Abstract
Background
Epithelial mesenchymal transition (EMT) is thought to be an essential feature of malignant tumor cells when they spread into the stroma. Despite the extracellular acidity of tumor tissues, the effect of acidic extracellular pH (pH e ) on EMT in carcinoma models, including the Lewis lung carcinoma (LLC) model, remains unclear.
Methods
High and low metastatic LLC variants were generated by repeated tail vein injection of metastatic cells. DMEM/F12 medium, which has been supplemented with 15 mM HEPES, 4 mM phosphoric acid, and 1 g/L NaHCO3 and adjusted to the desire pH with HCl or NaOH, was used for cell culture. EMT marker gene expression was determined by quantitative reverse transcription-polymerase chain reaction. Migration and invasion activities were analyzed by wound healing assay and the Boyden chamber assay through Matrigel®, respectively.
Results
Low metastatic variant LLCm1 cells showed a cobble-stone like morphology at pH e 7.4. At pH e 6.8, however, their morphology became fibroblastic, similar in shape to high metastatic variant LLCm4 cells. Steady state levels of matrix metalloproteinase-9 (Mmp9) mRNA were induced by acidic pH e , maximizing at pH 6.8, with the levels of Mmp9 mRNA higher in LLCm4 than in LLCm1 cells. Both variants showed decreased levels of E-cadherin and increased levels of vimentin at pH e 6.8. Acidic pH e also induced expression of mRNAs encoding the E-cadherin repressors, Zeb2, Twist1 and Twist2, as well as enhancing cell motility and in vitro invasion through Matrigel®.
Conclusions
Acidic pH e can induce EMT in some types of carcinoma.
Similar content being viewed by others
Background
Cancer metastasis involves complex steps. The invasion of cancer cells into surrounding tissues is accompanied by a change in properties from epithelial to mesenchymal, i.e., epithelial mesenchymal transition (EMT). EMT can be induced by cytokines and growth factors, such as epidermal growth factor (EGF), basic fibroblast growth factor (bFGF/FGF2), and transforming growth factor-β (TGF-β), either alone or in combinations. EMT is also accompanied by up-regulation of the expression of vimentin, N-cadherin, and E-cadherin repressors such as Snail, Slug, Twist1, Twist2, Zeb1, and Zeb2 (see [1] for review). Down-regulation of E-cadherin causes dysfunction at adherence junctions, leading to detachment of cancer cells from their primary site [2].
Acidic extracellular pH (pH e ) is an important feature of solid tumors [3] that induces tumor metastasis [4]. Indeed, acidic pH e is an important microenvironmental factor in metastasis induction [5]. Using mouse metastatic B16 melanoma cells, we found that acidic pH e induces cellular expression of matrix metalloproteinase-9 (MMP-9) and induces morphological changes to a fibroblastic phenotype [6]. In addition, acidic pH e was found to induce EMT-like changes in human melanoma cells [7]. Melanomas are tumors that are transformed from melanocytes derived from the neural crest, but do not show typical properties of EMT [8]. These results suggested that acidic pH e may act as a microenvironment inducing EMT in carcinoma models. Here, we tested this hypothesis using two variants of Lewis lung carcinoma (LLC) with different metastatic activities.
Results
Establishment of LLC variants with different metastatic abilities
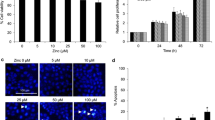
To compare the effect of acidic pH e on metastatic potential, we established two LLC variants (LLCm1 and LLCm4) by repeating cycles of the experimental pulmonary metastases. LLCm1 cells had a cobble-stone like morphology and were tightly attached to each other, whereas LLCm4 cells had a spindle shaped morphology and were scattered (Figure 1A). The metastatic potential of LLCm4 cells was 9 times higher than that of LLCm1 cells (Figure 1B).
Morphology and metastatic abilities of LLC variants. A. Photographs taken under an inverted phase contrast microscope of logarithmic phase cells grown in 10% FBS-containing medium. Bar, 100 μm. B. Cells were injected into the tail veins of syngeneic mice. Three weeks later, foci that had metastasized to the lungs were counted. Representative results were shown from two independent experiments. ***P<0.001.
Acidic pH e induces MMP-9 production
As shown previously, MMP-9 is an acidic pH e -signal target gene, with the acid-induced level of MMP-9, but not MMP-2, expression positively correlated with the metastatic potential of mouse B16 melanoma variants [6]. We therefore assessed whether acidic pH e induces MMP-9 production, and whether its level correlated with the metastatic potential of the two LLC variants. MMP-9 production by both variants was clearly pH e dependent (Figure 2), with maximal production at pH e 6.8. The acid pH e stimulated expression of MMP-9 was much higher by LLCm4 than by LLCm1 cells, while LLCm4 cells produced constitutively higher level of MMP-9 as the basal level at pH e 7.4. Although MMP-2 level was also slightly induced by acidic pH e , the highly metastatic LLCm4 cells produced much less MMP-2 than LLCm1 cells. This finding was in accordance with the mouse B16 melanoma model, in that MMP-2 levels are high in parental B16 cells but negligible in highly metastatic B16-F10 and B16-BL6 cells [6].
