of TACSTD2 verified that treatment with 5-Aza-dC, for either 48 or 72 h, decreases TACSTD2 promoter methylation in TMX2-28 but does not affect methylation in MCF7 (Fig. 3b). We next asked if the decrease in methylation in TMX2-28 results in increased expression. Figure 3c shows that treatment with 5-Aza-dC resulted in a significant increase in mRNA levels of TACSTD2 in TMX2-28, but not MCF7. The gene chosen for normalization (beta-actin) did not differ between treatment groups (Additional file 7: Figure S3). These results indicate that TACSTD2 expression is likely regulated by promoter methylation, and support a role of TROP2 in the proliferation differences observed between TMX2-28 and MCF7, as well as a possible role of TROP2 in hormone-resistant breast cancer.
5-Aza-dC decreases TACSTD2 DNA promoter methylation and is accompanied by increased mRNA expression in TMX2-28. DNA methylation was analyzed with the Illumina Human Methylation 450 BeadChip. Heatmap represents methylation of CpG sites (average beta-values, scale at right) in TACSTD2 (a), for MCF7-Control, TMX2-28-Control and TMX2-28-5-Aza-dC-treated cell lines. Left: Refgene Group location and MAPINFO number for each CpG site. Bottom: Mean average beta values for the 3 CpGs (orange box) identified as hypermethylated in TMX2-28 compared to MCF7 that have decreased methylation in TMX2-28 after 5-Aza-dC treatment. b Percent DNA methylation of six CpG sites in the promoter region of TACSTD2 determined by pyrosequencing (two of which were CpG sites identified as differentially methylated on the HM450 BeadChip, indicated by orange boxes, CpGs 2 and 6 correspond to MAPINFO numbers 59043280 and 59043255, respectively) for control and 5-Aza- -treated cells. Relative TACSTD2 mRNA expression was quantified by RT-qPCR (c) in control or 48-h or 72-h 5-Aza-treated cells, normalized to beta-actin (n = 3, *p < 0.01, **p < 0.001)
TACSTD2 methylation and TROP2 expression in primary and recurrent Tamoxifen-resistant breast cancers
Results from the cell lines suggest that TROP2 may be silenced by promoter methylation in recurrent Tamoxifen-resistant breast cancers, but to date, there are no published studies investigating TROP2 in Tamoxifen-resistant breast cancer. Therefore we used an available DNA methylation data set of 70 primary and recurrent tumors [12] to ask if recurrent breast tumors from women who received anti-estrogen therapy exhibited increased promoter methylation and if methylation was correlated with expression. The data set of 34 primary breast cancers (including 8 primaries that did not recur; 6 ERpos and 2 ERneg), 34 recurrent breast cancers (including 30 first recurrences and 4 second recurrences) and two metastatic breast cancers, included 18 women who had ERpos primary cancers, received anti-estrogen treatment, and had either ERpos (n = 12) or ERneg (n = 6), recurrences. Patient demographics for the 18 women are summarized in Table 1 and for all 70 women are summarized in Additional file 8: Table S5).
Limiting the analyses to those women who received anti-estrogen treatment, comparison of TACSTD2 promoter methylation (the 3 CpGs that were significantly methylated in TMX2-28) showed no difference between ERpos primary and recurrent tumors (mean beta = 0.142, n = 18 versus 0.138, n = 18 for the primary and recurrent, respectively; see Additional file 9: Table S6). Further stratifying the recurrent tumors by ER status showed no significant increase in methylation of ERneg recurrent tumors (Fig. 4a), or ERpos recurrent tumors (Fig. 4d).
