Abstract
Background
In Japan, epizootic arboviral infections have severely impacted the livestock industry for a long period. Akabane, Aino, Chuzan, bovine ephemeral fever and Ibaraki viruses have repeatedly caused epizootic abnormal births and febrile illness in the cattle population. In addition, Peaton, Sathuperi, Shamonda and D’Aguilar viruses and epizootic hemorrhagic virus serotype 7 have recently emerged in Japan and are also considered to be involved in abnormal births in cattle. The above-mentioned viruses are hypothesized to circulate in tropical and subtropical Asia year round and to be introduced to temperate East Asia by long-distance aerial dispersal of infected vectors. To watch for arbovirus incursion and assess the possibility of its early warning, monitoring for arboviruses was conducted in the Yaeyama Islands, located at the most southwestern area of Japan, between 1994 and 2014.
Results
Blood sampling was conducted once a year, in the autumn, in 40 to 60 healthy cattle from the Yaeyama Islands. Blood samples were tested for arboviruses. A total of 33 arboviruses including Akabane, Peaton, Chuzan, D’ Aguilar, Bunyip Creek, Batai and epizootic hemorrhagic viruses were isolated from bovine blood samples. Serological surveillance for the bovine arboviruses associated with cattle diseases in young cattle (ages 6–12 months: had only been alive for one summer) clearly showed their frequent incursion into the Yaeyama Islands. In some cases, the arbovirus incursions could be detected in the Yaeyama Islands prior to their spread to mainland Japan.
Conclusions
We showed that long-term surveillance in the Yaeyama Islands could estimate the activity of bovine arboviruses in neighboring regions and may provide a useful early warning for likely arbovirus infections in Japan. The findings in this study could contribute to the planning of prevention and control for bovine arbovirus infections in Japan and cooperative efforts among neighboring countries in East Asia.
Similar content being viewed by others
Background
Arthropod-borne viruses (arboviruses) are transmitted by hematophagous arthropod vectors, such as mosquitoes, ticks and Culicoides biting midges. Nearly 500 arboviruses have been recorded so far and some of them can seriously harm animal health [1]. Arbovirus infections often affect wide regions in a short period of time. The infected vectors are easily disseminated by air streams [2, 3], and their long-distance migrations (hundreds of kilometers) have been successfully estimated [4–7]. The recent emergence of bluetongue and Schmallenberg virus (SBV) infection in northern Europe demonstrated such rapid and wide expansion and caused huge economic damage to the livestock industry of countries that had been previously free from these arboviral infections [8, 9].
In Japan, epizootic abortion, stillbirth, premature birth and congenital malformations in cattle caused by arboviruses have severely impacted the livestock industry for a long period [10]. Akabane virus (AKAV) and Aino virus (AINOV) of the genus Orthobunyavirus in the family Bunyaviridae is principally associated with recurring epizootics of abnormal births [10–12]. It was estimated that approximately 42,000 abnormal calves caused by AKAV were born during the largest outbreak between 1972 and 1975. Chuzan virus (CHUV) of the genus Orbivirus in the family Reoviridae is also known to be an etiological agent of congenital abnormalities in cattle [13, 14]. In recent years, bovine encephalomyelitis caused by postnatal AKAV infection reemerged in Japan after a 21-year absence [15, 16]. Furthermore, incursions of Peaton virus (PEAV), Sathuperi virus (SATV) and Shamonda virus (SHAV) of the genus Orthobunyavirus were confirmed in Japan in the past 16 years [17–19]. Although these viruses potentially have teratogenicity in ruminants, little is known about their pathogenicity [20, 21]. As well as CHUV, D’Aguilar virus (DAGV) of the genus Orbivirus is a member of Palyam virus (PALV) group and has repeatedly been isolated in Japan since 1987 [22, 23]. Its etiological role in epizootic congenital abnormalities in cattle between 2001 and 2002 was serologically confirmed with colostrum-free calves. Bovine ephemeral fever virus (BEFV) of the genus Ephemerovirus in the family Rhabdoviridae causes a febrile illness in cattle and water buffalo, and is associated with reduction of milk production in dairy cattle and loss of condition in beef cattle [24]. The last occurrence of bovine ephemeral fever in mainland Japan was reported in 1991, but its periodic epizootics continue in the southwestern islands [25, 26]. Ibaraki virus (IBAV) is a strain of epizootic hemorrhagic disease virus (EHDV) serotype 2 of the genus Orbivirus, and difficulty swallowing is its main manifestation [27]. The contribution of EHDV serotype 7 (EHDV-7) to epizootic abortion and stillbirth in pregnant cows was also reported in the southern part of Japan in 1997 [28–30]. The above-mentioned arboviruses are principally transmitted by Culicoides biting midges [23, 31] while mosquitoes are strong candidates for BEFV vectors [24]. Because the vector activity ceases in winter, the overwintering of arboviruses is seemingly unrealistic in Japan. In fact, appearances of the same strain/genotype of arboviruses in consecutive years rarely happen in Japan [10, 32, 33]. Development of efficient commercial vaccines to AKAV, AINOV, CHUV, IBAV and BEFV has contributed to reducing diseases in cattle, but arbovirus infections still break out in unvaccinated cattle, and no effective prevention measures are prepared for newly emerged arboviruses.
