Abstract
Background
Sputum and blood eosinophil counts predict corticosteroid effects in COPD patients. Bacterial infection causes increased airway neutrophilic inflammation. The relationship of eosinophil counts with airway bacterial load in COPD patients is uncertain. We tested the hypothesis that bacterial load and eosinophil counts are inversely related.
Methods
COPD patients were seen at stable state and exacerbation onset. Sputum was processed for quantitative polymerase chain reaction detection of the potentially pathogenic microorganisms (PPM) H. influenzae, M. catarrhalis and S. pneumoniae. PPM positive was defined as total load ≥1 × 104copies/ml. Sputum and whole blood were analysed for differential cell counts.
Results
At baseline, bacterial counts were not related to blood eosinophils, but sputum eosinophil % was significantly lower in patients with PPM positive compared to PPM negative samples (medians: 0.5% vs. 1.25% respectively, p = 0.01). Patients with PPM positive samples during an exacerbation had significantly lower blood eosinophil counts at exacerbation compared to baseline (medians: 0.17 × 109/L vs. 0.23 × 109/L respectively, p = 0.008), while no blood eosinophil change was observed with PPM negative samples.
Conclusions
These findings indicate an inverse relationship between bacterial infection and eosinophil counts. Bacterial infection may influence corticosteroid responsiveness by altering the profile of neutrophilic and eosinophilic inflammation.
Similar content being viewed by others
Background
COPD is a heterogeneous condition, composed of different clinical and pathophysiological components that vary both in presence and severity between patients [1]. This heterogeneity causes variability in the responses to pharmacological treatments. Biomarkers that predict treatment responses to anti-inflammatory drugs may be useful for optimising the benefit versus risk ratio.
Sputum eosinophil counts in stable COPD patients predict the clinical response to corticosteroids [2, 3]. However, measuring sputum eosinophils is time consuming, and some patients do not provide adequate samples for analysis. Blood eosinophil measurements are more practical, and appear to be a surrogate biomarker for sputum eosinophils as these measurements show a degree of correlation within the same individual, both in stable COPD patients and during exacerbations [4,5,6,7]. Retrospective analysis of COPD clinical trials have shown that higher blood eosinophil counts predict a greater reduction in exacerbation rates with inhaled corticosteroid/long acting beta agonist (ICS/LABA) combinations compared to LABA [8,9,10]. Furthermore, oral corticosteroid treatment during exacerbations has a greater effect in COPD patients with higher blood eosinophil counts [11].
Chronic bacterial infection in COPD patients causes greater neutrophilic inflammation in the lungs [12,13,14]. Neutrophilic lung inflammation responds poorly to corticosteroids [15,16,17], implicating bacterial infection as a potential cause of corticosteroid insensitivity in COPD patients. The relationship between bacterial infection and eosinophil counts is not established in COPD. There may be an inverse relationship between these parameters, as blood eosinophil counts are known to be reduced during severe bacterial infection [18, 19]. The existence of such an inverse relationship would suggest that the interaction between bacterial infection and eosinophils determines the corticosteroid response in COPD patients.
The primary aim of the analysis reported here using data from the COPDMAP cohort was to test the hypothesis that the bacterial load and eosinophil counts are inversely related in COPD patients. We investigated the relationship between eosinophil counts (in the blood and sputum) and the bacterial load in the stable state and during exacerbations in COPD patients.
Methods
Subjects
COPD patients aged ≥ 40 years were recruited at 3 sites (Manchester, Leicester and London) and enrolled into the COPDMAP prospective observational cohort study (www.copdmap.org). Patients had a physician diagnosis of COPD, post-bronchodilator forced expiratory volume in 1 s (FEV1)/forced vital capacity (FVC) ratio <0.7, ≥10 pack year smoking history and no previous asthma diagnosis. The patients included in this analysis were those who provided a blood sample for eosinophil analysis and a sputum sample for bacterial quantification at the baseline visit. All patients provided written informed consent using protocols approved by the local Ethics Committees (11/L0/1630; 10/H/1003/108; 07/H0406/157).
Additional methods are in the online supplement (Additional file 1).
Study design
Patients were seen at baseline and at exacerbation onset. Symptoms were assessed using the modified MRC Scale (mMRC) and the COPD assessment test (CAT). Health related quality of life using the St George’s Respiratory Questionnaire (SGRQ-C) and fat free mass index (FFMI) were assessed. Lung function measurements and 6 min walk test (6MWT) were performed according to guidelines [20, 21]. Spontaneous and/or induced sputum was obtained as previously described [22] and blood samples sent to local hospital laboratories.
