Abstract
Background
Bovine tuberculosis has led to serious economic losses for Sika Deer producers in China. Strategies for controlling the spread of Mycobacterium bovis are often hampered by a lack of epidemiological data. Specifically, tracing infections requires the ability to trace back infections, which, in turn, requires the ability to determine isolates with a common source. This study was planned to assess the discriminatory power of each mycobacterial interspersed repetitive unit (MIRU)-variable number tandem repeats (VNTR) locus and evaluate the most appropriate combination of MIRU-VNTR loci for molecular epidemiological studies on Sika Deer in China.
Results
The discriminatory power of MIRU-VNTR typing based on 22 known loci (12 MIRUs, 2 ETRs, 4 QUBs, and 4 Mtubs) were assessed in 96 Mycobacterium bovis strains collected sequentially from Sika Deer at a slaughterhouse in northeastern China. We defined four loci (MIRU4, ETRA, QUB11b, and Mtub4) as highly discriminative, eight loci (MIRU2, MIRU23, MIRU27, MIRU31, MIRU39, MIRU40, QUB26, and Mtub21) as moderately discriminative, and three loci (MIRU16, Mtub30, and Mtub34) as poorly discriminative. The final locus showed no polymorphism between strains. MIRU-VNTR typing as a whole was highly discriminative, with an overall allelic diversity of 0.897. Of the loci tested, the four highly discriminative loci and eight moderately discriminative loci proved to be most appropriate for first line typing of M. bovis from Sika Deer, with the same resolving ability as all 22 loci (H = 0.897).
Conclusions
MIRU-VNTR typing is quick and effective for typing bovine tuberculosis isolates from Sika Deer in China.
Similar content being viewed by others
Background
Sika Deer tuberculosis is caused by the Mycobacterium tuberculosis complex (MTBC); most commonly, M. bovis. This disease has led to significant economic losses for deer producers in China. However, previous efforts in controlling MTBC have been hampered by a lack of data, as genotyping MTBC members is an important tool in epidemiological analysis for studying the spread of MTBC [1]. In 2001, VNTR genotyping method using MIRU units was developed [2] and used to study the molecular epidemiology of Mycobacterium tuberculosis [3]. MIRU-VNTR has high efficiency and reproducibility, because MIRU-VNTR typing results are easier to compare between different laboratories than restriction fragment length polymorphisms (RFLP), which were the previous “gold standard” for MTBC typing [4]. Another advantage of MIRU-VNTR is that it does not require large quantities of template DNA for typing [5].
In addition to M. tuberculosis, this technique has also been proven in M. bovis in different countries and epidemiologic scenarios [1,6-11]. Earlier studies frequently used the loci designated as mycobacterial interspersed repeat units (MIRU) [2,12] and exact tandem repeats (ETR) [13]. To obtain better resolution, other VTNR loci were investigated [14]. Recently, novel M. tuberculosis MIRU-VNTR loci groups, such as the Queen’s University Belfast (QUB) and Mtub groups [15], have been identified. Various combinations of MIRU-VNTR loci have been reported to give better differentiation of M. tuberculosis strains [16-19]. However, no specific set of loci have been agreed upon as a standard, and the allelic diversity of loci can vary from country to country and between M. tuberculosis complex species, requiring localized selection of suitable loci.
This study was therefore developed to determine the MIRU-VNTR loci most appropriate to use for typing MTBC organisms isolated from Sika deer in China.
Results
Analysis of individual MIRU-VNTR locus
MIRU-VNTR typing results of M.bovis Isolated in this study was described in Additional file 1. The copy numbers and allelic diversity (h) in MIRU-VNTR loci were recorded (Table 1). The diversity analysis showed that the discriminatory power of individual loci differed greatly, with h ranging from 0.000 to 0.739. The ETRA, MIRU4, QUB11b, and Mtub4 loci had high discriminatory power (h > 0.6). The MIRU2, MIRU23, MIRU27, MIRU31, MIRU39, MIRU40, QUB26, and Mtub21 loci were moderately discriminatory (0.3 ≤ h ≤ 0.6). MIRU16, Mtub30, and Mtub34 were poorly discriminatory (h < 0.3). Seven loci displayed no allelic diversity.