Acidic pH e induces MMP-9 production. Sub-confluent cells were cultured overnight in serum-free medium at pH 7.4 and stimulated with serum-free medium at the indicated pH. After 24 h, the conditioned medium was collected, concentrated, and analyzed by gelatin zymography. A. A clear zone indicates gelatinolytic activity. B. Intensity of zymogram was quantified by Scion Image (Scion corp., Frederick, MD, USA). Representative results were shown from three independent experiments. *P < 0.01; **P < 0.01; ***P < 0.001; NS, not significant.
MMP-9 induction by acidic pH e was also confirmed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Accordance with zymographic analysis (Figure 2), the basal level of Mmp9 mRNA at pH e 7.4 was higher in LLCm4 cells than LLCm1 cells. Acid pH e enhanced the steady state levels of Mmp9 mRNA 2-fold in LLCm1 cells and 6-fold in LLCm4 cells (Figure 3). In addition to Mmp9, Mmp3, and Mmp13 mRNA expressions were also stimulated by acidic pH e . Although induction of MMP-2 secretion was observed (see Figure 2), increase in Mmp2 mRNA expression was not statistically significant, suggesting a possibility that acidic pH e affects the efficiency of protein translation. Mmp14 mRNA, whose protein is critical for MMP-2 activation, was higher in LLCm1 cells than LLCm4 cells and was not affected by acidic pH e .
Acidic pH e induces not only Mmp9 but also Mmp3 and Mmp13 mRNA expressions but not obvious Mmp2 and Mmp14 mRNA expressions. Sub-confluent cell cultures were pretreated overnight with serum-free medium at pH 7.4 and stimulated by serum-free media at pH 6.8 or 7.4. After 24 h, total RNA was extracted, reverse-transcribed, and amplified by qPCR with specific primer sets. Data are shown as relative expression compared with LLCm1 cells cultured at pH 7.4. Representative results were shown from three or more independent experiments. **P < 0.01; ***P < 0.001; NS, not significant.
Acidic pH e induces morphological changes to a fibroblastic morphology
Since the optimal pH e value for MMP-9 induction was about pH 6.8, further experiments compared the effects of neutral (pH 7.4) and acidic (pH 6.8) pH e . At neutral pH e , LLCm1 cells had a cobble-stone appearance. When cultured at acidic pH e , however, their morphology became fibroblastic and of similar shape as LLCm4 cells (Figure 4). In contrast to LLCm1 cells, change in pH e did not induce morphological changes in LLCm4 cells. These morphological observations suggested that acidic pH e may induce EMT, however, the interesting lack of effect in the already mesenchymal cells should be mentioned.
Morphological changes of LLC variants in acidic medium. Cultures were pretreated overnight with serum-free DMEM/12 at pH 7.4 and incubated in serum-free medium at pH 6.8 or 7.4 for 24 h. Photographs were taken under an inverted phase contrast microscope. Representative results were shown from three or more independent experiments. Bar, 50 μm.
Acidic pH e induces EMT-related gene expression
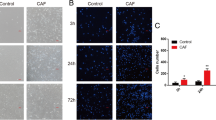
To determine whether acidic pH e induces EMT, we analyzed the expression of E-cadherin and vimentin using immunocytochemistry. In LLCm1 cells, E-cadherin and vimentin were significantly down- and up-regulated, respectively, by acidic pH e , findings typical of EMT (Figure 5). Interestingly, increase in ectodomain of E-cadherin was observed especially in LLCm1 cells (Figure 5E), suggesting that E-cadherin shedding was occurred by acidic pH e . Expression of Cdh1 (E-cadherin) mRNA in both LLCm1 and LLCm4 cells was significantly decreased by incubation at acidic pH e (Figure 6). Moreover, acidic pH e increased the expression of Vim (vimentin) mRNA in both variants, but the difference was significant only in LLCm1 cells. Because LLCm4 is highly metastatic, vimentin expression may have been increased by other stimulatory systems, resulting in acidic pH e having a weak effect on vimentin production by these cells.
Acidic pH e induces vimentin but reduces E-cadherin expression. Cultures were pretreated overnight with serum-free DMEM/12 at pH 7.4 and treated for 24 h with acidic serum-free medium at pH 6.8 and 7.4. The cells were subsequently fixed and incubated with antibodies to vimentin (A & B) and E-cadherin (C & D). FITC-labeled signals were detected by fluorescence microscopy. (A & C) Representative results were shown from three or more independent experiments. Immuno-positivity was caliculated and their positivity was shown as relative values compared to LLCm1 cells at pH e 7.4. Bar, 50 μm. *P < 0.05; **P < 0.01; NS, not significant. E. Western blot analysis for E-cadherin. After incubation of cells at pH 6.8, as described above, cells were lysed and analyzed by Western blotting. CM was concentrated and analyzed to detect shedded E-cadherin level in the CM.
