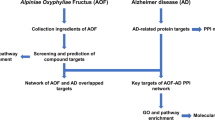
Abstract
Background
Shanzhuyu (the dried mature sarcocarp of Cornus officinalis Sieb. et Zucc., DMSCO) is a Chinese herb that can be used for the treatment of Alzheimer’s disease (AD), but its mechanism remains unknown. The present study aimed to investigate the active ingredients and effective mechanisms of DMSCO for the treatment of AD based on a network pharmacology approach.
Methods
The active components of DMSCO were collected from the TCMSP and ETCM databases and the target proteins of these compounds were predicted using TCMSP, SwissTargetPrediction and the STITCH database. The AD-related target proteins were identified from the OMIM, DisGeNet, GEO and GeneCards databases. The network interaction model of the compound-target-disease was established and was used to obtain the key targets of DMSCO on AD through network topology analysis. Subsequently, gene enrichment in Gene Ontology (GO) and KEGG pathways were conducted using the David 6.8 online tool.
Results
A total of 30 DMSCO effective compounds and 209 effective drug targets were obtained. A total of 172 AD-related genes and 37 shared targets of DMSCO and AD were identified. A total of 43 key targets for the treatment of AD were obtained from the topological analysis of the DMSCO-AD target network. These key targets were involved in a variety of biological processes, including amyloid deposition, apoptosis, autophagy, inflammatory response and oxidative stress and pathways, such as the PI3K-AKT, MAPK and TNF pathways. Three key compounds, namely ursolic acid, anethole and β-sitosterol were obtained from the analysis of the key targets.
Conclusions
Ursolic acid, anethole and β-sitosterol may be the main active components of DMSCO in the treatment of AD. DMSCO can treat AD by regulating amyloid deposition, apoptosis, autophagy, inflammatory response and oxidative stress via the PI3K-AKT, MAPK and other signaling pathways.
Similar content being viewed by others
Background
Alzheimer’s disease (AD) is a neurodegenerative disease with clinical manifestations of progressive memory loss, cognitive impairment and personality changes. Currently, approximately 50 million people worldwide have dementia and more than 35% of the population over the age of 80 has this disease [1]. Approximately 5 million new cases of dementia are reported each year and the population is expected to grow to 118 million by 2050 [2]. The pathological feature of AD is loss of neurons, formation of senile plaques and neurofibrillary tangles involving the amyloid β (Aβ) and the tau protein, as well as oxidative stress and inflammation [3]. Studies that target Aβ and the tau protein are under investigation. However, they have not yielded satisfactory results [4, 5]. Current drugs, such as Donepezil and Memantine, are not effective to meet clinical needs. Therefore, it is necessary to study the treatment of AD from the perspective of multi-target therapy.
Traditional Chinese medicine (TCM) has been used to treat AD related diseases for thousands of years and significant clinical evidences have been accumulated. Shanzhuyu (the dried mature sarcocarp of Cornus officinalis Sieb. et Zucc., DMSCO) is a commonly used Chinese herb for the treatment of AD, which has the ability to tonify the liver and kidney. DMSCO is an important component of the Chinese herbal formula for AD-related disease treatment, such as Di-Huang-Yin-Zi, Liu-Wei-Di-Huang pills [6, 7]. DMSCO can inhibit Aβ1–42-induced apoptosis and inflammation, tau hyperphosphorylation and aggregation [8,9,10]. It can also inhibit cholinesterase and beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) [11]. The active ingredients of DMSCO and their associated effective mechanism with regard to the treatment of AD require further investigation.
Network pharmacology is a new subject that has emerged recently. Network pharmacology can aid the exploration of the direct targets of the active ingredients of the Chinese herbs, define their functions in the context of molecular network [12]. In the present study, the active ingredients of DMSCO and its associated effective mechanism in the treatment of AD were systematically analyzed by establishing a “compound-target-pathway” network. The results indicated that DMSCO contained multiple active ingredients that could treat AD and that its mechanism was associated with the regulation of amyloid deposition, apoptosis, autophagy, inflammatory response and oxidative stress via the PI3K-AKT, MAPK and other signaling pathways.