Patients contacted the research team if they experienced a change in symptoms consistent with an acute exacerbation. Daily diary cards were used. Patients were assessed by a clinician and exacerbations defined as in increase in two respiratory symptoms (with at least one major symptom) for two consecutive days [23]. Blood and sputum sampling were performed prior to any systemic therapy including treatment with oral corticosteroids and/or antibiotics. Samples from the first exacerbation event per patient were used in this analysis.
Sputum and blood analysis
All patients attempted to produce a spontaneous sample at baseline and exacerbation. A sputum induction was attempted (safety permitting) if an insufficient spontaneous sample was produced. Spontaneous or induced sputum was processed for quantitative polymerase chain reaction (qPCR) detection of the common respiratory potentially pathogenic microorganisms (PPM) H. influenzae, M.catarrhalis and S. pneumoniae and for human rhinovirus (RV) as previously described [24, 25]. Patients were categorised as PPM positive if the total load was above 1 × 104copies/ml or RV positive if the load was greater than 1 × 101copies/ml [26, 27]. Sputum was also processed for differential cell counts (Additional file 2: Figure S1) [28]. However, small sputum samples (minimum weight of approximately 0.075 g) were preferentially processed for qPCR analysis only. Whole blood was analysed for differential leukocyte count and the blood eosinophil count measured at baseline and exacerbation were used in this analysis.
Statistical analysis
Statistical analysis for non-parametric data was performed using; Wilcoxon signed rank test for within individual comparison or the Mann–Whitney U test for between group comparison and chi-square test for categorical variables. Receiver operating characteristics (ROC) curves determined if blood eosinophil counts predict a sputum eosinophil count of ≥3%. Spearman’s correlation test assessed associations between variables. P < 0.05 was considered statistically significant. Analyses were performed using GraphPad Prism version 6.00 (San Diego, USA).
Results
Stable state microbiology and eosinophils
Blood eosinophil counts
One hundred sixty-eight patients had a blood eosinophil and sputum qPCR bacterial measurement at baseline. The demography of this population is shown in Table 1; the mean post-bronchodilator FEV1 was approximately 57% predicted, with a median exacerbation rate in the previous year of 1, and mean CAT and SGRQ scores of 18.7 and 47.8 respectively. 52% of patients had PPM positive samples (defined by a threshold value of ≥1 × 104). There were no differences in the blood eosinophil count (medians: 0.21 × 109/L vs. 0,18 × 109/L, p = 0.50), sputum cells counts (all p > 0.05) and total bacterial load (medians: 3.55 × 104 vs.5.38 × 104, p = 0.52) between ICS users and non-users respectively.
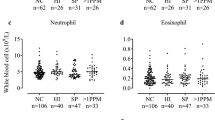
There were no significant associations between the blood eosinophil count and bacterial load (Fig. 1a), and no difference in bacterial load using different blood eosinophil threshold values i.e. ≥ and <150 cells/μl (Fig. 2a), 2% or 300 cell/μl (not shown). Additionally, there were no differences in blood eosinophil counts between patients with PPM positive and PPM negative samples (Fig. 2b).
The relationship between eosinophils and bacterial load at baseline; a the bacterial load in patients with blood eosinophil ≥ 150 cells/μl and < 150 cells/μl, b the blood eosinophil count between PPM negative and PPM positive groups, c the bacterial load in patients with sputum eosinophil count ≥ 3% and < 3%, d the sputum eosinophil % between PPM negative and PPM positive groups
Sputum eosinophil counts
Ninety-four of the 168 patients produced a sufficient sputum sample for differential cell count analysis; of which 75 and 19 were from spontaneous and induced samples respectively. Total cell count (x106/g) in the spontaneous samples were lower compared to induced samples (medians: 2.75 vs. 6.59 respectively, p = 0.003), but no differences were observed for sputum % differential counts including sputum eosinophil % (medians 0.75 vs. 0.6 respectively, p = 0.82).
Figure 2c shows that patients with a sputum eosinophil count ≥3% had a significantly lower bacterial load compared to those with a sputum eosinophil <3% (p = 0.046). The inflammatory cell profiles of the patients with sputum eosinophil count <3% and ≥3% are presented in Table 2; there was a higher neutrophil percentage (medians 80.1% vs. 68.4% respectively) and lower absolute eosinophil cell count (medians 0.2 × 106/g versus 0.0 × 106/g respectively) in the former group.
The sputum eosinophil % was significantly lower in patients with PPM positive samples compared to PPM negative samples (medians: 0.5% vs. 1.25%, p = 0.01, Fig. 2d). Sputum eosinophil % was inversely correlated with bacterial load (rho = −0.26, p < 0.01, Fig. 1b). This association was not evident with the sputum eosinophil absolute count (p = 0.5). There was a significant positive correlation between the bacterial load and both sputum neutrophil % and absolute cell count (rho = 0.25, p = 0.02 and rho = 0.22, p = 0.04 respectively, Fig. 3).