Acidic pH e induces Vim (vimentin) but reduces Cdh1 (E-cadherin) mRNA expression. Sub-confluent cultures were pretreated with serum-free medium at pH 7.4 and incubated in serum-free medium at pH 6.8 and 7.4. After 24 h, total RNA was extracted, reverse-transcribed, and amplified by qPCR with specific primer pairs. Data are shown as relative expression compared with their levels in LLCm1 cells cultured at pH 7.4. Representative results were shown from three independent experiments. *P < 0.05; NS, not significant.
Since several repressors of E-cadherin expression have been described, we tested the effects of acidic pH e on the expression of these repressor(s) by RT-qPCR technology. Incubation at acidic pH e induced steady state levels of Twist1, Twist2, and Zeb2 mRNAs (Figure 7), with these levels inversely correlated with E-cadherin expression, suggesting that Twist1, Twist2, and Zeb2 were involved in acidic pH e signaling that repressed E-cadherin expression. Zeb2 mRNA level showed that highest degree of induction, suggesting that Zeb2 was primarily responsible for acidic pH e -induced EMT. In contrast, acidic pH e had little effect on Snail, Slug, and Zeb1 mRNA levels, suggesting that these molecules may be little involved in acidic pH e -induced EMT.
Acidic pH e induces Twist1 , Twist2 and Zeb2 but not Snail , Slug, and Zeb1 mRNA expression. Sub-confluent cells were washed twice with PBS(−), pretreated with serum-free medium at pH 7.4, and incubated in serum-free medium at the indicated pH. After 24 h, total RNA was extracted, reverse-transcribed, and amplified by qPCR with specific primer sets. Data are shown as relative expression compared with their levels in LLCm1 cells cultured at pH 7.4. Representative results were shown from three or more independent experiments. *P < 0.05; ***P < 0.001; NS, not significant.
Acidic pH e induces cell migration and invasion
Migration activity was determined using wound healing assays. Cells that formed a confluent sheet were scratched with a plastic tip, and the distance between the wound edge and the migration front were estimated. As expected, LLCm4 cells showed higher migration (Figure 8) and in vitro invasion (Figure 9) activities than LLCm1 cells. Both LLCm1 and LLCm4 cells migrated significantly further under acidic than neutral conditions. Furthermore, acidic pH e stimulated the in vitro invasion activity of both cell lines.
Acidic pH e induces migration activity. Confluent cultures were scratched with micropipette tips and further cultured in 2% serum-containing medium adjusted to pH 7.4 or 6.8. Photographs were taken and cell migration was measured. A. Phase contrast micrograph. B. Migrated distance relative to LLCm1 cells at pH 7.4. Representative results were shown from three independent experiments (n = 8). *P < 0.05; **P < 0.01. C. Relative numbers of invasive cells compared with LLCm1 cells at pH 7.4.” between “B. Migrated distance relative to LLCm1 cells at pH 7.4.” and “Representative results were shown from three independent experiments (n = 8).
Acidic pH e induces invasive activity. Confluent cultures were serum-starved and treated with serum-free medium at pH 7.4 or pH 6.8 for 18 h. The conditioned medium was collected and the cells were harvested by trypsinization. The cells were subsequently incubated with 10% FBS for 30 min to inhibit trypsin activity, washed twice with PBS(−) and resuspended in their own conditioned medium. The chemoattractant was 20% FBS. A. Phase contrast micrograph. B. Relative numbers of invasive cells compared with LLCm1 cells at pH 7.4. Representative results were shown from three independent experiments. *P < 0.05; ***P < 0.001.
Discussion
Although reports using mouse [6] and human [7],[9] melanoma models have shown that acidic pH e induced EMT-like changes and increased invasive potential, the effects of acidic pH e on EMT in carcinoma models were unclear. We therefore examined the effect of an acidic microenvironment on EMT in a carcinoma model using the LLC cell line. We found that acidic pH e induces EMT in a carcinoma model, similar to findings in the melanoma model [6],[7].
Typically, EMT includes the down-regulation of E-cadherin and the up-regulation of vimentin, N-cadherin, fibronectin, and MMP-9 expression [10],[11]. The down-regulation of E-cadherin is driven by specific repressors, including Snail, Slug, Twist1, Twist2, Zeb1, and Zeb2 [1]. TGF-β induces EMT by increasing in Zeb1, Snail, Slug, and Twist1 mRNA expression through Smad2 and Erk1/2 signaling [12]. In contrast, we found that acidic pH e -induced EMT was accompanied by up-regulation of expression of Twist1, Twist2, and Zeb2 mRNAs, but not by changes in Snail, Slug, and Zeb1 mRNA levels. In contrast, we found that TGF-β induces EMT along with MMP-9 expression [11], findings observed in acidic pH e . Therefore, aside from MMP-9 induction, EMT induction by acidic pH e is via a somewhat different mechanism than TGF-β-triggered EMT.