The ideal climate for vector insects in the tropical zone permits arbovirus circulation through most of the year. Therefore, the lower-latitude regions are considered a potential source of arboviruses in the temperate zone, where vector insects are usually absent during the colder months. Culicoides- and mosquito-borne arboviruses are considered to be introduced from lower latitudes in Asia to Japan with the infected vectors carried by airstreams [10, 34]. Although former studies revealed the prevalence of several ruminant arboviruses in Southeast Asian countries [35–37], details of arbovirus activity remain unknown. The Yaeyama Islands in Okinawa Prefecture are located in the subtropical zone and are 100–200 km, 350–450 km, 700 km and 1000 km away from Taiwan, Southern China, Philippines and mainland Japan, respectively. Due to their geographic location, the islands may be well suited for playing a role as an early warning system for other parts of Japan. Indeed, it was suggested that previous epizootics of bovine ephemeral fever in the Yaeyama Islands were epidemiologically linked to those in Taiwan [25, 26, 38]. The enhanced surveillance of ruminant arboviruses in the bordering area could support the planning of preventive and control strategies for arbovirus infections in the remaining part of Japan. To monitor the annual incursion of arboviruses into the Yaeyama Islands, we conducted virus isolation from sentinel cattle and serosurveillance for the above-mentioned arboviruses in the cattle population for 20 years. This long-term monitoring indicates a high risk of incursion by multiple arboviruses from overseas to the Yaeyama Islands. We also discuss whether the monitoring in the islands could contribute to earlier detection of arbovirus activities prior to their spread in mainland Japan.
Methods
Study area
The Yaeyama Islands comprise an archipelago located in the most southwestern part of Japan (24.00-24.67° N, 122.75-124.50°E) from Ishigaki Island to Yonaguni Island (Fig. 1). Approximately 36,000 beef cattle are reared through the islands, and over 70 % of this cattle population exists on Ishigaki Island, which is the second largest island in the archipelago. The climate of the Yaeyama Islands is subtropical; the annual mean temperature was 24 °C between 1994 and 2014; mean temperatures of the hottest and coldest months are about 29 °C and 19 °C, respectively. Annual precipitation was about 2200 mm and humidity ranged from 70 to 80 % during the study period. Blood sampling was continuously conducted in Ishigaki, Iriomote and Yonaguni Islands, but was optional in Taketomi, Kohama, Kuroshima and Hateruma Islands (Table 1).
Blood collection
Blood sampling was conducted once a year between the middle of October and beginning of December in 1994–2003, 2005–2009 and 2011–2014 (Table 1). Heparinized blood and serum samples were obtained from 40–60 healthy cattle. The blood samples were separated into plasma and blood cells by centrifugation and the blood cells were washed three times with phosphate buffered saline to eliminate the antibodies. The plasmas and blood cells were stored at −80 °C until virus isolation. The serum samples were kept at −20 °C until they were used for virus neutralization tests (VNTs). A total of 977 blood samples were subjected to arbovirus screening. To detect antibodies against arboviruses, VNTs were conducted for 813 serum samples harvested after 1995. The tested sera were selectively collected from cattle that were 6–12 months old and had only been alive for one summer. The blood samples were voluntarily collected by local veterinary officers for monitoring of arbovirus infections and were permitted to be used in this study from the animal health authority of Okinawa Prefecture.
Virus isolation
The processed plasmas and blood cells were inoculated into monolayer cultures of baby hamster kidney (BHK-21) and hamster lung (HmLu-1) cells as described previously [23]. Briefly, cells were incubated into test tubes with Eagle’s minimum essential medium (MEM) (Nissui, Tokyo, Japan) supplemented with 0.295 % tryptose phosphate broth (Becton Dickinson and Company, Franklin Lakes, NJ, USA), 0.015 % sodium bicarbonate and 10 % of bovine serum overnight at 37 °C and were washed three times with Earl’s solution before inoculation. The cell cultures inoculated with blood samples were maintained in serum-free medium by rotation at 37 °C for 7 days and were collected if a cytopathic effect (CPE) was observed. Two further blind passages were conducted in the same manner if CPE was not observed.
Dot immunobinding assay
Cytopathic agents in the cultured cells were characterized by a dot immunobinding assay (DIA) as described previously [23, 39]. Briefly, the supernatants of cell cultures showing CPE were blotted onto the Immobilon PVDF transfer membrane (Millipore, Billerica, MA, USA) using a slot blotting apparatus. The membrane was immersed in blocking buffer [8 % skim milk in Tris-buffered saline (TBS; 20 mM Tris–HCl pH 7.5, 0.15 M NaCl)] for more than 2 h and then reacted with monoclonal antibodies to the nucleocapsid protein and viral surface glycoprotein (Gc) of AKAV, the Gc protein of AINOV and PEAV, and mouse antisera to CHUV, IBAV and bluetongue virus for 1 h. The membrane was reacted with horseradish peroxidase-conjugated goat antibody to mouse IgG in blocking buffer for 30 min, and the immune complex was detected by color development with 0.027 % 3,3-diaminobenzidine tetrahydrochloride and 0.016 % H2O2 in TBS.
RT-PCR and sequence analysis
Viral RNA was extracted from the supernatant of cell cultures showing CPE with the High Pure Viral RNA Kit (Roche Diagnostics, Mannheim, Germany). Group-specific RT-PCRs targeting the S RNA segment of orthobunyaviruses, segment 3 of PALV group viruses and segment 7 of EHDV were performed using the Titan One tube RT-PCR Kit (Roche Diagnostics) in accordance with previous studies [40, 41]. Specific detections of segment 2 of CHUV and DAGV by RT-PCR were conducted with the primer sets CHUVL2F-2/CHUVL2R-2 and DAGVL2F/DAGVL2R-2, respectively (Additional file 1) [23]. An isolate of EHDV was tested in an RT-PCR assay with the serotype-specific primer sets targeting segment 2 [41]. Sample denaturation at 94 °C for 4 min was performed for the orbivirus detection before cDNA synthesis. The cDNA synthesis was conducted at 50 °C for 30 min followed by 94 °C for 2 min. The PCR profile was 10 cycles of 94 °C for 30 s, 55 °C for 30 s and 68 °C for 45 s, followed by 25 cycles of 94 °C for 30 s, 55 °C for 30 s and 68 °C for 45 s, with the latter time increased by 5 s per cycle.