Evaluation of MIRU-VNTR loci combinations
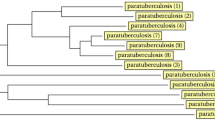
Based on all 15 MIRU-VNTR loci with non-zero allelic diversity, 12 different types (H = 0.897) were identified; of these, 1 was unique and 11 were clustered. Clusters contained 1 (n = 1) to 17 (n = 1) identical strains (Table 2), and the 96 clinical isolates were classified into two groups (Figure 1). Group 1, which included 7 genotypes, was more complex. This group included isolates from four different regions—ShuangYang, LiaoYuan, TongHua, SongYuan and SiPing—strongly suggesting interregional transmission.
The discrimination ability was compared with different combinations of loci (Table 3). The four highly discriminatory loci and the eight moderately discriminatory loci, also identified 12 genotypes (H = 0.897). The MIRU loci with moderate discriminatory power (MIRU2, MIRU23, MIRU27, MIRU31, MIRU39, and MIRU40) resolved the 96 isolates into 10 types (H = 0.877). If the MIRU4 and MIRU16 loci were also used, 11 types (H = 0.893) were resolved. With the Mtub4, Mtub21, Mtub30, and Mtub34 loci, 8 types (H = 0.836) were resolved. Based on these results, we consider ETRA, MIRU4, QUB11b, and Mtub4 to be robust enough for initial molecular epidemiological studies when rapid results are of paramount importance.
Discussion
M. bovis is the primary pathogen for tuberculosis in Sika Deer. Because it also infects humans, molecular epidemiologic studies on M. bovis are of major importance in tuberculosis prevention. Genotyping of MTBC isolates is a useful tool, not only for regional investigations, such as tracebacks, but to allow comparisons of tuberculosis isolates worldwide [20]. Different typing methods published for M. bovis include IS6110, RFLP, VNTR, and spoligotyping [21]. While IS6110 has been the most common, the discriminatory power of IS6110 is generally low for M. bovis typing [22,23]. Because the resolution provided by MIRU-VNTR is adequate for most situations and this method is relatively cheap and rapid, it is quickly becoming the standard as the initial step in M. bovis epidemiology [14].
The individual discriminatory power of 22 MIRU-VNTR loci were analyzed. The QUB11b locus produced the highest individual discriminatory index (h = 0.739), followed by Mtub4, ETRA, and MIRU4. This correlates well with previous studies [14,15].
Mtub loci exhibit good application potential; however, the discriminatory power of Mtub4, Mtub21, Mtub30, and Mtub34 differed greatly, with h ranging from 0.108 to 0.613. These loci exhibited a high enough discrimination ability to be useful in early research [24,25], and may prove useful in future M. bovis typing studies. The ETRC, MIRU10, MIRU20, MIRU24, MIRU26, QUB1895, and QUB3336 loci showed no allelic diversity in this test. MIRU10, MIRU20, MIRU26 and QUB3336 had similar results in studies on M. bovis isolated from Xinjiang region [16]. The published data on M. tuberculosis from Ghana [24] and other locations [6,13,14] showed higher allelic diversities than our study, which suggests that not all MIRU-VNTR loci are informative for M. bovis strains form Sika Deer in China.
The allelic diversity of MIRU-VNTR loci are higher in our study than previous studies in China [26,27], Published data on M. tuberculosis from France [28], South Africa [29], and the United States [30] found that the discriminatory power of MIRU-VNTR loci in Sika Deer is higher than in humans. Although these MIRU-VNTR loci showed enough discrimination ability for genotyping current M. bovis isolates, additional studies with new MIRU-VNTRs are needed to ensure accurate genotyping and epidemiological resolution for field studies.
The discriminatory power of MIRU-VNTR loci may vary between countries, geographical regions, and epidemiological scenarios. Analysis of 15 VNTR loci showed the expected results: when compared with previous studies the test had a relatively high Hunter–Gaston discriminatory index (HGDI) and low clustering rates [31-33]. The subset of 15 loci provides increased resolution compared to the original 12 MIRU loci.
The discriminatory power of MIRU-VNTR loci also varies with high allelic diversity. The ETRA, MIRU4, QUB11b, and Mtub4 loci provide the greatest resolution (H = 0.897) for individual loci, while the MIRUs alone (MIRU2, 23, 31, 39, 40, 27, 16, and 4) provided less resolution (H = 0.893). The moderately discriminatory MIRUs (MIRU2, MIRU23, MIRU31, MIRU39, MIRU40, MIRU27) had less resolution alone (H = 0.877) than when combined with the other moderately loci (QUB26 and Mtub21), which had the greatest amount (H = 0.897). Therefore, the discriminatory power of VNTR loci appears greatly improved by applying novel combinations of VNTR loci.