Exacerbation microbiology and eosinophils
There were 109 exacerbation events that had blood eosinophil counts at both baseline and exacerbation, and qPCR bacterial counts at exacerbation. The exacerbations occurred a mean 171 days after the baseline visit; 78 exacerbations were PPM positive and 31 were PPM negative. A subset of 70 patients also had a baseline qPCR bacterial count. Sputum eosinophil % counts at baseline and exacerbation were available in 30 of these patients; 22 were PPM positive and 8 were PPM negative (Additional file 3: Figure S2 shows a summary of the number of patients with blood/sputum eosinophil counts and qPCR data at exacerbation alone, or coupled to baseline measurements).
The differences in blood counts, sputum counts and bacterial load between baseline and exacerbation in the whole population are presented in Table 3. White blood cell count (WBC), blood neutrophil and monocyte counts were significantly increased at exacerbation, while there was no difference for the blood eosinophil count. Bacterial load, total sputum cell count (x106/g) and the absolute and % sputum neutrophil count were higher at exacerbation. The absolute and % sputum macrophage counts and sputum eosinophil % were significantly lower at exacerbation.
Blood eosinophil counts
Table 4 presents a comparison of the blood counts between baseline and exacerbation for the PPM positive and negative groups. Patients with PPM positive samples (using a threshold value of ≥1 × 104) during an exacerbation had significantly lower blood eosinophil counts at exacerbation compared to baseline (medians: 0.17 × 109/L vs. 0.23 × 109/L respectively, p = 0.008, Fig. 4). There was no change in the blood eosinophil count at exacerbation compared to baseline in patients with PPM negative samples at exacerbation (medians: 0.23 × 109/L vs. 0.22 × 109/L respectively). The median decrease in blood eosinophil count between the baseline and exacerbation visit was significantly different in PPM positive compared to PPM negative patients (medians: −0.02 × 109/L vs. 0.04 × 109/L respectively, p = 0.004, Fig. 4). A similar pattern was observed using a PPM threshold of ≥1 × 106 at exacerbation (Additional file 4: Figure S3).
The proportion of individuals with a decrease in blood eosinophil counts at exacerbation from baseline was significantly higher in PPM positive patients compared to PPM negative patients (53% vs 29%, p = 0.02). 4/78 (5%) of the PPM positive and 1/31 (3%) of the PPM negative patients had blood eosinopenia (counts ≤0.04 × 109/L).
A significant increase in the blood neutrophil count was observed at exacerbation compared to baseline in patients with PPM positive (medians: 6.13 × 109/L vs. 4.95 × 109/L respectively, p < 0.0001) and negative samples (5.50 × 109/L vs 4.38 × 109/L respectively, p = 0.004) during exacerbation (Table 4). There was no significant difference in the median change between the 2 groups (p > 0.99). In addition, the total WBC and blood monocyte count were significantly increased at exacerbation compared to baseline in both the PPM positive and negative groups (Table 4).
Seventy-five of the 109 patients had rhinovirus load measured at exacerbation; 16 were rhinovirus positive and 59 were rhinovirus negative. 75% and 71% of the rhinovirus positive and negative samples were PPM positive respectively. There were no changes in blood eosinophil counts between the baseline and exacerbation visit in the rhinovirus positive (medians: 0.26 vs. 0.20 respectively, p = 0.87) and negative (medians: 0.22 vs. 0.17 respectively, p = 0.34) groups (Additional file 5: Table S1).
Sputum eosinophil counts
Sputum eosinophil % decreased significantly at exacerbation compared to baseline in patients who were PPM positive (medians: 0.23% vs. 0.75% respectively, p = 0.046 & Fig. 5). No significant change was observed in patients who were PPM negative at exacerbation (Fig. 5). The comparison of the eosinophil count change from baseline to exacerbation revealed no difference between PPM positive and PPM negative patients. When repeating the analysis using sputum neutrophil % in both the PPM positive and negative groups, we observed a numerical but non-significant increase in both groups (Additional file 6: Figure S4). Comparisons of the other sputum cell counts are presented further in Table 4.