Genome RNA of an orbivirus that was unidentified by the above-mentioned methods was extracted from the infected cells using the TRIZOL LS Reagent (Life Technologies, Carlsbad, CA, USA). Amplification of the cDNA of segment 2 was performed by Full-Length Amplification of cDNAs (FLAC) as described previously [42]. In brief, double-stranded RNA was ligated to the anchor primer (5-15-1) with T4 RNA ligase (New England Bio Labs, Ipswich, MA, USA) at 4 °C overnight and separated in 1 % agarose gel by electrophoresis. Segment 2 was recovered from the gel and purified using the RNaid Kit with SPIN (Bio 101, Vista, CA, USA). First-strand cDNA synthesis was performed using the Superscript III First-Strand Synthesis System for RT-PCR (Life Technologies) in accordance with the manufacturer’s instructions. The cDNA was amplified using KOD-plus-ver.2 (TOYOBO, Osaka, Japan) in a reaction mixture under the following conditions: 30 cycles of 98 °C for 10 s, 62 °C for 30 s and 68 °C for 3.5 min.
The PCR products were purified with the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and directly sequenced with the BigDye Terminator Cycle sequencing Kit v3.1 (Life Technologies) on the ABI 3100-Avanti Genetic Analyzer (Life Technologies). The nucleotide (nt) sequences were edited by DNASIS Pro Ver. 3.0 (Hitachi Solutions, Tokyo, Japan), and a sequence similarity search was conducted with the basic local alignment search tool (BLAST). Pairwise nt sequence identities were calculated with GENETYX software ver. 10 (GENETYX, Tokyo, Japan).
Virus neutralization test
Virus neutralization tests with AKAV OBE-1, AINOV JaNAr28, PEAV KSB-1/P/06, SATV KSB-2/C/08, SHAV KSB-6/C/02, BEFV YHL, CHUV C31, DAGV KSB-29/E/01, IBAV No.2 and EHDV-7 KSB-14/E/97 were performed. After heat inactivation at 56 °C for 30 min, bovine sera were serially diluted twofold in serum-free Eagle’s MEM containing 10 μg/ml gentamicin sulfate from 1:2 to 1:64 in the 96-well microplates. Fifty microliters of serum dilution was mixed with an equal volume of virus inoculum containing 100 × the 50 % tissue culture infective doses and incubated at 37 °C under 5 % CO2 for 1 h. Then, 100 μl of the suspension of HmLu-1 cells in GIT medium (Wako Pure Chemical Industries, Ltd., Osaka, Japan) was added into each well and incubated at 37 °C under 5 % CO2 for 7 days. The antibody titer was calculated as the reciprocal of the highest serum dilution inhibiting the CPE. Samples were deemed positive if they had neutralizing antibodies to the viruses in at least a dilution of 1/8.
Results
Isolation and identification of arboviruses from bovine blood samples
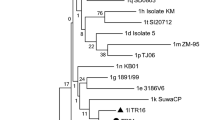
A total of 33 arbovirus isolates were obtained from bovine blood samples during the study period (Table 2). Some of the isolated viruses had been genetically analyzed in previous studies [22, 32, 33]. Nine isolates obtained in 1994, 1998 and 2001 were identified as AKAV by DIA. Four PEAV isolates were also found in the viruses obtained in 2001 and 2009. Eighteen isolates were reacted with the polyclonal antibodies against CHUV in DIAs and the group-specific and strain-specific RT-PCRs sorted themselves into CHUV (6 isolates in 1998, 2002 and 2006), DAGV (5 isolates in 2000, 2006 and 2012) and an unidentified PALV group virus (7 isolates in 2008 and 2009). Segment 2 of ON-1/E/08, which is an isolate of the unidentified PALV group viruses, was amplified by the FLAC method. The whole sequence of segment 2 (GenBank accession no. AB973440) showed the highest identity with Bunyip Creek virus (BCV) CSIRO58 (90.8 % nt and 94.9 % aa identities). The other 6 isolates of the PALV group viruses tested positive by RT-PCR with the BCV segment 2-specific primer pair. DIA and the group-specific RT-PCRs revealed that a 2003 isolate designated as ON-1/E/03 belongs to EHDV. High levels of similarity (82–84 % nt identity) in segment 7 between ON-1/E/03 (GenBank accession No. LC066302) and other EHDV strains were also revealed. However, ON-1/E/03 tested negative by RT-PCRs with the serotype-specific primer sets. The remaining isolate obtained in 1994 could not be identified by the serological tests. However, detailed genetic analyses in a previous study determined the isolate as Batai virus (BATV) of the genus Orthobuyavirus [43].
Detection of antibodies against bovine arboviruses
All arboviruses that were investigated in this study were observed as present in cattle blood in the Yaeyama Islands between 1995 and 2014 (Fig. 2). Neutralizing antibodies to the above-mentioned arboviruses were often detected in the 4-fold diluted serum. These results may indicate infection by another closely related virus [44–46] or the presence of maternally derived antibodies included in colostrum [47, 48]. Also, non-specific reactivity due to the low dilution of the sera in the VNTs could not be ruled out. Therefore, neutralizing antibody titers < 1:8 were defined as negative. Neutralizing antibodies to AKAV, AINOV, CHUV and IBAV were detected in tested cattle for more than 10 years during the study period. The seroprevalence varied a great deal according to the tested viruses (4.4–22.2 % for AKAV; 2.0–29.2 % for AINOV; 2.0–57.8 % for CHUV and 2.6–38.5 % for IBAV) and the year (Additional file 2). PEAV, BEFV, DAGV and EHDV-7 were each observed in 7–9 years. Neutralizing antibodies against SHAV and SATV were detected in 2–6 years. Serum samples collected in the Yaeyama Islands in 1994 were attempted to detect the antibodies against AKAV and BATV. A certain level of seroprevalence for both viruses (10.3 % for AKAV and 50.0 % for BATV) was observed in examined cattle (data not shown).