TACSTD2 methylation and TROP2 staining in primary and recurrent breast tumors. Mean beta values for each of three CpGs in the promoter region of TACSTD2 (MAPINFO numbers 59043370, 59043280 and 59043255), which were previously identified as hypermethylated in TMX2-28, are shown for 6 sets of ERpos primary to ERneg recurrent breast cancers (a), and 12 sets of ERpos primary to ERpos recurrent breast cancers (d). There was no significant change in methylation between primary and recurrent tumors when the recurrent tumor was ERneg (a: p = 0.25) or ERpos (d: p = 0.27). Membrane TROP2 and Ki67 expression, determined by IHC, for same sets of primary and recurrent breast tumors shown in (b, e) and (c, f). There was no significant difference in membrane TROP2 expression between primary and recurrent tumor when the recurrent tumor was ERneg (b: p = 0.891) and or ERpos (e: p = 0.067; n = 10 because staining failed in two recurrent tumors). There was no significant difference in Ki67 expression between primary and recurrent tumor when the recurrent tumor was ERneg (c: p = 0.182) or ERpos (f: p = 0.463)
We next asked if the DNA methylation was associated with protein expression. Protein expression and localization (cytoplasmic and/or membrane) of TROP2 were determined in 66 tumor samples by IHC. Examples of cytoplasmic and membrane TROP2 staining are shown in Fig. 5. Analysis of all 66 tumors showed that there was no correlation between the TACSTD2 methylation (average of the three CpGs) with either cytoplasmic or membrane TROP2 staining (r2 = 0.0033 and 0.00063, respectively); nor was there a correlation between methylation and protein staining when the data were stratified by ER status of the primary and recurrent tumors (ERpos primary to ERpos recurrent; n = 12; r2 = 0.023 for cytoplasmic; r2 = 0.040 for membrane; ERpos primary to ERneg recurrent; n = 6; r2 = 0.044 for cytoplasmic; r2 = 0.062 for membrane).
Stratifying by ER status of the recurrent tumors, and limiting our analyses to just those women who received anti-estrogen treatment, there was slightly greater TROP2 expression in the ERpos recurrent as compared to the ERneg recurrent (Fig. 4b, e). Considering the small sample size in the subgroups of tumors and the slight increased methylation and decreased expression in ERneg recurrent cancers, it remains feasible that TROP2 plays a role in hormone-resistant breast cancer. Therefore, we further investigated the role of TROP2 in proliferation in the cell lines.
Proliferation in TMX2-28-TACSTD2-expressing and MCF7-TACSTD2-knockdown cell lines
To examine the role of TROP2 in regulating proliferation, stable cell lines were generated with reduced (MCF7-TACSTD2-Kd) and increased (TMX2-28-TACSTD2) expression of TACSTD2. Control cell lines with a vector containing either the scrambled control shRNA (MCF7-Control) or lacking the TACSTD2 coding sequence (TMX2-28-Control) were also generated. As determined by RT-qPCR, transfection with the shRNA vector against TACSTD2 resulted in an 80% knockdown of TACSTD2 mRNA compared to MCF7-Control, and TACSTD2 mRNA expression increased by more than 100% in TMX2-28-TACSTD2 (Fig. 6a). Examination of stained cultures by IHC showed decreased TROP2 expression in the MCF7-TACSTD2-Kd cell line, and increased expression of TROP2 in the TMX2-28-TACSTD2 cell line (Fig. 6c and d). If TROP2 plays a major role in the phenotype observed in response to 5-Aza-dC treatment, TMX2-28-TACSTD2 should exhibit decreased proliferation compared to TMX2-28-Control. In contrast to our expectation, proliferation was not decreased in TMX2-28-TACSTD2. Also surprising was the finding that knockdown of TACSTD2 in MCF7 resulted in a slight increase in proliferation (25%), compared to MCF7-Control (Fig. 6b).
TACSTD2 expression and proliferation and stable expression of TROP2 in TMX2-28 and knockdown of TROP2 in MCF7. (a) TACSTD2 mRNA expression determined by RT-qPCR run in technical triplicate for MCF7 (non-transfected), MCF7-Control, MCF7-TACSTD2-Knockdown, TMX2-28 (non-transfected), TMX2-28-Control and TMX2-28-TACSTD2 cell lines (**p < 0.001). b The MTS assay was used to quantify cell proliferation in MCF7-TACSTD2-Kd and TMX2-28-TACSTD2 cell lines (n = 16), represented as percent of control for each respective cell line (***p < 0.0001). c Expression of TROP2 determined by IHC for MCF7-Control-Clone and MCF7-TACSTD2-Knockdown-Clone. d Expression of TROP2 determined by IHC for TMX2-28-Control-Clone and TMX2-28-TACSTD2-Clone. Magnification = 200×
Discussion
Several studies, including one from our lab, indicate that DNA methylation may be involved in the development of endocrine-resistant breast cancer [13,14,15,16]. Therefore, epigenetic therapies that reverse aberrant DNA methylation may prove to be promising therapy for Tamoxifen-resistant disease. However, it is unclear how DNMT inhibitors such as 5-Aza-dC affect methylation and cell behavior, specifically in ERneg, acquired Tamoxifen resistance.