Relationship between blood and sputum eosinophils
One hundred ten patients had both a blood and sputum eosinophil count available at baseline, while 62 patients had a blood and sputum eosinophil count available during their first exacerbation event. The baseline blood eosinophil % and absolute counts were weakly correlated with sputum eosinophil % (rho = 0.24, p = 0.01 and rho = 0.19, p = 0.05 respectively) and baseline blood eosinophil % and absolute counts did not predict sputum eosinophil % (area under the ROC curve (AUC) 0.59, p = 0.21 and AUC 0.52, p = 0.79 respectively, Fig. 6a). In contrast, at exacerbation there was a stronger correlation for blood eosinophil % (rho = 0.41, p = 0.001) and absolute counts (rho = 0.43, p < 0.001) with sputum eosinophil %. Both blood eosinophil % and absolute count were good predictors of sputum eosinophil % at exacerbation (AUC 0.80, p = 0.005 and AUC 0.76, p = 0.01 respectively, Fig. 6b).
Discussion
We observed evidence of an inverse relationship between bacterial infection and eosinophil counts in COPD. Firstly, sputum eosinophil counts were lower in COPD patients with bacterial infection in the stable state. Secondly, COPD patients with bacterial infection during exacerbations had a significant decrease in the blood eosinophil absolute count compared to the stable state, while no blood eosinophil count changes were observed in patients without bacterial infection. These observations during the stable state and exacerbations showing an inverse relationship between bacterial infection and eosinophil counts may be relevant to corticosteroid responsiveness in COPD; the increased corticosteroid responsiveness observed with higher eosinophil counts could be due, at least partly, to lower levels of bacterial infection..
This inverse relationship between bacterial counts and blood eosinophils was present during exacerbations but not in the stable state. This is likely to be due to the stronger association between blood and sputum eosinophils at exacerbation compared to the stable state. Blood eosinophils have been proposed as a surrogate biomarker of sputum eosinophils, but the weak relationship between these measurements in the stable state, demonstrated here and in previous studies [6, 7], highlights a limitation of blood eosinophil counts in this context.
The prevalence of bacterial colonisation at baseline in our study (52%) was comparable to previously published studies that used qPCR to detect PPM presence; Bafadhel et al., [12] reported a 51% prevalence while, Barker et al., [26] reported a 77% prevalence. In the stable state, higher bacterial counts were associated with higher sputum neutrophil % and lower eosinophil %. This relationship between higher sputum bacterial counts and increased sputum neutrophil % is known [12, 13, 29]. The lower sputum eosinophil % in COPD patients with bacterial colonisation is partly due to the bacterial driven increase in absolute and percentage neutrophil counts causing a lower calculated eosinophil %. However, the absolute sputum eosinophil count is not dependent on any calculation involving the neutrophil count, and we observed that the absolute sputum eosinophil count was also lower in patients with sputum eosinophils <3%, indicating a genuine reduction in eosinophil numbers in the airways of patients with higher bacterial counts.
The inverse relationships between bacterial counts and sputum eosinophils provide a potential mechanism that determines corticosteroid responsiveness in COPD patients. Individuals with lower sputum eosinophil counts have higher levels of bacterial infection; perhaps the presence of more bacteria, with associated neutrophilic inflammation [12,13,14], contributes to the lower corticosteroid response previously reported [16, 17]. Neutrophilic airway inflammation appears to be poorly responsive to corticosteroid treatment [15, 17, 30], and the absence of bacterial inflammation may therefore favour a greater corticosteroid response through a shift in the balance of airway inflammation away from neutrophilic inflammation towards more eosinophilic inflammation.
Previous studies have failed to show a significant difference in sputum eosinophil % between stable COPD patients with bacteria present versus those without, although a numerical decrease in the former group was observed [12, 26]. Compatible with our results is the previous finding of a significantly lower level of sputum CCL13 (an eosinophil associated cytokine) in PPM positive COPD patients [26].
It has been reported that the effects of oral corticosteroids are dependent on the blood eosinophil count during COPD exacerbations, with more likelihood of treatment failure or a longer hospital stay with lower blood eosinophil counts [11, 31, 32]. The data reported here show lower eosinophil counts during acute bacterial infections, and it could be inferred that treatment failure with oral corticosteroids in such cases is related to the presence of airway bacteria. During exacerbations both sputum and blood eosinophil measurements showed similar relationships to bacterial counts. However, the sputum data were less robust due to smaller sample size.
Blood neutrophil counts are known to increase during infection, irrespective of bacterial presence [33]; we observed the same during COPD exacerbations. The decrease in blood eosinophil absolute count during exacerbation cannot be due to the increase in blood neutrophil counts for two reasons. First, there was a similar increase in blood neutrophil counts in both PPM positive and negative patients; second, we measured blood eosinophil absolute numbers which are not influenced by calculations of percentage relative to neutrophils.
In COPD patients hospitalised for exacerbations, blood eosinopenia is an independent predictor of mortality and length of stay [34, 35]. The exacerbations in this study were treated in the outpatient setting (only 3/109 exacerbations resulted in hospitalisation). These less severe events had few cases of eosinopenia (5/109 patients). Nevertheless, our results demonstrate an effect of airway bacteria on blood eosinophil counts even in patients without overt bacterial sepsis. Similarly, blood eosinophil counts decrease in asthma patients with bacterial infections [36].