Methods
Identification and screening of chemical ingredients of DMSCO
At present, several databases are available with regard to TCM ingredients. Traditional Chinese Medicine System Pharmacology (TCMSP, http://lsp.nwu.edu.cn/browse.php) is a comprehensive TCM platform containing 499 herbs and more than 2.9 × 104 chemical components, providing comprehensive information on Chinese herbal ingredients, including chemical structure, oral bioavailability (OB), intestinal epithelial permeability, half-life, drug similarity and drug targets [13]. The Encyclopedia of Traditional Chinese Medicine (ETCM, http://www.nrc.ac.cn:9090/ETCM/) is also a commonly used TCM database, containing comprehensive information on Chinese herbs, TCM formulations and their ingredients [14]. TCMSP uses authoritative algorithms to predict the pharmacokinetic properties of compounds, such as absorption, distribution, metabolism and drug excretion (ADME) in order to provide comprehensive scores. In the present study, the chemical components of DMSCO were collected through literature research and via the TCMSP and ETCM databases. The ADME parameters OB ≥ 30% and Drug-likeness (DL) ≥ 0.18 were used to screen the potential active ingredients from the TCMSP database [15]. In addition, a Drug-likeness Weight ≥ 0.49 was used to retrieve the active ingredients of DMSCO from the ETCM database [14].
Investigation and prediction of compound-related targets
Based on the chemical similarity and pharmacophore model, the present study used TCMSP, STITCH (http://stitch.embl.de/) and SwissTargetPrediction (http://www.swisstargetprediction.ch/) to retrieve and predict the related targets of compounds in DMSCO. STITCH is a database containing various structural and predictive interactions of compounds that support target prediction based on structural similarity [16]. In the present study, a confidence score ≥ 0.7 was used as the screening criterion. SwissTargetPrediction is a database used for predicting compound targets based on 2D and 3D structures of known compounds [17]. The probability value ≥ 0.5 served as the target screening standard in the present study.
Identification of AD-related targets
AD-related genes were screened using online mendelian inheritance in man (OMIM, https://omim.org/) [18], DisGeNET (http://www.disgenet.org/) [19], GeneCards (https://www.genecards.org/) [20], and Gene Expression Omnibus (GEO) databases (http://www.ncbi.nlm.nih.gov/geo). DisGeNET is a comprehensive platform developed to solve problems regarding the genetic basis of human disease. The platform was searched using the keyword “Alzheimer’s Disease” and disease related genes were identified based on a score ≥ 0.4. AD-related genes with GeneCards Inferred Functionality Score (GIFtS) ≥ 52 were selected from the GeneCards database. We further used the GEO2R online tool (http://www.ncbi.nlm.nih.gov/geo/geo2r/) to select AD-related genes from the GSE36980 dataset (15 AD patients and 33 healthy subjects). The criteria for screening differentially expressed genes were P ≤ 0.05, fold change (FC) ≥ 1.5. The target ID was converted to the gene symbol by retrieving either the UniProtKB (https://www.uniprot.org/) or the STRING (https://string-db.org/) databases.
Network construction and topological analysis
PPI (protein-protein network) was constructed via the STRING database and the targets with a confidence score ≥ 0.7 were selected. The following network and topology analyses were performed using the Cytoscape 3.6.0 software: 1. The compound-target network of DMSCO; 2. The AD-related target network; 3. The DMSCO potential target-AD target interaction network; 4. The networks of shared targets between DMSCO and AD targets. Degree centrality (DC), betweenness centrality (BC) and closeness centrality (CC) are the most common topology parameters used to evaluate the central properties of nodes in a network. In the DMSCO potential target-AD target interaction network, the parameter settings of DC ≥ 3 × median DC, BC ≥ median BC and CC ≥ median CC were used to screen the key targets of DMSCO.