Treatment with 5-Aza-dC inhibited the proliferation of the Tamoxifen-selected, TMX2-28, but not the parental MCF7. TMX2-28 is hypermethylated compared to MCF7 ([12, 16] and Fig. 2) and was more sensitive to inhibition of DNA methylation by treatment with 5-Aza-dC (Fig. 2b and c) suggesting that epigenetic treatment might be particularly beneficial for women with endocrine-resistant breast cancer. The genes identified as methylated in TMX2-28 and demethylated and potentially re-expressed after treatment with 5-Aza-dC may be good biomarkers of sensitivity to epigenetic therapy. We focused on one of these genes, TACSTD2, and its product, TROP2, because of its importance as a potential target for treatment.
TACSTD2 is overexpressed in many cancers and has been associated with disease progression, migration, recurrence, and increased proliferation [27, 33,34,35,36]. In published cell culture experiments, knockdown of TACSTD2 inhibits growth in MCF7 (contrary to our findings) and colon cancer cells [20], fetal rat lung cells (fibroblasts) [28], fetal lung fibroblasts [27], cervical cancer cells [26], and laryngeal carcinoma cells [30]. However, there also is evidence of decreased TACSTD2 expression in cancer [20, 23, 43, 44]. Lin and colleagues [23] reported that knockdown of TACSTD2 in lung cancer cells expressing high levels of endogenous TACSTD2, increased AKT activation and promoted growth. They further showed that promoter methylation is correlated with decreased TACSTD2 expression in lung cancer cell lines and tumors [23]. In breast cancer, high levels of membrane TROP2 have been associated with poor prognosis while cytoplasmic staining indicated better survival [21]. Analysis of 2061 ERpos breast cancers using the Kaplan–Meier Plotter indicates that low levels of TACSTD2 mRNA are associated with decreased survival (p = 0.018; Additional file 10: Figure S4), while analysis of 801 ERneg breast cancers provides the opposite result: low levels of TACSTD2 are associated with increased survival (p = 0.083; Additional file 10: Figure S4). [37] The extent to which TACSTD2 mRNA levels in these 2862 ERpos and ERneg breast cancers is associated with membrane versus cytoplasmic TROP2 is unknown. Despite the discrepancies, the evidence for over-expression of TACSTD2 in cancer is sufficiently compelling to support clinical trials targeting membrane TROP2 [38,39,40,41,42,43,44,45].
Conclusions
Our results contribute to the growing literature examining the extent to which TROP2 acts as an oncogene or a tumor suppressor. These results also illustrate the complexity of the TROP2 signaling network. Although we found no correlation between promoter methylation and TROP2 expression in patients who received anti-hormone therapy, it would be important to investigate promoter methylation and protein expression in a larger population, as methylation may still play a role in regulating gene expression. Accuracy of IHC scoring may be improved by the use of antibodies specific to cytoplasmic-localized TROP2 and membrane-localized TROP2. Interestingly, we found that primary tumors that did not recur during the time of patient follow-up (approximately 7 years) had higher expression of membrane TROP2 than the ERpos tumors that recurred (data not shown). This also would be important to examine in a larger sample size, as therapies targeted against TROP2 may be unnecessary treatment for some patients. TROP2 may be a valid therapeutic target for some cancers, however, further studies are needed to identify biomarkers that indicate how TROP2 signaling affects tumor growth and whether targeting TROP2 would be beneficial to the patient.