NF-κB has been regarded as a key molecule for acidic pH e signaling. NF-κB not only induces the expression of Snail, Twist1, Slug, and Zeb2 mRNAs [13] but also blocks ubiquitination followed by the stabilization of Snail [14]. In some cases, NF-κB-induced EMT was accompanied by MMP-9 expression [15],[16]. Metastatic cells of mesenchymal origin secrete abundant amounts of MMP-9 [17]. In addition, as we reported previously, acidic pH e induces MMP-9 expression in mouse melanoma cells through NF-κB activation with change to a fibroblastic morphology [6],[18],[19]. These observations suggested that induction of MMP-9 expression through NF-κB was deeply associated with EMT.
TGF-β was shown to induce the expression of MMP9 mRNA but decrease the expression of MMP2 mRNA by human oral squamous cell carcinoma cell lines [20]. We have shown that acidic pH e induces abundant MMP-9 production by the highly metastatic B16 variants, B16-F10 and B16-BL6 [6]. Although MMP-2 was expressed in parental B16 cell, little was expressed in the low and high metastatic variants, B16-F1, B16-F10 and B16-BL6. Using an LLC model, we observed findings similar to those in the B16 melanoma lines. Reduction of MMP-2 activation was reported due to increased intracellular Ca2+[21]. In contrast, we previously showed that elevation of intracellular Ca2+ induced MMP-9 expression [19]. Although acidic pH e induces a Ca2+ influx, its mechanism of expression of MMP-2 is still unclear.
EMT includes the suppression of the epithelial marker E-cadherin. Immunocytochemical staining showed that incubation in acidic pH e markedly reduced E-cadherin expression, but had little effect on the expression of Cdh1 (E-cadherin) mRNA. Acidic pH e did not affect the half life of Cdh1 mRNA level (data not shown). Interestingly, E-cadherin was increased in CM, especially in LLCm2 cells, by acidic pH e . Because MMP-9 catalyzes E-cadherin ectodomain shedding during EMT [22],[23], acidic pH e -induced MMP-9 production may not only promote extracellular matrix degradation but also E-cadherin shedding to induce EMT.
Signal cross talk is also important in EMT. For example, in an oral squamous cell carcinoma model, the combination of TGF-β1 and EGF, but not either alone, induced cell scattering activity [24]. Similarly, TGF-β sensitizes cells to basic FGF/FGF-2 stimulation to induce EMT [25]. Thus, intracellular signaling pathways seem to be complex. Acidic pH e can arise through the production of lactate and/or the generation of excess amounts of CO2 through the pentose-phosphate pathway [26]. Because hypoxia induces EMT [27], acidic pH e -induced EMT may be modulated by hypoxia.
Acidic pH e induces the expression of many genes, including MMP-9 [6],[28], VEGF-A [29]-[31], VEGF-C [32], interleukin-8 [33]-[35], the inducible isoform of nitric oxide synthase (iNOS) [36], platelet-derived endothelial cell growth factor (PDGF)/thymidine phosphorylase [37], and acidic sphyngomyelinase [19], as well as Twist1, Twist2, Zeb2, vimentin, MMP-3, and MMP-13, which were shown in this study. In addition, acidic pH e can affect cell migration, stress fiber formation, invasion and metastasis [4],[9]. Thus, acidic pH e acts as a microenvironmental factor that promotes malignant phenotypes, such as metastatic ability.
LLC is thought to originate from a cancer derived from granular pneumocytes and to be equivalent to a human alveolar cell carcinoma (squamous cell carcinoma) [38]. Therefore, LLC is regarded as a mouse model of non-small cell lung cancer [39],[40]. Acidic pH e -induced EMT may also be involved in the origins of other types of squamous cell carcinoma such as head and neck and esophageal cancers.
Conclusion
These findings suggest that acidic pH e constitutes an important microenvironment that induces EMT in some types of carcinoma.
Methods
Reagents
Dulbecco’s modified Eagle medium (DMEM), Ham’s F12 medium, and High Capacity RNA-to-cDNA kit were purchased from Life Technologies (Grand Island, NY, USA). Realtime PCR master mix was from Takara (Tokyo, Japan). Anti-vimentin polyclonal antibodies was from ImmunoResearch Laboratories (Grove, PA, USA). Anti-E-cadherin monoclonal antibody was from Santa Cruz (G-10, Santa Cruz, CA, USA) Avidin-conjugated fluorescein isothiocyanate (FITC) was from Vector Laboratories (Burlingame, CA, USA), and N102 blocking reagent was from NOF Corporation (Tokyo, Japan). Fetal bovine serum (FBS) was from Hyclone (South Logan, UT, USA). DC protein assay kits (based on the Lowry method) were from Bio-Rad Laboratories (Hercules, CA, USA), and Isogen RNA extraction kits were from Nippon Gene (Tokyo, Japan). Transwell chambers with 8 μm pores were from BD Bioscience (Franklin Lakes, NJ, USA). Matrigel® was from Corning (Tewksbury, MA, USA).