The 15 VNTR loci methodology is a highly discriminatory method for first line typing of M. bovis isolated from Sika deer and should be considered as a replacement for the original 12 MIRU loci method. However, for in-depth studies, we recommend using ETRA, MIRU4, QUB11b, and Mtub4 loci as a first set to obtain initial molecular epidemiology data quickly before proceeding on to the full set of loci.
Conclusions
The results of this study will be used in a molecular epidemiology study to identify risk factors for recent transmission of Sika Deer tuberculosis in China. VNTR studies will also help in tracing infection sources during Sika deer tuberculosis outbreaks and help control transmission between Sika Deer and humans. Finally, this will improve Chinese Sika Deer cultivation by reducing losses due to MTBC infections.
Methods
Bacterial strains
A total of 96 M. bovis strains were used in this study. These strains were isolated from samples collected at the slaughterhouse for Sika Deer in China covering five different regions: Shuang Yang, Liao Yuan, Tong Hua, Si Ping, and Song Yuan between 2008 and 2012. All 96 strains were characterized by real-time PCR [34].
Ethical considerations
This research was performed according to the international, national and institutional rules regarding animal experimentation. The research was approved by the Institutional Animal Care and Use Committee of Jilin Agricultural University (reference number:2008627).
Preparetion of chromosomal DNA
DNA was extracted with the method followed. Cells were suspended in 400 μL TE buffer (10 mM Tris-CI and 1 mM EDTA at pH 8.0) and inactivated at 80°C for 30 min followed by centrifugation at 5000 × g. The supernatant was discarded and the pellet resuspended in 400 μL TE buffer and 45 μL 10% SDS. This was heated in a 37°C water bath for 2 h after incubation with 3 μL of 20 μg/mL proteinase K. The suspension was re-centrifuged and the supernatant extracted using an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1 [v/v]). The resulting supernatant was diluted with a double volume of anhydrous ethanol for 10 min and precipitated at −20°C after centrifugation at 12000 × g for 20 min. The precipitate was washed with 70% ethanol twice, and the ethanol evaporated at room temperature for 20 min. The final pellet was dissolved in 30 μL sterile deionized water and stored at −20°C until use.
PCR amplification
The primers and designations used in this study for each MIRU-VNTR locus are described by Supply et al. [14]. The primer sequences and PCR product sequencing in this study was described in Additional file 2. PCR reactions were performed using the method described in Table 4. The reaction mixture for all loci contained 2 μL M. bovis DNA, 0.4 μΜ of each primer pair, 25 μL EmeraldAmp MAX PCR Master Mix (Takara Biomedical Technology, Beijing, China) and 22.6 μL ddH2O.
Allelic diversity and VNTR typing
The individual allelic diversity (h) of each VNTR locus was calculated using the following equation: h = 1-∑xi 2[n/(n − 1)], where n is the number of isolates and xi the frequency of the ith allele at the locus [35]. We considered h > 0.6 as highly discriminative, 0.3 ≤ h ≤ 0.6 as moderately discriminative, and h < 0.3 as poorly discriminative [36]. The HGDI was calculated as previously described [37]. The HGDI varies between 0.00 and 1.00 and represents the discriminatory power for VNTR loci in combination. Clustering analyses were performed using the online tool at http://www.MIRU-VNTRplus.org.
Availability of supporting data
Dataset doi:10.6070/H4HX19PG
Abbreviations
- MIRU-VNTR:
-
Mycobacterial Interspersed Repetitive Units - variable number of tandem repeat
- PCR:
-
Polymerase chain reaction
- HGDI:
-
Hunter–Gaston discriminatory index
References
Roring S, Scott AN, Glyn Hewinson R, Neill SD, Skuce RA. Evaluation of variable number tandem repeat (VNTR) loci in molecular typing of Mycobacterium bovis isolates from Ireland. Vet Microbiol. 2004;101:65–73.
Supply P, Lesjean S, Savine E, Kremer K, van Soolingen D, Locht C. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J Clin Microbiol. 2001;39:3563–71.
Neill SD, Skuce RA, Pollock JM. Tuberculosis-new light from an old window. J Appl Microbiol. 2005;98:1261–9.
Yokoyama E, Kishida K, Uchimura M, Ichinohe S. Improved differentiation of Mycobacterium tuberculosis strains, including many Beijing genotype strains, using a new combination of variable number of tandem repeats loci. Infect Genet Evol. 2007;7:499–508.
Hilty M, Diguimbaye C, Schelling E, Baggi F, Tanner M, Zinsstag J. Evaluation of the discriminatory power of variable number tandem repeat (VNTR) typing of Mycobacterium bovis strains. Vet Microbiol. 2005;109:217–22.