Arboviruses were frequently detected in the blood of cattle on Ishigaki and Iriomote Islands, but only occasionally in those of the other islands (Table 3). However, the seroprevalence in Yonaguni Island was sometimes independent of those in Ishigaki and Iriomote Islands (e.g. against PEAV in 2001 and 2006). Synchronization of seroprevalence between Ishigaki Island and surrounding islands, Taketomi, Kohama, Kuroshima and Hateruma, was generally observed during the study period.
Discussion
In Japan, bovine diseases associated with AKAV, AINOV, CHUV, IBAV and BEFV are notifiable diseases under the domestic animal infectious disease control law, and nationwide serosurveillance for these arboviruses with sentinel cattle has been conducted since 1985 [47, 49, 50]. AKAV activity has been observed on mainland Japan almost every year. Also, on the Yaeyama Islands, AKAV was frequently observed and was sometimes linked with occurrences of disease. The isolated viruses in 1994 were genetically undistinguishable from AKAVs isolated on mainland Japan the following year [32, 33]. The finding indicated that the earlier detection of AKAV activity in the Yaeyama Islands could be available in some cases prior to its spread in the mainland. Although the AINOV isolation was not made in this study, its prevalence in the Yaeyama Islands was observed by serological surveillance. The epizootics of abnormal births caused by AINOV occurred in mainland Japan, in 1998–1999, 2002–2003 and 2005–2006 seasons. In contrast, the seroprevalence in the Yaeyama Islands was concurrently found in 1998 and 2002. Although AINOV has not been detected in the mainland since 2006, neutralizing antibodies against AINOV were continuously detected in the islands, indicating that AINOV is still active in the lower latitudes in Asia and its reemergence in the mainland is highly possible.
On mainland Japan, the last occurrence of congenital abnormalities by CHUV was reported in 2000 and, since then, CHUV has not been detected. However, the CHUV activity in the neighboring regions could be estimated from the frequent sero-detection and the virus isolations in the Yaeyama Islands. Although no clinical case was reported, the activity of DAGV had been detected several times in the Yaeyama Islands since 1991. It is noteworthy that a DAGV isolate in Ishigaki Island in 2000 shared 99.5 % nt identity in segment 2 with DAGV isolated in mainland Japan during the 2001 outbreak [22]. Furthermore, DAGV returned to the mainland in 2013 after its activity was observed in the Yaeyama Islands in the previous year [23].
Neutralizing antibodies to BEFV were present in the tested cattle in 2001 and 2012 when outbreaks of bovine ephemeral fever occurred. Low seroprevalences (2.0–5.1 %) of BEFV were also detected without clinical cases in 1995, 1997, 1998, 2000 and 2002, suggesting that the incursion and small-scale circulation of BEFV often occurs in the Yaeyama Islands. This also reflects the pattern of frequent epizootics of BEFV in neighboring regions, such as Taiwan and mainland China, in recent years [38, 51–53].
The resurgence of IBAV in the mainland was confirmed in 2013 after a 26-year absence [30]. However, the neutralizing antibodies to IBAV have been frequently detected in the Yaeyama Islands during the study period. Although the clinical evidence and the virus isolation were reported in Taiwan and Korea, respectively [54, 55], there is currently little information on this virus outside of Japan. It should be noted that IBAV caused an extensive outbreak of disease in cattle in Japan in 1959–1960 and resulted in the deaths of over 4000 cattle [27]. The epizootic of EHDV-7 once occurred in the mainland in 1997 [28]. Relatively high seroprevalences (24.5 and 31.1 %) of the virus were detected in the Yaeyama Islands in both 1997 and 2006. Although the serological surveillance suggests the continuous circulation of EHDV-7 in Asian countries, the epizootic abortion and still birth in cattle caused by the virus has not been recorded outside of Japan. However, this virus poses a serious potential threat to the livestock industry because no effective vaccine is currently available. Lower neutralization antibodies (≤1:8) to each of IBAV and EHDV-7 are often detected, probably due to the cross-reactivities with heterogeneous serotypes of EHDV [46]. The possible incursions of other EHDV serotypes into the cattle population in the Yaeyama Islands should be investigated in the future.
The pathogenicities of PEAV, SATV, and SHAV in cattle remain uncertain. However, colostrum-free calves with congenital abnormalities sometimes carry neutralizing antibodies to these viruses, suggesting their teratogenicity [17–19]. Although the detection of PEAV in Japan was first reported in 1999 [17], retrospective analysis for an unassigned virus from Ishigaki Island revealed that PEAV was already introduced as early as 1987 [56]. Repeated isolation of PEAV in the Yaeyama Islands, other islands in Okinawa Prefecture and mainland Japan also may link to its widespread circulation in Asian countries. Seroprevalences for SATV and SHAV were lower than those of the other arboviruses. However, the surveillance in the Yaeyama Island agreed with the recent incursion of these viruses to mainland Japan [18, 19, 21]. Although suspected clinical cases caused by SATV and SHAV have been very few in Japan, their close relationship with SBV has been clearly indicated by previous studies [21, 57, 58]. It is uncertain whether Japanese strains of SATV and SHAV, as well as SBV, maintain high pathogenicity to ruminants. However, the possible presence of SATV and SHAV should be considered to avoid misdiagnosis of SBV infection in the regions commonly affected by insect-borne arboviruses.