Cells and cell culture
The LLC cell line, a squamous cell carcinoma cell line derived from granular pneumocytes and equivalent to human alveolar cell carcinoma [38], was the kind gift of Dr. Ryu-Ichro Hata (Kanagawa Dental University, Yokosuka, Japan). LLC cells were cultured in DMEM/F12 (a 1:1 mixture of DMEM and Ham’s F12 media, adjusted to pH 7.4 with 15 mM HEPES, 4 mM phosphorus, 1 mg/ml NaHCO3), supplemented with 10% FBS at 37°C in a humidified atmosphere of 5% CO2 and 95% air. The cells were passaged with 0.05% trypsin/0.02% EDTA. The pH of the medium was adjusted with NaOH or HCl and measured after incubation in the CO2 incubator for 3 h.
Experimental metastasis and in vivo selection of metastatic variants
Cells (3 × 105 cells) were resuspended in 200 μl Ca2+- and Mg2+-free Dulbecco’s phosphate-buffered saline (PBS(−)) and injected into the tail vein of a C57BL/6 mouse (Clea Japan, Tokyo, Japan). Three weeks later, the mice were sacrificed under anesthesia with an excess dose of pentobarbital and the metastasized foci in the lungs were counted.
Variants with different metastatic potential were established as described [41]. Briefly, LLC parental cells were injected into the tail vein of a syngeneic C57BL/6 mouse. Three weeks later, the mice were sacrificed, and metastasized foci were taken from the lungs, treated with trypsin/EDTA and cultured in DMEM/F12 + 10% FBS. These cells, referred to as LLCm1 cells, were again injected into mouse tail veins and metastasized foci in the lungs were cultured. The cells obtained after 4 cycles were the high metastatic variant, referred to as LLCm4 cells.
All animal protocols were approved by the Animal Use Committee of Ohu University.
Preparation of conditioned medium for zymography
Conditioned media were prepared essentially as described [6], with modifications. Briefly, serum-free conditioned media were taken from cultured cells, and proteins were precipitated by addition of 2.5 volumes of acetone. The samples were reconstituted in PBS(−) or 0.5 mM Tris–HCl (pH 6.8) supplemented with 1% SDS and 0.2% glycerol.
Zymography
Zymography on gelatin-containing sodium dodecyl sulfate (SDS)-7.5% polyacrylamide gels was performed as described [19]. Briefly, concentrated samples were electrophoresed in 0.1% gelatin-containing 7.5% polyacrylamide gels. The gels were washed with 2.5% Triton X-100 at room temperature with gentle shaking for 1 h and incubated for 20 h in reaction buffer (50 mM Tris–HCl (pH 7.5), 10 mM CaCl2) at 37°C. Gelatinolytic activity was visualized by Coomassie Brilliant Blue R250 staining. Activity was normalized relative to cell number or protein concentration, with the latter determined by the Lowry method using bovine serum albumin as the standard.
RT-qPCR
Total RNA was extracted by Isogen and transcribed into cDNA using reverse transcriptase. Quantitative PCR was performed using specific primer sets (Table 1) according to the manufacturer’s protocol.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde and blocked with 20% N102 blocking reagent in Tris-buffered saline (20 mM Tris, pH 7.5, 150 mM NaCl, supplemented with 0.05% Tween 20 (TBS-T). The cells were incubated overnight at 4°C with primary antibody, washed extensively, incubated with biotin-conjugated secondary antibody, washed, and treated with avidin-conjugated fluorescein isothiocyanate (FITC). The cells were viewed by fluorescence microscopy (EVOS® FLoid® Cell Imaging Station, Life Technologies, Carlsbad, CA, USA).
Wound healing (scratch) assay
Wound healing assays were performed as described, with slight modifications [42]. Briefly, confluent cultures were serum-starved for 24 h and scratched off with a micropipette tip. The cells were cultured in medium containing 0.2% FBS at pH 7.4 or pH 6.8. After indicated time period, photographs were taking and estimated wound distance.
In vitro invasion assay
In vitro invasive activity was determined using Matrigel®-coated filter mounted transwell chambers (Corning, Tewksbury, MA, USA) as described [43]. Briefly, cells were serum-starved overnight at pH 7.4 and then further maintained in serum-free media at pH 7.4 or 6.8 for 18 h. The cells were harvested with 0.05% trypsin/0.02% EDTA and incubated at 37°C for 30 min in medium containing 10% FBS to inhibit trypsin activity. The cells were washed twice with PBS(−), re-suspended in serum-free medium at pH 7.4 or 6.8, and inoculated at density of 5 × 105 cells/100 μl/chamber onto a Matrigel® (37.9 μg/cm2)-coated filter in the insert, which had been mounted onto a well of a 24 well plate, with each well filled with 600 μl 10% FBS-containing medium adjusted to each pH. After incubation for 18 h, non-invasive cells were scraped off with a cotton swab and invasive cells were fixed in 100% methanol, stained with hematoxylin, and counted under light microscope (×200).