Abbreviations
- TROP2:
-
trophoblast antigen protein 2
- TACSTD2 :
-
tumor-associated calcium signal transducer 2
- 5-Aza-dC:
-
5-aza-2′ deoxycytidine (or decitabine)
- ERneg/ERpos:
-
estrogen receptor-negative/positive
- DNMT1:
-
DNA methyltransferase 1
- FFPE:
-
formalin-fixed paraffin-embedded
- IHC:
-
immunohistochemistry
References
Azad N, Zahnow CA, Rudin CM, Baylin SB. The future of epigenetic therapy in solid tumours—lessons from the past. Nat Rev Clin Oncol. 2013;10(5):256–66.
Widschwendter M, Jones P. DNA methylation and breast carcinogenesis. Oncogene. 2002;21(35):5462–82.
Jones P. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–92.
Lay FD, Liu Y, Kelly TK, Witt H, Farnham PJ, Jones PA, Berman BP. The role of DNA methylation in directing the functional organization of the cancer epigenome. Genome Res. 2015;25(4):467–77.
Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31(1):27–36.
Yang X, Yan L, Davidson NE. DNA methylation in breast cancer. Endocr Relat Cancer. 2001;8(2):115–27.
Figueroa JD, Yang H, Garcia-Closas M, Davis S, Meltzer P, Lissowska J, Horne HN, Sherman ME, Lee M. Integrated analysis of DNA methylation, immunohistochemistry and mRNA expression, data identifies a methylation expression index (MEI) robustly associated with survival of ER-positive breast cancer patients. Breast Cancer Res Treat. 2015;150(2):457–66.
Bediaga NG, Acha-Sagredo A, Guerra I, Viguri A, Albaina C, Ruiz Diaz I, Rezola R, Alberdi MJ, Dopazo J, Montaner D, Renobales M, Fernandez AF, Field JK, Fraga MF, Liloglou T, de Pancorbo MM. DNA methylation epigenotypes in breast cancer molecular subtypes. Breast Cancer Res. 2010;12(5):R77.
Benevolenskaya EV, Islam AB, Ahsan H, Kibriya MG, Jasmine F, Wolff B, Al-Alem U, Wiley E, Kajdacsy-Balla A, Macias V, Rauscher GH. DNA methylation and hormone receptor status in breast cancer. Clin Epigenetics. 2016;8:17.
Fackler MJ, Umbricht CB, Williams D, Argani P, Cruz LA, Merino VF, Teo WW, Zhang Z, Huang P, Visvananthan K, Marks J, Ethier S, Gray JW, Wolff AC, Cope LM, Sukumar S. Genome-wide methylation analysis identifies genes specific to breast cancer hormone receptor status and risk of recurrence. Cancer Res. 2011;71(19):6195–207.
Fang F, Turcan S, Rimner A, Kaufman A, Giri D, Morris LG, Shen R, Seshan V, Mo Q, Heguy A, Baylin SB, Ahuja N, Viale A, Massague J, Norton L, Vahdat LT, Moynahan ME, Chan TA. Breast cancer methylomes establish an epigenomic foundation for metastasis. Sci Transl Med. 2011;3(75):75ra25.
Williams KE. DNA-based epigenetic changes in recurrent and tamoxifen-resistant breast cancer. University of Massachusetts Amherst, Ph.D. Dissertation, 2016.
Stone A, Valdés-Mora F, Gee JM, Farrow L, McClelland R, Fiegl H, Dutkowski C, McCloy R, Sutherland R, Musgrove E, Nicholson R. Tamoxifen-induced epigenetic silencing of oestrogen-regulated genes in anti-hormone resistant breast cancer. PLoS ONE. 2012;7(7):e40466.
Fan M, Yan P, Hartman-Frey C, Chen L, Paik H, Oyer S, Salisbury J, Cheng ASL, Li L, Abbosh P, Huang TH, Nephew K. Diverse gene expression and DNA methylation profiles correlate with differential adaptation of breast cancer cells to the antiestrogens Tamoxifen and fulvestrant. Cancer Res. 2006;66(24):11954–66.