Rhinovirus infection had no effect on blood eosinophil counts. Experimental rhinovirus infection causes an increase in the airway bacterial load, demonstrating the complexity of the relationship between virus and bacterial infection during COPD exacerbations [37]. Our results indicate that bacterial, rather than viral, infection modulates blood eosinophil counts. However, it is worth noting that due to insufficient sputum samples being available, rhinovirus detection was performed in a smaller proportion of patients (75/109 patients). Consequently, the lack of effect of rhinovirus on eosinophil counts could potentially be attributed to limited power in the analysis.
The mechanism responsible for the decrease in eosinophil counts during bacterial infection is unclear. Eosinophils can trigger innate immune responses to pathogens through the release of extracellular DNA traps and the expression of specific pattern recognition receptors including Toll like receptor 4 [38, 39]. Furthermore, eosinophil granule proteins have bactericidal activity [40]. A decrease in circulating eosinophils may be a result of adrenal glucocorticoid stimulation in response to the stress of bacterial infection or the rapid accumulation of eosinophils at the inflammatory site [41, 42].
A limitation of this study is that we defined a bacteria positive sample based on the total load of bacteria measured rather than a species specific count. This was done to increase statistical power to address the relationship between eosinophils and the common pathogenic bacteria in COPD. However, it is important to note that the total bacterial load from each bacteria positive sample had at least one of the three quantified pathogens above the 1 × 104 threshold used. The presence of other important bacteria, such as Pseudomonas aeruginosa, was not assessed using qPCR. Additionally, we cannot discount the possibility of selection bias as a proportion of our patients failed to provide an adequate, high quality sputum sample for differential cell count analysis. The majority of the sputum samples used in our study were spontaneously produced and although cell viability can be lower in spontaneous samples [43], we observed no differences in % sputum counts between spontaneous and induced samples, including the sputum eosinophil % count as previously reported [43]. Finally, the majority of our patients were using ICS, but we found no difference in sputum or blood cell counts or bacterial loads due to ICS use. This is in keeping with studies that have shown no effect of ICS on blood or sputum eosinophil counts [2, 44].
Future studies of the effects of ICS during the stable state could measure sputum (and blood) eosinophil counts and bacterial loads to test the hypothesis that increased bacterial load associated with reduced sputum eosinophil counts predicts reduced therapeutic response. Similar studies using oral corticosteroids during acute exacerbations measuring blood eosinophil counts and bacterial loads could test the hypothesis that increased bacterial load associated with reduced blood eosinophil counts predicts reduced therapeutic response.
Conclusion
In conclusion, our results show an inverse relationship between sputum eosinophils and airway bacterial load during the stable state, while a decrease in blood eosinophil counts occurs in COPD exacerbations with bacterial presence. These inverse relationships between eosinophil measurements and bacterial counts are potentially important determinants of individual responses to corticosteroid treatment.
Abbreviations
- COPD:
-
Chronic obstructive pulmonary disease
- GOLD:
-
Global initiative for chronic obstructive
- ICS:
-
Inhaled corticosteroid
- LABA:
-
Long acting beta agonist
- FEV1 :
-
Forced expiratory volume in 1 s
- FVC:
-
Forced vital capacity
- mMRC:
-
modified MRC scale
- CAT:
-
COPD assessment test
- SGRQ-C:
-
St George’s respiratory questionnaire
- FFMI:
-
Fat free mass index
- 6MWT:
-
6 min walk test
- qPCR:
-
quantitative polymerase chain reaction
- PPM:
-
Potentially pathogenic microorganisms
- RV:
-
Rhinovirus
- ROC:
-
Receiver operating curves
- AUC:
-
Area under the curve
References
Singh D, Roche N, Halpin D, Agusti A, Wedzicha JA, Martinez FJ. Current Controversies in the Pharmacological Treatment of Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med. 2016;194:541–9.
Brightling CE, McKenna S, Hargadon B, Birring S, Green R, Siva R, Berry M, Parker D, Monteiro W, Pavord ID, Bradding P. Sputum eosinophilia and the short term response to inhaled mometasone in chronic obstructive pulmonary disease. Thorax. 2005;60:193–8.
Brightling CE, Monteiro W, Ward R, Parker D, Morgan MD, Wardlaw AJ, Pavord ID. Sputum eosinophilia and short-term response to prednisolone in chronic obstructive pulmonary disease: a randomised controlled trial. Lancet. 2000;356:1480–5.