Attempts at virus isolation proved the incursions of BATV, BCV and an EHDV isolate to the Yaeyama Islands. Serological surveillance in 1994 and 1995 also revealed the transient incursion of BATV in 1994 (data not shown). It was reported that BATV occasionally causes a febrile illness in humans and ruminants [59]. It was noteworthy that BATV probably provided a genome segment to Ngari virus, associated with severe hemorrhagic disease in humans [43, 60]. Epizootic activity of BATV is supposed to continue in Asian countries [43, 61] and thus its involvement in human and animal diseases should be monitored hereafter. Although BCV infection in the cattle population was evident in this and previous studies [62, 63], its contribution to cattle illness is not yet clear. The BCV isolation was made in mainland Japan in 2009, one year after the initial isolation in the Yaeyama Islands [23]. The co-circulation of BCV and other PALVs, such as CHUV and DAGV, potentially generate reassortants between these viruses. Because the reassortment may cause changes in viral pathogenicity as well as viral genetic and serological properties, the emergence of reassortants among PALVs should be monitored. The EHDV isolate was obtained from bovine blood in 2003. Unfortunately, serotyping for the isolated EHDV could not be concluded by the current serotype-specific RT-PCRs [41]. The existence of multiple serotypes of EHDV makes the diagnostics of EHDV-related disease more complicated. To our knowledge, at least three serotypes of EHDV have been detected in Japan so far [30]. Further investigation, such as the use of FLAC, should be conducted to investigate the detailed properties of the virus to develop proper diagnostic systems in this region.
Annual seroprevalence patterns differed among the islands. Several factors could contribute to this result. The sampling size and cattle population on Ishigaki Island are significantly larger than those on other islands, probably resulting in the higher frequency of the virus and detection of neutralizing antibodies. Because the distance between Ishigaki Island and surrounding islands including Iriomote, Taketomi, Kohama and Kuroshima Islands is not so far (maximally 15 km apart), wind dispersal of infected vectors likely occurs between the islands. However, Yonaguni Island is approximately 65–110 km from the other islands in the archipelago. This geographical feature might cause independent epizootics there. In addition, Yonaguni Island is the part of Japan closest to Taiwan and mainland China, and thus more sensitive detection of arbovirus activities in adjacent regions could be conducted in some cases. Although the source of arboviruses remains unknown, long-distance migration of the infected insect vectors by air flows is the most probable route by which bovine arboviruses are introduced to the Yaeyama Islands. Atmospheric dispersal modeling would be useful to estimate plausible incursion events and possible source sites of infected vectors.
It is of interest that fewer clinical cases have been reported in the Yaeyama Islands, despite the frequent incursion of ruminant-pathogenic arboviruses. Most cows kept for reproduction in the islands probably gain humoral immunity to the viruses by vaccination or natural infection before their pregnancy. Therefore, the abnormal births might be prevented in most cases if the teratogenic arboviruses were introduced to the islands. Also, constant and high seroprevalence in the cattle population might limit acute and massive epizootics of arboviruses.
Because viremia of bovine arboviruses is generally short, the occasion of isolation is highly limited from sentinel cattle [23]. Therefore, the frequency of blood sampling in this study might not be enough for effective virus isolation. Culicoides biting midges are still active in the Yaeyama Islands between October and December [64] and thus the viral transmission possibly occurred after the blood samplings. To obtain the conclusive seroprevalence in each season, additional sampling from the sentinel cattle should have be conducted few months later. It would be necessary to modify the frequency and period of blood sampling for future monitoring in this area.
Too often ruminant arbovirus diseases are ignored and neglected in tropical countries because they have not yet impacted the livestock industry on the surface. However, these diseases might have caused continuous, but unrecognized losses in these countries. Further investigation regarding the ruminant arboviruses will be necessary to assess their impact on the livestock industry in the regions where they persist. Moreover, environmental change through global warming will enhance the virus spread from the tropical to the temperate zone [65]. Monitoring for exotic arboviruses increases in importance in the high-latitude regions where such viruses would do more serious damage to the livestock industry. The Yaeyama Islands were identified as a high-risk zone for arbovirus incursions in this study and thus would be an ideal point for monitoring.
Conclusions
We showed that long-term monitoring in the Yaeyama Islands could estimate the activity of bovine arboviruses in neighboring regions and may provide a useful early warning of risk of arbovirus infections in Japan. The findings in this study could contribute to planning of prevention and control for bovine arbovirus infections in Japan and also to beneficial information-sharing on monitoring efforts among neighboring countries. Enhancement of collaborative work in this region should be essential to reduce the livestock loss caused by arbovirus infections.
Abbreviations
AINOV, Aino virus; AKAV, Akabane virus; BATV, Batai virus; BCV, Bunyip Creek virus; BEFV, bovine ephemeral fever virus; CHUV, Chuzan virus; CPE, cytopathic effects; DAGV, D’Aguilar virus; DIA, dot immunobinding assay; EHDV, epizootic hemorrhagic disease virus; FLAC, Full-length Amplification of cDNAs; IBAV, Ibaraki virus; MEM, minimum essential medium; PALV, Palyam virus; PEAV, Peaton virus; SATV, Sathuperi virus; SBV, Schmallenberg virus; SHAV, Shamonda virus; VNT, virus neutralization test
References
Hubálek Z, Rudolf I, Nowotny N. Arboviruses pathogenic for domestic and wild animals. Adv Virus Res. 2014;89:201–15.
Hendrickx G, Gilbert M, Staubach C, Elbers A, Mintiens K, Gerbier G, Ducheyne E. A wind density model to quantify the airborne spread of Culicoides species during north-western Europe bluetongue epidemic, 2006. Prev Vet Med. 2008;87:162–81.