Lin X, Li J, Yin G, Zhao Q, Elias D, Lykkesfeldt A, Stenvang J, Brünner N, Wang J, Yang H, Bolund L, Ditzel H. Integrative analyses of gene expression and DNA methylation profiles in breast cancer cell line models of Tamoxifen-resistance indicate a potential role of cells with stem-like properties. Breast Cancer Res. 2013;15(6):R119.
Williams KE, Anderton DL, Lee MP, Pentecost BT, Arcaro KF. High-density array analysis of DNA methylation in Tamoxifen-resistant breast cancer cell lines. Epigenetics. 2014;9(2):297–307.
Fornaro M, Dell’Arciprete R, Stella M, Bucci C, Nutini M, Capri MG, Alberti S. Cloning of the gene encoding Trop-2, a cell-surface glycoprotein expressed by human carcinomas. Int J Cancer. 1995;62(5):610–8.
Shvartsur A, Bonavida B. Trop2 and its overexpression in cancers: regulation and clinical/therapeutic implications. Genes Cancer. 2015;6(3–4):84–105.
McDougall AR, Tolcos M, Hooper SB, Cole TJ, Wallace MJ. Trop2: from development to disease. Dev Dyn. 2014;244(2):99–109.
Trerotola M, Cantanelli P, Guerra E, Tripaldi R, Aloisi AL, Bonasera V, Lattanzio R, de Lange R, Weidle UH, Piantelli M, Alberti S. Upregulation of Trop-2 quantitatively stimulates human cancer growth. Oncogene. 2013;32(2):222–33.
Ambrogi F, Fornili M, Boracchi P, Trerotola M, Relli V, Simeone P, La Sorda R, Lattanzio R, Querzoli P, Pedriali M, Piantelli M, Biganzoli E, Alberti S. Trop-2 is a determinant of breast cancer survival. PLoS ONE. 2014;9(5):e96993.
Sawanyawisuth K, Tantapotinan N, Wongkham C, Riggins GJ, Kraiklang R, Wongkham S, Puapairoj A. Suppression of trophoblast cell surface antigen 2 enhances proliferation and migration in liver fluke-associated cholangiocarcinoma. Ann Hepatol. 2016;15(1):71–81.
Lin J, Wu Y, Wu J, Lin T, Wu C, Chang Y, Jou Y, Hong T, Yang P. TROP2 is epigenetically inactivated and modulates IGF-1R signalling in lung adenocarcinoma. EMBO Mol Med. 2012;4(6):472–85.
Kanai Y, Ushijima S, Saito Y, Nakanishi Y, Sakamoto M, Hirohashi S. MRNA expression of genes altered by 5-azacytidine treatment in cancer cell lines is associated with clinicopathological parameters of human cancers. J Cancer Res Clin Oncol. 2001;127(12):697–706.
Cubas R, Zhang S, Li M, Chen C, Yao Q. Trop2 expression contributes to tumor pathogenesis by activating the ERK MAPK pathway. Mol Cancer. 2010;9:253.
Liu T, Liu Y, Bao X, Tian J, Liu Y, Yang X. Overexpression of TROP2 predicts poor prognosis of patients with cervical cancer and promotes the proliferation and invasion of cervical cancer cells by regulating ERK signaling pathway. PLoS ONE. 2013;8(9):e75864.
McDougall AR, Hooper SB, Zahra VA, Sozo F, Lo CY, Cole TJ, Doran T, Wallace MJ. The oncogene Trop2 regulates fetal lung cell proliferation. Am J Physiol Lung Cell Mol Physiol. 2011;301(4):L478–89.
McDougall AR, Hooper SB, Zahra VA, Cole TJ, Lo CY, Doran T, Wallace MJ. Trop2 regulates motility and lamellipodia formation in cultured fetal lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. 2013;305(7):L508–21.
Trerotola M, Jernigan DL, Liu Q, Siddiqui J, Fatatis A, Languino LR. Trop-2 promotes prostate cancer metastasis by modulating beta(1) integrin functions. Cancer Res. 2013;73(10):3155–67.