Statistical analysis
Between group differences were compared using Student’s t-tests or ANOVA. Statistical significance was defined as a p value less than 0.05.
Abbreviations
- EMT:
-
Epithelial mesenchymal transition
- FGF:
-
Basic fibroblast growth factor
- TGF-β:
-
Transforming growth factor-β
- pH e :
-
Extracellular pH
- MMP-9:
-
Matrix metalloproteinase-9
- LLC:
-
Lewis lung carcinoma
- RT-qPCR:
-
Reverse transcription-quantitative polymerase chain reaction
- DMEM:
-
Dulbecco’s modified Eagle medium
- FBS:
-
Fetal bovine serum
- PBS(−):
-
Ca2+- and Mg2+-free Dulbecco’s phosphate-buffered saline
References
Gheldof A, Berx G: Cadherins and epithelial-to-mesenchymal transition. Prog Mol Biol Transl Sci. 2013, 116: 317-336. 10.1016/B978-0-12-394311-8.00014-5.
Fujimoto J, Ichigo S, Hirose R, Sakaguchi H, Tamaya T: Suppression of E-cadherin and α- and β-catenin mRNA expression in the metastatic lesions of gynecological cancers. Eur J Gynaecol Oncol. 1997, 18: 484-487.
Warburg O, Posener K, Negelein E: Über den Stoffwechsel der Tumoren (On metabolism of tumors). Biochem Z. 1924, 152: 319-344.
Rofstad EK, Mathiesen B, Kindem K, Galappathi K: Acidic extracellular pH promotes experimental metastasis of human melanoma cells in athymic nude mice. Cancer Res. 2006, 66: 6699-6707. 10.1158/0008-5472.CAN-06-0983.
Kato Y, Ozawa S, Miyamoto C, Maehata Y, Suzuki A, Maeda T, Baba Y: Acidic extracellular microenvironment and cancer. Cancer Cell Int. 2013, 13: 89-10.1186/1475-2867-13-89.
Kato Y, Nakayama Y, Umeda M, Miyazaki K: Induction of 103-kDa gelatinase/type IV collagenase by acidic culture conditions in mouse metastatic melanoma cell lines. J Biol Chem. 1992, 267: 11424-11430.
Peppicelli S, Bianchini F, Torre E, Calorini L: Contribution of acidic melanoma cells undergoing epithelial-to-mesenchymal transition to aggressiveness of non-acidic melanoma cells. Clin Exp Metastasis. 2014, 31: 423-433. 10.1007/s10585-014-9637-6.
Kim JE, Leung E, Baguley BC, Finlay GJ: Heterogeneity of expression of epithelial-mesenchymal transition markers in melanocytes and melanoma cell lines. Front Genet. 2013, 4: 97-
Martínez-Zaguilán R, Seftor EA, Seftor RE, Chu YW, Gillies RJ, Hendrix MJ: Acidic pH enhances the invasive behavior of human melanoma cells. Clin Exp Metastasis. 1996, 14: 176-186. 10.1007/BF00121214.
Chen X, Xiao W, Wang W, Luo L, Ye S, Liu Y: The complex interplay between ERK1/2, TGFβ/Smad, and Jagged/Notch signaling pathways in the regulation of epithelial-mesenchymal transition in retinal pigment epithelium cells. PLoS One. 2014, 9: e96365-10.1371/journal.pone.0096365.
Islam SS, Mokhtari RB, El Hout Y, Azadi MA, Alauddin M, Yeger H, Farhat WA: TGF-β1 induces EMT reprogramming of porcine bladder urothelial cells into collagen producing fibroblasts-like cells in a Smad2/Smad3-dependent manner. J Cell Commun Signal. 2014, 8: 39-58. 10.1007/s12079-013-0216-4.
Liu LC, Tsao TC, Hsu SR, Wang HC, Tsai TC, Kao JY, Way TD: EGCG inhibits transforming growth factor-β-mediated epithelial-to-mesenchymal transition via the inhibition of Smad2 and Erk1/2 signaling pathways in nonsmall cell lung cancer cells. J Agric Food Chem. 2012, 60: 9863-9873. 10.1021/jf303690x.
Kumar M, Allison DF, Baranova NN, Wamsley JJ, Katz AJ, Bekiranov S, Jones DR, Mayo MW: NF-κB regulates mesenchymal transition for the induction of non-small cell lung cancer initiating cells. PLoS One. 2013, 8: e68597-10.1371/journal.pone.0068597.
Wu Y, Deng J, Rychahou PG, Qiu S, Evers BM, Zhou BP: Stabilization of snail by NF-κB is required for inflammation-induced cell migration and invasion. Cancer Cell. 2009, 15: 416-428. 10.1016/j.ccr.2009.03.016.
Wang F, He W, Fanghui P, Wang L, Fan Q: NF-κBP65 promotes invasion and metastasis of oesophageal squamous cell cancer by regulating matrix metalloproteinase-9 and epithelial-to-mesenchymal transition. Cell Biol Int. 2013, 37: 780-788. 10.1002/cbin.10089.