Bafadhel M, McKenna S, Terry S, Mistry V, Reid C, Haldar P, McCormick M, Haldar K, Kebadze T, Duvoix A, et al. Acute exacerbations of chronic obstructive pulmonary disease: identification of biologic clusters and their biomarkers. Am J Respir Crit Care Med. 2011;184:662–71.
Schleich F, Corhay JL, Louis R. Blood eosinophil count to predict bronchial eosinophilic inflammation in COPD. Eur Respir J. 2016;47:1562–4.
Singh D, Kolsum U, Brightling CE, Locantore N, Agusti A, Tal-Singer R. Eosinophilic inflammation in COPD: prevalence and clinical characteristics. Eur Respir J. 2014;44:1697–700.
Negewo NA, McDonald VM, Baines KJ, Wark PA, Simpson JL, Jones PW, Gibson PG. Peripheral blood eosinophils: a surrogate marker for airway eosinophilia in stable COPD. Int J Chron Obstruct Pulmon Dis. 2016;11:1495–504.
Pascoe S, Locantore N, Dransfield MT, Barnes NC, Pavord ID. Blood eosinophil counts, exacerbations, and response to the addition of inhaled fluticasone furoate to vilanterol in patients with chronic obstructive pulmonary disease: a secondary analysis of data from two parallel randomised controlled trials. Lancet Respir Med. 2015;3:435–42.
Siddiqui SH, Guasconi A, Vestbo J, Jones P, Agusti A, Paggiaro P, Wedzicha JA, Singh D. Blood Eosinophils: A Biomarker of Response to Extrafine Beclomethasone/Formoterol in Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med. 2015;192:523–5.
Pavord ID, Lettis S, Locantore N, Pascoe S, Jones PW, Wedzicha JA, Barnes NC. Blood eosinophils and inhaled corticosteroid/long-acting beta-2 agonist efficacy in COPD. Thorax. 2016;71:118–25.
Bafadhel M, McKenna S, Terry S, Mistry V, Pancholi M, Venge P, Lomas DA, Barer MR, Johnston SL, Pavord ID, Brightling CE. Blood eosinophils to direct corticosteroid treatment of exacerbations of chronic obstructive pulmonary disease: a randomized placebo-controlled trial. Am J Respir Crit Care Med. 2012;186:48–55.
Bafadhel M, Haldar K, Barker B, Patel H, Mistry V, Barer MR, Pavord ID, Brightling CE. Airway bacteria measured by quantitative polymerase chain reaction and culture in patients with stable COPD: relationship with neutrophilic airway inflammation, exacerbation frequency, and lung function. Int J Chron Obstruct Pulmon Dis. 2015;10:1075–83.
Sethi S, Maloney J, Grove L, Wrona C, Berenson CS. Airway inflammation and bronchial bacterial colonization in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173:991–8.
Soler N, Ewig S, Torres A, Filella X, Gonzalez J, Zaubet A. Airway inflammation and bronchial microbial patterns in patients with stable chronic obstructive pulmonary disease. Eur Respir J. 1999;14:1015–22.
Singh D, Siew L, Christensen J, Plumb J, Clarke GW, Greenaway S, Perros-Huguet C, Clarke N, Kilty I, Tan L. Oral and inhaled p38 MAPK inhibitors: effects on inhaled LPS challenge in healthy subjects. Eur J Clin Pharmacol. 2015;71:1175–84.
Bourbeau J, Christodoulopoulos P, Maltais F, Yamauchi Y, Olivenstein R, Hamid Q. Effect of salmeterol/fluticasone propionate on airway inflammation in COPD: a randomised controlled trial. Thorax. 2007;62:938–43.
Boorsma M, Lutter R, van de Pol MA, Out TA, Jansen HM, Jonkers RE. Long-term effects of budesonide on inflammatory status in COPD. COPD. 2008;5:97–104.
Terradas R, Grau S, Blanch J, Riu M, Saballs P, Castells X, Horcajada JP, Knobel H. Eosinophil count and neutrophil-lymphocyte count ratio as prognostic markers in patients with bacteremia: a retrospective cohort study. PLoS One. 2012;7:e42860.
Abidi K, Khoudri I, Belayachi J, Madani N, Zekraoui A, Zeggwagh AA, Abouqal R. Eosinopenia is a reliable marker of sepsis on admission to medical intensive care units. Crit Care. 2008;12:R59.
ATS statement: guidelines for the six-minute walk test. Am J Respir Crit Care Med. 2002;166:111–117.
Miller MR, Hankinson J, Brusasco V, Burgos F, Casaburi R, Coates A, Crapo R, Enright P, van der Grinten CP, Gustafsson P, et al. Standardisation of spirometry. Eur Respir J. 2005;26:319–38.