Wang XD, Wang Q, Chen XL, Huang JF, Yin Y, Da P, Wu H. Trop2 inhibition suppresses the proliferation and invasion of laryngeal carcinoma cells via the extracellular signal-regulated kinase/mitogen-activated protein kinase pathway. Mol Med Rep. 2015;12(1):865–70.
Gozgit J, Pentecost B, Marconi S, Otis C, Wu C, Arcaro K. Use of an aggressive MCF7 cell line variant, TMX2-28, to study cell invasion in breast cancer. Mol Cancer Res. 2006;4(12):905–13.
Gozgit JM, Pentecost BT, Marconi SA, Ricketts-Loriaux RS, Otis CN, Arcaro KF. PLD1 is overexpressed in an ER-negative MCF7 cell line variant and a subset of phospho-Akt-negative breast carcinomas. Br J Cancer. 2007;97(6):809–17.
Fang YJ, Lu ZH, Wang GQ, Pan ZZ, Zhou ZW, Yun JP, Zhang MF, Wan DS. Elevated expressions of MMP7, TROP2, and survivin are associated with survival, disease recurrence, and liver metastasis of colon cancer. Int J Colorectal Dis. 2009;24(8):875–84.
Ning S, Liang N, Liu B, Chen X, Pang Q, Xin T. TROP2 expression and its correlation with tumor proliferation and angiogenesis in human gliomas. Neurol Sci. 2013;34(10):1745–50.
Ohmachi T, Tanaka F, Mimori K, Inoue H, Yanaga K, Mori M. Clinical significance of TROP2 expression in colorectal cancer. Clin Cancer Res. 2006;12(10):3057–63.
Zhao P, Yu HZ, Cai JH. Clinical investigation of TROP-2 as an independent biomarker and potential therapeutic target in colon cancer. Mol Med Rep. 2015;12(3):4364–9.
Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–31.
Lin H, Zhang H, Wang J, Lu M, Zheng F, Wang C, Tang X, Xu N, Chen R, Zhang D, Zhao P, Zhu J, Mao Y, Feng Z. A novel human Fab antibody for Trop2 inhibits breast cancer growth in vitro and in vivo. Int J Cancer. 2014;134(5):1239–49.
Liu D, Cardillo TM, Wang Y, Rossi EA, Goldenberg DM, Chang CH. Trop-2-targeting tetrakis-ranpirnase has potent antitumor activity against triple-negative breast cancer. Mol Cancer. 2014;13:53.
Liu T, Tian J, Chen Z, Liang Y, Liu J, Liu S, Li H, Zhan J, Yang X. Anti-TROP2 conjugated hollow gold nanospheres as a novel nanostructure for targeted photothermal destruction of cervical cancer cells. Nanotechnology. 2014;25(34):345103.
Sharkey RM, van Rij CM, Karacay H, Rossi EA, Frielink C, Regino C, Cardillo TM, McBride WJ, Chang CH, Boerman OC, Goldenberg DM. A new Tri-Fab bispecific antibody for pretargeting Trop-2-expressing epithelial cancers. J Nucl Med. 2012;53(10):1625–32.
Starodub AN, Ocean AJ, Shah MA, Guarino MJ, Picozzi VJ Jr, Vahdat LT, Thomas SS, Govindan SV, Maliakal PP, Wegener WA, Hamburger SA, Sharkey RM, Goldenberg DM. First-in-human trial of a novel anti-trop-2 antibody-SN-38 conjugate, sacituzumab govitecan, for the treatment of diverse metastatic solid tumors. Clin Cancer Res. 2015;21(17):3870–8.
van Rij CM, Lutje S, Frielink C, Sharkey RM, Goldenberg DM, Franssen GM, McBride WJ, Rossi EA, Oyen WJ, Boerman OC. Pretargeted immuno-PET and radioimmunotherapy of prostate cancer with an anti-TROP-2 x anti-HSG bispecific antibody. Eur J Nucl Med Mol Imaging. 2013;40(9):1377–83.