Hang’ombe MB, Munyeme M, Nakajima C, Fukushima Y, Suzuki H, Matandiko W, et al. Mycobacterium bovis infection at the interface between domestic and wild animals in Zambia. BMC Vet Res. 2012;8:221.
Roring S, Scott A, Brittain D, Walker I, Hewinson G, Neill S, et al. Development of variable-number tandem repeat typing of Mycobacterium bovis: comparison of results with those obtained by using existing exact tandem repeats and spoligotyping. J Clin Microbiol. 2002;40:2126–33.
Skuce RA, McDowell SW, Mallon TR, Luke B, Breadon EL, Lagan PL, et al. Discrimination of isolates of Mycobacterium bovis in Northern Ireland on the basis of variable numbers of tandem repeats (VNTRs). Vet Res. 2005;157:501–4.
Allix C, Walravens K, Saegerman C, Godfroid J, Supply P, FauvilleDufaux M. Evaluation of the epidemiological relevance of variable-number tandem-repeat genotyping of Mycobacterium bovis and comparison of the method with IS6110 restriction fragment length polymorphism analysis and spoligotyping. J Clin Microbiol. 2006;44:1951–62.
Michel AL, Hlokwe TM, Coetzee ML, Mare L, Connoway L, Rutten VPMG, et al. High Mycobacterium bovis genetic diversity in a low prevalence setting. Vet Microbiol. 2008;126:151–9.
Romero B, Aranaz A, Sandoval N, Alvarez J, de Juan L, Bezos J, et al. Persistence and molecular evolution of Mycobacterium bovis population from cattle and wildlife in Doňana National Park revealed by genotype variation. Vet Microbiol. 2008;132:87–95.
Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol. 2000;36:762–71.
Duarte EL, Domingos M, Amado A, Cunha MV, Botelho A. MIRU-VNTR typing adds discriminatory value to groups of Mycobacterium bovis and Mycobacterium caprae strains defined by spoligotyping. Vet Microbiol. 2010;143:299–306.
Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44:4498–510.
Le Fleche P, Fabre M, Denoeud FJ, Koec LK, Vergnaud G. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 2002;2:37.
Sun ZG, Cao R, Tian M, Zhang XX, Zhang XY, Li Y, et al. Evaluation of Spoligotyping and MIRU-VNTR for Mycobacterium bovis in Xinjiang, China. Res Vet Sci. 2012;92:236–9.
Kremer K, Au BK, Yip PC, Skuce R, Supply P, Kam KM, et al. Use of variable-number-tandem-repeat typing to differentiate Mycobacterium tuberculosis Beijing family isolates from Hong Kong and comparison with IS6110 restriction fragment length polymorphism typing and spoligotyping. J Clin Microbiol. 2005;43:314–20.
Smittipat N, Billamas P, Palittapongarnpim M, Thong-On A, Temu MM, Thanakijcharoen P, et al. Polymorphism of variable number tandem repeats at multiple loci in Mycobacterium tuberculosis. J Clin Microbiol. 2005;43:5034–43.
Surikova OV, Voitech DS, Kuzmicheva G, Tatkov SI, Mokrousov IV, Narvskaya OV, et al. Efficient differentiation of Mycobacterium tuberculosis strains of the W-Beijing family from Russia using highly polymorphic VNTR loci. Eur J Epidemiol. 2005;20:963–74.
Christianson S, Wolfe J, Orr P, Karlowsky J, Levett PN, Horsman GB, et al. Evaluation of 24 locus MIRU-VNTR genotyping of Mycobacterium tuberculosis isolates in Canada. Tuberculosis. 2010;90:31–8.
Navarro Y, Herranz M, Romero B, Bouza E, Domínguez L, De Juan L, et al. High-throughput multiplex MIRU-VNTR typing of Mycobacterium bovis. Res Vet Sci. 2014;96:422–225.
Kremer K, van Soolingen D, Frothingham R, Haas WH, Hermans PWM, Martin C, et al. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol. 1999;37:2607–18.
Serraino A, Marchetti G, Sanguinetti V, Rossi MC, Zanoni RG, Catozzi L, et al. Monitoring of transmission of tuberculosis between wild boars and cattle: genotypical analysis of strains by molecular epidemiology techniques. J Clin Microbiol. 1999;37:2766–71.