Pizzichini MM, Popov TA, Efthimiadis A, Hussack P, Evans S, Pizzichini E, Dolovich J, Hargreave FE. Spontaneous and induced sputum to measure indices of airway inflammation in asthma. Am J Respir Crit Care Med. 1996;154:866–9.
Anthonisen NR, Manfreda J, Warren CP, Hershfield ES, Harding GK, Nelson NA. Antibiotic therapy in exacerbations of chronic obstructive pulmonary disease. Ann Intern Med. 1987;106:196–204.
Garcha DS, Thurston SJ, Patel AR, Mackay AJ, Goldring JJ, Donaldson GC, McHugh TD, Wedzicha JA. Changes in prevalence and load of airway bacteria using quantitative PCR in stable and exacerbated COPD. Thorax. 2012;67:1075–80.
George SN, Garcha DS, Mackay AJ, Patel AR, Singh R, Sapsford RJ, Donaldson GC, Wedzicha JA. Human rhinovirus infection during naturally occurring COPD exacerbations. Eur Respir J. 2014;44:87–96.
Barker BL, Haldar K, Patel H, Pavord ID, Barer MR, Brightling CE, Bafadhel M. Association between pathogens detected using quantitative polymerase chain reaction with airway inflammation in COPD at stable state and exacerbations. Chest. 2015;147:46–55.
Singh R, Mackay AJ, Patel AR, Garcha DS, Kowlessar BS, Brill SE, Donnelly LE, Barnes PJ, Donaldson GC, Wedzicha JA. Inflammatory thresholds and the species-specific effects of colonising bacteria in stable chronic obstructive pulmonary disease. Respir Res. 2014;15:114.
Bafadhel M, McCormick M, Saha S, McKenna S, Shelley M, Hargadon B, Mistry V, Reid C, Parker D, Dodson P, et al. Profiling of sputum inflammatory mediators in asthma and chronic obstructive pulmonary disease. Respiration. 2012;83:36–44.
Siva R, Bafadhel M, Monteiro W, Brightling CE, Pavord ID. Effect of levofloxacin on neutrophilic airway inflammation in stable COPD: a randomized, double-blind, placebo-controlled trial. Int J Chron Obstruct Pulmon Dis. 2014;9:179–86.
Wang M, Gao P, Wu X, Chen Y, Feng Y, Yang Q, Xu Y, Zhao J, Xie J. Impaired anti-inflammatory action of glucocorticoid in neutrophil from patients with steroid-resistant asthma. Respir Res. 2016;17:153.
Bafadhel M, Greening NJ, Harvey-Dunstan TC, Williams JE, Morgan MD, Brightling CE, Hussain SF, Pavord ID, Singh SJ, Steiner MC. Blood eosinophils and outcomes in severe hospitalised exacerbations of COPD. Chest. 2016;150(2):320–8.
Serafino-Agrusa L, Scichilone N, Spatafora M, Battaglia S. Blood eosinophils and treatment response in hospitalized exacerbations of chronic obstructive pulmonary disease: A case–control study. Pulm Pharmacol Ther. 2016;37:89–94.
Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, Schein RM, Sibbald WJ. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101:1644–55.
Holland M, Alkhalil M, Chandromouli S, Janjua A, Babores M. Eosinopenia as a marker of mortality and length of stay in patients admitted with exacerbations of chronic obstructive pulmonary disease. Respirology. 2010;15:165–7.
Steer J, Gibson J, Bourke SC. The DECAF Score: predicting hospital mortality in exacerbations of chronic obstructive pulmonary disease. Thorax. 2012;67:970–6.
Spallarossa D, Sacco O, Girosi D, Rossi GA. Blood eosinophil counts and arterial oxygen tension in acute asthma. Arch Dis Child. 1995;73:333–7.
Mallia P, Footitt J, Sotero R, Jepson A, Contoli M, Trujillo-Torralbo MB, Kebadze T, Aniscenko J, Oleszkiewicz G, Gray K, et al. Rhinovirus infection induces degradation of antimicrobial peptides and secondary bacterial infection in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;186:1117–24.
Kvarnhammar AM, Cardell LO. Pattern-recognition receptors in human eosinophils. Immunology. 2012;136:11–20.
Yousefi S, Gold JA, Andina N, Lee JJ, Kelly AM, Kozlowski E, Schmid I, Straumann A, Reichenbach J, Gleich GJ, Simon HU. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med. 2008;14:949–53.
Lehrer RI, Szklarek D, Barton A, Ganz T, Hamann KJ, Gleich GJ. Antibacterial properties of eosinophil major basic protein and eosinophil cationic protein. J Immunol. 1989;142:4428–34.