Sedda L, Rogers DJ. The influence of the wind in the Schmallenberg virus outbreak in Europe. Sci Rep. 2013;3:3361.
Ritchie SA, Rochester W. Wind-blown mosquitoes and introduction of Japanese encephalitis into Australia. Emerg Infect Dis. 2001;7:900–3.
Gloster J, Burgin L, Witham C, Athanassiadou M, Mellor PS. Bluetongue in the United Kingdom and northern Europe in 2007 and key issues for 2008. Vet Rec. 2008;162:298–302.
Aziz-Boaron O, Klausner Z, Hasoksuz M, Shenkar J, Gafni O, Gelman B, David D, Klement E. Circulation of bovine ephemeral fever in the Middle East ― strong evidence for transmission by winds and animal transport. Vet Microbiol. 2012;158:300–7.
Eagles D, Melville L, Weir R, Davis S, Bellis G, Zalucki MP, Walker PJ, Durr PA. Long-distance aerial dispersal modelling of Culicoides biting midges: case studies of incursions into Australia. BMC Vet Res. 2014;10:135.
Wilson AJ, Mellor PS. Bluetongue in Europe: past, present and future. Philos Trans R Soc Lond B Biol Sci. 2009;364:2669–81.
Wernike K, Conraths F, Zanella G, Granzow H, Gache K, Schirrmeier H, Valas S, Staubach C, Marianneau P, Kraatz F, Böntgen HD, Reimann I, Zientara S, Beer M. Schmallenberg virus-two years of experiences. Prev Vet Med. 2014;116:423–34.
Forman S, Hungerford N, Yamakawa M, Yanase T, Tsai HJ, Joo YS, Yang DK, Nha JJ. Climate change impacts and risks for animal health in Asia. Rev Sci Tech. 2008;27:581–97.
Kurogi H, Inaba Y, Goto Y, Miura Y, Takahashi H. Serologic evidence for etiologic role of Akabane virus in epizootic abortion-arthrogryposis-hydranencephaly in cattle in Japan, 1972–1974. Arch Virol. 1975;47:71–83.
Tsuda T, Yoshida K, Ohashi S, Yanase T, Sueyoshi M, Kamimura S, Misumi K, Hamana K, Sakamoto H, Yamakawa M. Arthrogryposis, hydranencephaly and cerebellar hypoplasia syndrome in neonatal calves resulting from intrauterine infection with Aino virus. Vet Res. 2004;35:531–8.
Goto Y, Miura Y, Kono Y. Serologic evidence for the etiologic role of Chuzan virus in an epizootic of congenital abnormalities with hydranencephaly-cerebellar hypoplasia syndrome of calves in Japan. Am J Vet Res. 1988;49:2026–9.
Miura Y, Kubo M, Goto Y, Kono Y. Hydranencephaly-cerebellar hypoplasia in a newborn calf after infection of its dam with Chuzan virus. Jpn J Vet Sci. 1990;52:689–94.
Miyazato S, Miura Y, Hase M, Kubo M, Goto Y, Kono Y. Encephalitis of cattle caused by Iriki isolate, a new strain belonging to Akabane virus. Jpn J Vet Sci. 1989;51:128–36.
Kono R, Hirata M, Kaji M, Goto Y, Ikeda S, Yanase T, Kato T, Tanaka S, Tsutsui T, Imada T, Yamakawa M. Bovine epizootic encephalomyelitis caused by Akabane virus in southern Japan. Vet Rec. 2008;4:20.
Matsumori Y, Inai K, Yanase T, Ohashi S, Kato T, Yoshida K, Tsuda T. Serological and genetical characterization of newly isolated Peaton virus in Japan. Arch Virol. 2002;147:401–10.
Yanase T, Fukutomi T, Yosida K, Kato T, Ohashi S, Yamakawa M, Tsuda T. The emergence in Japan of Sathuperi virus, a tropical Simbu serogroup virus of the genus Orthobunyavirus. Arch Virol. 2004;149:1007–13.
Yanase T, Maeda K, Kato T, Nyuta S, Kamata H, Yamakawa M, Tsuda T. The resurgence of Shamonda virus of the genus Orthobunyavirus, in Japan. Arch Virol. 2005;150:361–9.
Parsonson IM, Della-Porta AJ, McPhee DA. Pathogenesis and virulence studies of Australian simbu serogroup bunyaviruses. In: Mackenzie JS, editor. The viral diseases in Southeast Asia and the western pacific. Sydney: Academic Press; 1982. p. 644–7.
Yanase T, Kato T, Aizawa M, Shuto Y, Shirafuji H, Yamakawa M, Tsuda T. Genetic reassortment between Sathuperi and Shamonda viruses of the genus Orthobunyavirus in nature: implications for their genetic relationship to Schumallenberg virus. Arch Virol. 2012;157:1611–6.
Ohashi S, Matsumori Y, Yanase T, Yamakawa M, Kato T, Tsuda T. Evidence of an antigenic shift among Palyam serogroup orbiviruses. J Clin Microbiol. 2004;42:4610–4.
Kato T, Shirafuji H, Tanaka S, Sato M, Yamakawa M, Tsuda T, Yanase T. Bovine arboviruses in Culicoides biting midges and sentinel cattle in Southern Japan from 2003 to 2013. Transbound Emerg Dis. 2015. doi:10.1111/tbed.12324.
Walker PJ. Bovine ephemeral fever in Australia and the world. Curr Top Microbiol Immunol. 2005;292:57–80.
Kato T, Aizawa M, Takayoshi K, Kokuba T, Yanase T, Shirafuji H, Tsuda T, Yamakawa M. Phylogenetic relationships of the G gene sequence of bovine ephemeral fever virus isolated in Japan, Taiwan and Australia. Vet Microbiol. 2009;137:217–23.