Wang Y, Yue B, Yu X, Wang Z, Wang M: SLUG is activated by nuclear factor kappa B and confers human alveolar epithelial A549 cells resistance to tumor necrosis factor-alpha-induced apoptosis. World J Surg Oncol. 2013, 11: 12-10.1186/1477-7819-11-12.
Kawashima A, Nakanishi I, Tsuchiya H, Roessner A, Obata K, Okada Y: Expression of matrix metalloproteinase 9 (92-kDa gelatinase/type IV collagenase) induced by tumour necrosis factor alpha correlates with metastatic ability in a human osteosarcoma cell line. Virchows Arch. 1994, 424: 547-552. 10.1007/BF00191442.
Kato Y, Lambert CA, Colige AC, Mineur P, Noël A, Frankenne F, Foidart JM, Baba M, Hata RI, Miyazaki K, Tsukuda M: Acidic extracellular pH induces matrix metalloproteinase-9 expression in mouse metastatic melanoma cells through the phospholipase D-mitogen-activated protein kinase signaling. J Biol Chem. 2005, 280: 10938-10944. 10.1074/jbc.M411313200.
Kato Y, Ozawa S, Tsukuda M, Kubota E, Miyazaki K, St-Pierre Y, Hata R: Acidic extracellular pH increases calcium influx-triggered phospholipase D activity along with acidic sphingomyelinase activation to induce matrix metalloproteinase-9 expression in mouse metastatic melanoma. FEBS J. 2007, 274: 3171-3183. 10.1111/j.1742-4658.2007.05848.x.
Qiao B, Johnson NW, Gao J: Epithelial-mesenchymal transition in oral squamous cell carcinoma triggered by transforming growth factor-β1 is Snail family-dependent and correlates with matrix metalloproteinase-2 and −9 expressions. Int J Oncol. 2010, 37: 663-668.
Yu M, Sato H, Seiki M, Spiegel S, Thompson EW: Calcium influx inhibits MT1-MMP processing and blocks MMP-2 activation. FEBS Lett. 1997, 412: 568-572. 10.1016/S0014-5793(97)00849-1.
Symowicz J, Adley BP, Gleason KJ, Johnson JJ, Ghosh S, Fishman DA, Hudson LG, Stack MS: Engagement of collagen-binding integrins promotes matrix metalloproteinase-9-dependent E-cadherin ectodomain shedding in ovarian carcinoma cells. Cancer Res. 2007, 67: 2030-2039. 10.1158/0008-5472.CAN-06-2808.
Zuo JH, Zhu W, Li MY, Li XH, Yi H, Zeng GQ, Wan XX, He QY, Li JH, Qu JQ, Chen Y, Xiao ZQ: Activation of EGFR promotes squamous carcinoma SCC10A cell migration and invasion via inducing EMT-like phenotype change and MMP-9-mediated degradation of E-cadherin. J Cell Biochem. 2011, 112: 2508-2517. 10.1002/jcb.23175.
Richter P, Umbreit C, Franz M, Berndt A, Grimm S, Uecker A, Bohmer FD, Kosmehl H, Berndt A: EGF/TGFβ1 co-stimulation of oral squamous cell carcinoma cells causes an epithelial-mesenchymal transition cell phenotype expressing laminin 332. J Oral Pathol Med. 2011, 40: 46-54. 10.1111/j.1600-0714.2010.00936.x.
Shirakihara T, Horiguchi K, Miyazawa K, Ehata S, Shibata T, Morita I, Miyazono K, Saitoh M: TGF-β regulates isoform switching of FGF receptors and epithelial-mesenchymal transition. EMBO J. 2011, 30: 783-795. 10.1038/emboj.2010.351.
Helmlinger G, Sckell A, Dellian M, Forbes NS, Jain RK: Acid production in glycolysis-impaired tumors provides new insights into tumor metabolism. Clin Cancer Res. 2002, 8: 1284-1291.
Salnikov AV, Liu L, Platen M, Gladkich J, Salnikova O, Ryschich E, Mattern J, Moldenhauer G, Werner J, Schemmer P, Buchler MW, Herr I: Hypoxia induces EMT in low and highly aggressive pancreatic tumor cells but only cells with cancer stem cell characteristics acquire pronounced migratory potential. PLoS One. 2012, 7: e46391-10.1371/journal.pone.0046391.
Kato Y, Ozono S, Shuin T, Miyazaki K: Slow induction of gelatinase B mRNA by acidic culture conditions in mouse metastatic melanoma cells. Cell Biol Int. 1996, 20: 375-377. 10.1006/cbir.1996.0044.
Fukumura D, Xu L, Chen Y, Gohongi T, Seed B, Jain RK: Hypoxia and acidosis independently up-regulate vascular endothelial growth factor transcription in brain tumors in vivo. Cancer Res. 2001, 61: 6020-6024.