Bass DA. Behavior of eosinophil leukocytes in acute inflammation. II. Eosinophil dynamics during acute inflammation. J Clin Invest. 1975;56:870–9.
Hills AG, Forsham PH, Finch CA. Changes in circulating leukocytes induced by the administration of pituitary adrenocorticotrophic hormone in man. Blood. 1948;3:755–68.
Khurana S, Ravi A, Sutula J, Milone R, Williamson R, Plumb J, Vestbo J, Singh D. Clinical characteristics and airway inflammation profile of COPD persistent sputum producers. Respir Med. 2014;108:1761–70.
Kreindler JL, Watkins ML, Lettis S, Tal-Singer R, Locantore N. Effect of inhaled corticosteroids on blood eosinophil count in steroid-naive patients with COPD. BMJ Open Respir Res. 2016;3:e000151.
Acknowledgements
The authors thank staff and participants of the COPD MAP consortium study.
Funding
This work was supported by the Medical Research Council (MRC); and COPD MAP (MRC/ABPI Inflammation and Immunology Initiative); National Institute for Health Research Respiratory and Allergy Clinical Research Facility at University Hospital of South Manchester NHS Foundation Trust; National Institute for Health Research (NIHR) Leicester Respiratory Biomedical Research Unit and National Institute for Health Research (NIHR) Respiratory Biomedical Research Unit at the Royal Brompton and Harefield NHS Foundation Trust and Imperial College, London UK. The views expressed in this publication are those of the author(s) and not necessarily those of the NHS, the National Institute for Health Research or the Department of Health.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article (and its additional files).
Authors’ contributions
UK and DS were responsible for the concept and design of study. UK, GD, RS, BB, VG, LG, AW, ST, TMcH and AB were involved in data acquisition. UK analysed the data and DS oversaw all analyses. UK and DS were responsible for data interpretation and drafting the manuscript. GD, RS, BB, VG, LG, AW, ST, TMcH, AB, CB and JAW revised the manuscript critically for intellectual content. All authors have approved the final version to be published and are jointly accountable for all aspects of the work.
Competing interests
UK, VG, RS, BB,,LG, AW, ST, TMcH, AB have no competing interests; GD received personal fees from MiCom SRL; CEB has received grants and or consultancy paid via his Institution from GSK, AZ/MedImmune, Novartis, Chiesi, BI, Pfizer, Theravance, Vectura; JW reports personal fees and non-financial support from Novartis, Pfizer, Astra Zeneca, Boehringer Ingelheim, grants, personal fees and non-financial support from GlaxoSmithKline, Takeda, grants and personal fees from Johnson and Johnson and Vifor Pharma, personal fees from Bayer, Chiesi, Napp; DS has received sponsorship to attend international meetings, honoraria for lecturing or attending advisory boards and research grants from various pharmaceutical companies including Almirall, AstraZeneca, Boehringer Ingelheim, Chiesi, Genentech, GlaxoSmithKline, Glenmark, Merck, NAPP, Novartis, Pfizer, Respivert, Skypharma, Takeda, Teva, Therevance and Verona.
Ethics approval and consent to participate
All patients provided written informed consent using protocols approved by the local Ethics Committees at each site (Hampstead, 11/L0/1630; Greater Manchester South, 10/H/1003/108; East Midlands, 07/H0406/157).
Consent for publication
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1:
Additional methods supplement. (DOCX 38.7 kb)
Additional file 2: Figure S1.
Outline of the methods used for the processing of sputum qPCR and differential cell count samples. (PPTX 64.1 kb)
Additional file 3: Figure S2.
A summary of the number of patients with blood / sputum eosinophil counts and qPCR data at exacerbation alone, or coupled to baseline measurements. PPM=potentially pathogenic microorganisms. (PPTX 51.6 kb)
Additional file 4: Figure S3.
The blood eosinophil count at baseline and exacerbation in patients defined as PPM positive or PPM negative at exacerbation (PPM threshold value of 1x10 ). PPM=potentially pathogenic microorganisms. Dotted lines represent median values. (PPTX 274 kb)
Additional file 5: Table S1.
The change in blood eosinophil count between RV negative and RV positive exacerbations. (PPTX 18.8 kb)
Additional file 6: Figure S4.
The sputum neutrophil % at baseline and exacerbation in patients defined as PPM positive or PPM negative at exacerbation. PPM=potentially pathogenic microorganisms. Dotted lines represent median values. (PPTX 189 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kolsum, U., Donaldson, G.C., Singh, R. et al. Blood and sputum eosinophils in COPD; relationship with bacterial load. Respir Res 18, 88 (2017). https://doi.org/10.1186/s12931-017-0570-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12931-017-0570-5