Niwa T, Shirafuji H, Ikemiyagi K, Nitta Y, Suzuki M, Kato T, Yanase T. Occurrence of bovine ephemeral fever in Okinawa Prefecture, Japan, in 2012 and development of a reverse-transcription polymerase chain reaction assay to detect bovine ephemeral fever virus gene. J Vet Med Sci. 2015;77:455–60.
Omori T, Inaba Y, Morimoto T, Tanaka Y, Ishitani R. Ibaraki virus, an agent of epizootic disease of cattle resembling bluetongue. I. Epidemiologic, clinical and pathologic observations and experimental transmission to calves. Jpn J Microbiol. 1969;13:139–57.
Ohashi S, Yoshida K, Watanabe Y, Tsuda T. Identification and PCR-restriction fragment length polymorphism analysis of a variant of the Ibaraki virus from naturally infected cattle and aborted fetuses in Japan. J Clin Microbiol. 1999;37:3800–3.
Ohashi S, Yoshida K, Yanase T, Tsuda T. Analysis of intratypic variation evident in an Ibaraki virus strain and its epizootic hemorrhagic disease virus serogroup. J Clin Microbiol. 2002;40:3684–8.
Hirashima Y, Kato T, Yamakawa M, Shirafuji H, Okano R, Yanase T. Reemergence of Ibaraki disease in southern Japan in 2013. J Vet Med Sci. 2015;77:1253–9.
Yanase T, Kato T, Kubo T, Yoshida K, Ohashi S, Yamakawa M, Miura Y, Tsuda T. Isolation of bovine arboviruses from Culicoides biting midges (Diptera: Ceratopogonidae) in southern Japan: 1985–2002. J Med Entomol. 2005;42:63–7.
Yamakawa M, Yanase T, Kato T, Tsuda T. Chronological and geographical variations in the small RNA segment of the teratogenic Akabane virus. Virus Res. 2006;121:84–92.
Kobayashi T, Yanase T, Yamakawa M, Kato T, Yoshida K, Tsuda T. Genetic diversity and reassortments among Akabane virus field isolates. Virus Res. 2007;130:162–71.
Morita K. Molecular epidemiology of Japanese encephalitis in East Asia. Vaccine. 2009;27:7131–2.
Miura Y, Inaba Y, Tsuda T, Tokuhisa S, Sato K, Akashi H, Matumoto M. A survey of antibodies to arthropod-borne viruses in Indonesian cattle. Jpn J Vet Sci. 1982;44:857–63.
Daniels PW, Sendow I, Soleha E, Sukarsih, Hunt NT, Bahri S. Australian-Indonesian collaboration in veterinary arbovirology-a review. Vet Microbiol. 1995;46:151–74.
Bryant JE, Crabtree MB, Nam VS, Yen NT, Duc HM, Miller BR. Isolation of arboviruses from mosquitoes collected in northern Vietnam. Am J Trop Med Hyg. 2005;73:470–3.
Ting LJ, Lee MS, Lee SH, Tsai HJ, Lee F. Relationships of bovine ephemeral fever epizootics to population immunity and virus variation. Vet Microbiol. 2014;173:241–8.
Yoshida K, Tsuda T. Rapid detection of antigenic diversity of Akabane virus isolates by dot immunobinding assay using neutralizing monoclonal antibodies. Clin Diagn Lab Immunol. 1998;5:192–8.
Ohashi S, Yoshida K, Yanase T, Kato T, Tsuda T. Simultaneous detection of bovine arboviruses using single-tube multiplex reverse transcription-polymerase chain reaction. J Virol Methods. 2004;120:79–85.
Maan NS, Maan S, Nomikou K, Johnson DJ, El Harrak M, Madani H, Yadin H, Incoglu S, Yesilbag K, Allison AB, Stallknecht DE, Batten C, Anthony SJ, Mertens PP. RT-PCR assays for seven serotypes of epizootic haemorrhagic disease virus & their use to type strains from the Mediterranean region and North America. PLoS One. 2010;5:e10323.
Maan S, Rao S, Maan NS, Anthony SJ, Attoui H, Samuel AR, Mertens PP. Rapid cDNA synthesis and sequencing techniques for the genetic study of bluetongue and other dsRNA viruses. J Virol Methods. 2007;143:132–9.
Yanase T, Kato T, Yamakawa M, Takayoshi K, Nakamura K, Kokuba T, Tsuda T. Genetic characterization of Batai virus indicates a genomic reassortment between orthobunyaviruses in nature. Arch Virol. 2006;151:2253–60.
Kinney RM, Calisher CH. Antigenic relationships among Simbu serogroup (Bunyaviridae) viruses. Am J Trop Med Hyg. 1981;30:1307–18.
Knudson DL, Tesh RB, Main AJ, St George TD, Digoutte JP. Characterization of the Palyam serogroup viruses (Reoviridae: Orbivirus). Intervirology. 1984;22:41–9.
Anthony SJ, Maan S, Maan N, Kgosana L, Bachanek-Bankowska K, Batten C, Darpel KE, Sutton G, Attoui H, Mertens PP. Genetic and phylogenetic analysis of the outer-coat proteins VP2 and VP5 of epizootic haemorrhagic disease virus (EHDV): comparison of genetic and serological data to characterise the EHDV serogroup. Virus Res. 2009;145:200–10.
Tsutsui T, Yamamoto T, Hayama Y, Akiba Y, Nishiguchi A, Kobayashi S, Yamakawa M. Duration of maternally derived antibodies against Akabane virus in calves: survival analysis. J Vet Med Sci. 2009;71:913–8.