Shi Q, Le X, Wang B, Abbruzzese JL, Xiong Q, He Y, Xie K: Regulation of vascular endothelial growth factor expression by acidosis in human cancer cells. Oncogene. 2001, 20: 3751-3756. 10.1038/sj.onc.1204500.
Xu L, Fukumura D, Jain RK: Acidic extracellular pH induces vascular endothelial growth factor (VEGF) in human glioblastoma cells via ERK1/2 MAPK signaling pathway: mechanism of low pH-induced VEGF. J Biol Chem. 2002, 277: 11368-11374. 10.1074/jbc.M108347200.
Peppicelli S, Bianchini F, Contena C, Tombaccini D, Calorini L: Acidic pH via NF-κB favours VEGF-C expression in human melanoma cells. Clin Exp Metastasis. 2013, 30: 957-967. 10.1007/s10585-013-9595-4.
Shi Q, Abbruzzese JL, Huang S, Fidler IJ, Xiong Q, Xie K: Constitutive and inducible interleukin 8 expression by hypoxia and acidosis renders human pancreatic cancer cells more tumorigenic and metastatic. Clin Cancer Res. 1999, 5: 3711-3721.
Shi Q, Le X, Wang B, Xiong Q, Abbruzzese JL, Xie K: Regulation of interleukin-8 expression by cellular pH in human pancreatic adenocarcinoma cells. J Interferon Cytokine Res. 2000, 20: 1023-1028. 10.1089/10799900050198471.
Xu L, Fidler IJ: Acidic pH-induced elevation in interleukin 8 expression by human ovarian carcinoma cells. Cancer Res. 2000, 60: 4610-4616.
Bellocq A, Suberville S, Philippe C, Bertrand F, Perez J, Fouqueray B, Cherqui G, Baud L: Low environmental pH is responsible for the induction of nitric-oxide synthase in macrophages. Evidence for involvement of nuclear factor-κB activation. J Biol Chem. 1998, 273: 5086-5092. 10.1074/jbc.273.9.5086.
Griffiths L, Dachs GU, Bicknell R, Harris AL, Stratford IJ: The influence of oxygen tension and pH on the expression of platelet-derived endothelial cell growth factor/thymidine phosphorylase in human breast tumor cells grown in vitro and in vivo. Cancer Res. 1997, 57: 570-572.
Garcia-Sanz M, Simón-Marín R, Hilario E: Morphological characterization of Lewis lung carcinoma (3LL). A light and electron microscopic study. Tumori. 1989, 75: 23-27.
Sharma S, Zhu L, Yang SC, Zhang L, Lin J, Hillinger S, Gardner B, Reckamp K, Strieter RM, Huang M, Batra RK, Dubinett SM: Cyclooxygenase 2 inhibition promotes IFN-γ-dependent enhancement of antitumor responses. J Immunol. 2005, 175: 813-819. 10.4049/jimmunol.175.2.813.
Liu R, Khullar OV, Griset AP, Wade JE, Zubris KA, Grinstaff MW, Colson YL: Paclitaxel-loaded expansile nanoparticles delay local recurrence in a heterotopic murine non-small cell lung cancer model. Ann Thorac Surg. 2011, 91: 1077-1083. 10.1016/j.athoracsur.2010.12.040. discussion 1083–1074
Fidler IJ: Selection of successive tumour lines for metastasis. Nat New Biol. 1973, 242: 148-149. 10.1038/newbio242148a0.
Baba Y, Kato Y, Mochimatsu I, Nagashima Y, Kurihara M, Kawano T, Taguchi T, Hata R, Tsukuda M: Inostamycin suppresses vascular endothelial growth factor-stimulated growth and migration of human umbilical vein endothelial cells. Clin Exp Metastasis. 2004, 21: 419-425. 10.1007/s10585-005-0041-0.
Baba Y, Tsukuda M, Mochimatsu I, Furukawa S, Kagata H, Nagashima Y, Sakai N, Koshika S, Imoto M, Kato Y: Inostamycin, an inhibitor of cytidine 5′-diphosphate 1,2-diacyl-sn-glycerol (CDP-DG): inositol transferase, suppresses invasion ability by reducing productions of matrix metalloproteinase-2 and −9 and cell motility in HSC-4 tongue carcinoma cell line. Clin Exp Metastasis. 2000, 18: 273-279. 10.1023/A:1006798900743.
Acknowledgements
We acknowledge Dr. Satoshi Yuzawa for his constructive criticism.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AS conceived the study, performed experiments, analyzed data, and drafted the manuscript. TM analyzed data and gave constructive criticism throughout the study. YB designed the wound and invasion assays and helped in drafting the manuscript. KS made critical suggestions. YK jointly conceived the study and participated in design, coordination, and manuscript editing. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Suzuki, A., Maeda, T., Baba, Y. et al. Acidic extracellular pH promotes epithelial mesenchymal transition in Lewis lung carcinoma model. Cancer Cell Int 14, 129 (2014). https://doi.org/10.1186/s12935-014-0129-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-014-0129-1