Wang J, Zhang K, Grabowska D, Li A, Dong Y, Day R, Humphrey P, Lewis J, Kladney RD, Arbeit JM, Weber JD, Chung CH, Michel LS. Loss of Trop2 promotes carcinogenesis and features of epithelial to mesenchymal transition in squamous cell carcinoma. Mol Cancer Res. 2011;9(12):1686–95.
Zhang K, Jones L, Lim S, Maher CA, Adkins D, Lewis J, Kimple RJ, Fertig EJ, Chung CH, Van Tine BA, Ellis MJ, Herrlich A, Michel LS. Loss of Trop2 causes ErbB3 activation through a neuregulin-1-dependent mechanism in the mesenchymal subtype of HNSCC. Oncotarget. 2014;5(19):9281–94.
Williams KE, Jawale RM, Schneider SS, Otis CN, Pentecost BT, Arcaro KF. DNA methylation in breast cancers: differences based on estrogen receptor status and recurrence. J Cell Biochem. 2018 (in press).
Authors’ contributions
SZ, KA, KW and SS designed the study. SZ carried out the cell culture experiments with the help of EB. CO and RJ obtained the FFPE tissues and analyzed the IHC data. KW isolated DNA from FFPE tissues. KA, SZ and EB reviewed the patient data, and analyzed the HM450 K data. SZ, SS, EB and KA wrote the paper and made the figures. All authors read and approved the final manuscript.
Acknowledgements
The authors thank Elizabeth Punska and Ana Caballero for helping to maintain cell lines, Brooke Bentley for staining the tissue block samples, Dr. Saverio Alberti and Dr. Marco Trerotola for generously providing the pSUPER, hTrop-2siRNA#19, pΔEYFP-N1 and hTrop2 in pΔEYFP-N1 vectors. Special thanks to Dr. Brian Pentecost for helpful discussions on DNA methylation and breast cancer, and to Dr. Katerina Fagan-Solis for helpful discussions on transfection optimization.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Data from the cell line and tumor sample DNA methylation microarray experiments will be made available on the National Center for Biotechnology Information’s gene expression omnibus after the publication of [46].
Consent for publication
Not applicable.
Ethics approval and consent to participate
The collection and use of FFPE tissue was approved by the Institutional Review Board of Baystate Medical Center. (IRB REFERENCES #: BH13-013).
Funding
The design and research of this publication were funded by a Department of Defense CDMRP Breast Cancer Predoctoral grant to KW, by awards from the AVON Foundation for Women and the Rays of Hope Foundation to KA, and by a Baystate Medical Center Incubator award to CO.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1:Table S1.
Changes in Global Methylation Between Tamoxifen-Resistant Cell Lines and MCF-7.
Additional file 2: Table S2.
Changes in Global Methylation Between 5-Aza-dC- and Control-Treated Cell Lines.
Additional file 3: Table S3.
Hypermethylated CpG Sites in TMX2-28 compared to MCF-7 that have decreased methylation after 5-Aza-dC treatment.
Additional file 4: Table S4.
List of CpGs that are Hypermethylated in TMX2-28 compared to MCF-7 and have Decreased Methylation after 5-Aza-dC Treatment.
Additional file 5: Figure S1.
Functional genomic location and neighborhood distribution of hypermethylated CpG Sites in TMX2-28 compared to MCF-7 that have decreased methylation after 5-Aza-dC treatment.
Additional file 6: Figure S2.
Overview of selection process for target genes.
Additional file 7: Figure S3.
Raw Ct values for β‐actin RT‐qPCR using two different cell lines (MCF7 and TMX2‐28) and different lengths of 5‐Aza‐dC treatment (5‐aza).
Additional file 8: Table S5.
Patient (n= 70) and tumor characteristics.
Additional file 9: Table S6.
Beta values for TACSTD2 methylation in clinical samples (ERposprimaries that recur, mean of 3CpGs).
Additional file 10: Figure S4.
Kaplan Meier plots stratified by ER status.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zimmers, S.M., Browne, E.P., Williams, K.E. et al. TROP2 methylation and expression in tamoxifen-resistant breast cancer. Cancer Cell Int 18, 94 (2018). https://doi.org/10.1186/s12935-018-0589-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-018-0589-9