Elbers AR, Stockhofe-Zurwieden N, van der Poel WH. Schmallenberg virus antibody persistence in adult cattle after natural infection and decay of maternal antibodies in calves. BMC Vet Res. 2014;10:103.
Ministry of Agriculture, Forestry and Fisheries (MAFF). Statics on animal hygiene 2014. Tokyo: MAFF; 2015.
National Institute of Animal Health. Serological surveillance for arbovirus infections with sentinel cattle. 2015. http://www.naro.affrc.go.jp/niah/arbo/index.html. Accessed 22 Jun 2015 (in Japanese).
Hsieh YC, Chen SH, Chou CC, Ting LJ, Itakura C, Wang FI. Bovine ephemeral fever in Taiwan (2001–2002). J Vet Med Sci. 2005;67:411–6.
Zheng F, Qiu C. Phylogenetic relationships of the glycoprotein gene of bovine ephemeral fever virus isolated from mainland China, Taiwan, Japan, Turkey, Israel and Australia. Virol J. 2012;9:268.
Li Z, Zheng F, Gao S, Wang S, Wang J, Liu Z, Du J, Yin H. Large-scale serological survey of bovine ephemeral fever in China. Vet Microbiol. 2015;176:155–60.
Bak UB, Cheong CK, Choi HI, Lee CW, Oh HS. An outbreak of Ibaraki disease in Korea. Korean J Vet Res. 1983;23:81–9.
Liao YK, Inaba Y, Li NJ, Chain CY, Lee SL, Liou PP. Epidemiology of bovine ephemeral fever virus infection in Taiwan. Microbiol Res. 1998;153:289–95.
Yanase T, Aizawa M, Kato T, Yamakawa M, Shirafuji H, Tsuda T. Genetic characterization of Aino and Peaton virus field isolates reveals a genetic reassortment between these viruses in nature. Virus Res. 2010;153:1–7.
Hoffmann B, Scheuch M, Höper D, Jungblut R, Holsteg M, Schirrmeier H, Eschbaumer M, Goller KV, Wernike K, Fischer M, Breithaupt A, Mettenleiter TC, Beer M. Novel orthobunyavirus in Cattle, Europe, 2011. Emerg Infect Dis. 2012;18:469–72.
Goller KV, Höper D, Schirrmeier H, Mettenleiter TC, Beer M. Schmallenberg virus as possible ancestor of Shamonda virus. Emerg Infect Dis. 2012;18:1644–6.
Hubálek Z. Mosquito-borne viruses in Europe: vector-borne diseases and climate change. Parasitol Res. 2008;103 Suppl 1:29–43.
Briese T, Bird B, Kapoor V, Nichol ST, Lipkin WI. Batai and Ngari viruses: M segment reassortment and association with severe febrile disease outbreaks in East Africa. J Virol. 2006;80:5627–30.
Liu H, Shao XQ, Hu B, Zhao JJ, Zhang L, Zhang HL, Bai X, Zhang RX, Niu DY, Sun YG, Yan XJ. Isolation and complete nucleotide sequence of a Batai virus strain in Inner Mongolia, China. Virol J. 2014;11:138.
Cybinski DH, St George TD. Preliminary characterization of D’Aguilar virus and three Palyam group viruses new to Australia. Aust J Biol Sci. 1982;35:343–51.
Littlejohns IR, Burton RW, Sharp JM. Bluetongue and related viruses in New South Wales: isolations from, and serological tests on samples from sentinel cattle. Aust J Biol Sci. 1988;41:579–87.
Hoshino C. Notes on biting midges collected by light traps at a cowshed in Ishigaki-jima, Ryukyu Islands. Med Entomol Zool. 1985;36:55–8 (in Japanese with English summary).
Purse BV, Carpenter S, Venter GJ, Bellis G, Mullens BA. Bionomics of temperate and tropical Culicoides midges: knowledge gaps and consequences for transmission of Culicoides-borne viruses. Annu Rev Entomol. 2015;60:373–92.
Acknowledgements
We thank all staff of Yaeyama Livestock Hygiene Service Center and Okinawa Prefectural Animal Health Institute for blood sample collection and data acquisition. We are also grateful to Dr. Yoko Hayama for providing the supporting illustrating figures in this manuscript. This work was financially supported by the Research Project for Improving Food Safety and Animal Health of the Ministry of Agriculture, Forestry and Fisheries of Japan.
Funding
This work was financially supported by the Research Project for Improving Food Safety and Animal Health of the Ministry of Agriculture, Forestry and Fisheries of Japan.
Availability of data and materials
All row data are available in Kyushu Research Station of National Institute of Animal Health upon official request.
Authors’ contributions
TK performed the virus isolation and serological tests in the study and drafted the manuscript; TY participated in the design of the study and the interpretation, and contribute to the writing of the manuscript; MS, YK, KI and KT participated in the data acquisition and contributed to the improvement of the manuscript; HS, SO, KY and MY participated in the interpretation and contributed to the improvement of the manuscript; TT participate in the design and coordination of the study and contributed to the improvement of the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent to publish
Not applicable.
Ethics approval and consent to participate
This study was performed under guidelines provided by National Institute of Animal Health, NARO. This study did not include animal experiments.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1:
Primers used for RT-PCR and sequencing in this study. (XLSX 12 kb)
Additional file 2:
Seropositive rate to arboviruses in the Yaeyama Islands from 1995 to 2014. (XLSX 11 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kato, T., Yanase, T., Suzuki, M. et al. Monitoring for bovine arboviruses in the most southwestern islands in Japan between 1994 and 2014. BMC Vet Res 12, 125 (2016). https://doi.org/10.1186/s12917-016-0747-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-016-0747-z