Asante-Poku A, Nyaho MS, Borrell S, Comas I, Gagneux S, Yeboah-Manu D. Evaluation of customised lineage-specific sets of MIRU-VNTR Loci for genotyping Mycobacterium tuberculosis complex isolates in Ghana. PLoS One. 2014;9:e92675.
Ali A, Hasan Z, Jafri S, Inayat R, Hasan R. Mycobacterium tuberculosis Central Asian Strain(CAS) lineage strains in Pakistan reveal lower diversity of MIRU loci than other strains. Int J Mycobacteriology. 2014;3:108–16.
Cao R, Zhang XY, Sun ZG, Hu XD, Tian M, Li Y, et al. The genotyping of Mycobacterium bovis isolated from Xinjiang region with MIRU-VNTR. Chin J Prev Vet Med. 2010;32:524–8.
Yanfen D, Lin J, Liu S, Wang C, Liu H, Si W, et al. Application of Spoligotyping and VNTR-MIRU in genotype studies of Mycobacterium bovis in China. Chin J Prev Vet Med. 2009;31:114–6.
Mazars E, Lesjean S, Banuls AL, Gilbert M, Vincent V, Gicquel B, et al. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc Natl Acad Sci, USA. 2001;98(4):1901–6.
Savine E, Warren RM, van der Spuy GD, Beyers N, van Helden PD, Locht C, et al. Stability of variable number tandem repeats of mycobacterial interspersed repetitive units from 12 loci in serial isolates of Mycobacterium tuberculosis. J Clin Microbiol. 2002;40:4561–6.
Cowan LS, Mosher L, Diem L, Massey JP, Crawford JT. Variable-number-tandem repeat typing of mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J Clin Microbiol. 2002;40:1592–602.
Oelemann MC, Diel R, Vatin V, Haas W, Rüsch-Gerdes S, Locht C, et al. Assessment of an optimized mycobacterial interspersed repetitive- unit-variable-number tandem-repeat typing system combined with spoligotyping for population-based molecular epidemiology studies of tuberculosis. J Clin Microbiol. 2007;45:691–7.
Allix-Béguec C, Fauville Dufaux M, Supply P. Three-year population- based evaluation of standardized mycobacterial interspersed repetitive-unit- variable-number tandem-repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2008;46:1398–406.
Maes M, Kremer K, van Soolingen D, Takiff H, De W, De Waard JH. 24-locus MIRU-VNTR genotyping is a useful tool to study the molecular epidemiology of tuberculosis among Warao Amerindians in Venezuela. Tuberculosis (Edinb). 2008;88:490–4.
Wang CY, Wang ZG, Liu JH, Song ZY, Zhou L, Gao HW, et al. Detection of Mycobacterium bovis with TaqMan real-time PCR. Chin J Prev Vet Med. 2010;33:133–6.
Selander RK, Caugant DA, Ochman H, Musser JM, Gilmour MN, Whittam TS. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–84.
Sola C, Filliol I, Legrand E, Lesjean S, Locht C, Supply P, et al. Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect Genet Evol. 2003;3:125–33.
Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–6.
Acknowledgements
The authors would like thank Jinhua Liu, Zhanyun Song and Liang Zhou (Technology Center of Jilin Entry-Exit Inspection and Quadrant Bureau) for their valuable technical support. We would also like to thank the employees of the slaughter facility for their support and providing valuable insight about M. bovis infections in deer. This work was supported by Natural Science for Youth Foundation of China (No.31200241); International Science and Technology Cooperation Project of China (2011DFA32900); The Development Plan Project of Science and Technology of Jilin Province (20100718).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LY: conceived of the study, participated in the final study design, performed statistical analyses, and drafted the manuscript. QW, QM: responsible for sample collection, and M. bovis isolation. HW: performed PCR amplification and analysis of the amplification products. CW: performed the identification of M. bovis, coordinated research, and critically reviewed the manuscript. All authors read and approved the final manuscript.
Additional files
Additional file 1:
MIRU-VNTR type of M. bovis Isolated in this study. Additional file 1 including the MIRU-VNTR typing results of M.bovis Isolated which was used in this study.
Additional file 2:
PCR primer sequences and PCR product sequencing in this study. Additional file 2 including PCR primer sequences of MIRU-VNTR loci, and the result of PCR product sequencing in this study.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Yang, L., Wang, C., Wang, H. et al. Evaluation of MIRU-VNTR for typing of Mycobacterium bovis isolated from Sika deer in Northeast China. BMC Vet Res 11, 93 (2015). https://doi.org/10.1186/s12917-015-0402-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-015-0402-0