GO and KEGG pathway enrichment analysis
DAVID 6. 8 (https://david.ncifcrf.gov/) is an online biological information repository and analysis tool for extracting biological information regarding gene functional annotation and pathways enrichment [21]. Drug targets and key targets of DMSCO acting on AD were imported into the DAVID 6.8 database and the species were defined as “Homo sapiens”, whereas the target genes were identified as “official gene symbol”. Gene Ontology (GO) and KEGG pathway analysis were performed.
Results
DMSCO compound-target network
According to the search results of TCMSP, DMSCO exhibited a total of 226 chemical components, including mainly iridoids, pentacyclic triterpenoid acids and their corresponding esters, polysaccharides and tannins. A total of 20 compounds were screened by OB ≥ 30% and DL ≥ 0.18. A total of 55 types of compounds were retrieved from ETCM and 20 compounds were filtered by Drug-likeness Weight ≥ 0.49. A thorough literature search using Chinese and international references resulted in the identification of 7 important active ingredients that fulfilled the aforementioned standards. Therefore, 47 compounds were obtained in total. TCMSP, STITCH and SwissTargetPrediction were used to collect and predict targets and 17 compounds were identified without a corresponding target. Therefore, 30 DMSCO effective compounds were obtained, corresponding to 209 targets.
The main information of the compounds with the number of targets > 10, namely the main active components of DMSCO are listed in Table 1. Among these major active ingredients, ursolic acid exhibited a total of 72 targets, whereas β-sitosterol exhibited 39 targets, stigmasterol 38 targets, retinol 33 targets and tetrahydroalstonine 30 targets. Safrole, methyleugenol, oleanolic acid, elemicin, anethole, sitosterol and poriferast-5-en-3beta-ol (clionasterol) demonstrated 27, 26, 25, 22, 21, 19 and 12 targets, respectively. The corresponding association between compounds and targets is shown in Fig. 1a.
Potential targets of DMSCO compounds. a DMSCO Compound-target network was generated by Cytoscape 3.6.0 software. It consists of 239 nodes and 435 edges. Pink diamond nodes represent compounds in DMSCO, while lavender round nodes represent potential targets of DMSCO. b GO enrichment analysis for potential targets of DMSCO (Count number > 15). c KEGG pathway enrichment analysis for potential targets of DMSCO (Count number ≥ 12), q value refers to -log10(P value). d DMSCO compound-target-pathway (Count number > 20) network were generated by Cytoscape 3.6.0 software. Lavender eclipse nodes stand for potential targets of DMSCO, while yellow round rectangle nodes stand for active compounds in DMSCO and pink diamond nodes stand for the pathways whose count number > 20
In order to clarify the characteristics of the relevant targets of the key active components of DMSCO, GO and KEGG pathway enrichment analyses were performed (Fig. 1b and c). These targets existed in the nucleus, cytoplasm and plasma membrane of cell and were involved in biological processes, such as transcriptional regulation, drug response, signal transduction, cell proliferation and senescence. The molecular function of these genes was involved in binding proteins, enzymes and zinc ions. KEGG enrichment analysis indicated that 127 pathways were affected by the active components of DMSCO (P < 0.05). The top 13 (Count Number ≥ 12) pathways included cancer, hepatitis B, PI3K-AKT, neuroactive ligand-receptor interactions, HTLV-I infection, TNF, influenza A, cAMP, proteoglycans in cancer, MAPK, non-alcoholic fatty liver disease, viral carcinogenesis and microRNAs in cancer signaling pathways. Based on this information, the network association of DMSCO compound-target-pathway was established (Fig. 1d).
AD-target network
A total of 218 AD related genes were screened from OMIM, DisGeNET, GeneCards and GEO databases, among which 172 targets exhibited high interaction in the protein-protein interaction (PPI) graph generated from the STRING database (confidence score ≥ 0.7). The AD disease target network was constructed using Cytoscape 3.6.0, consisting of 172 nodes and 1410 edges and the central properties of each node were evaluated using topology analysis (Fig. 2). The size of nodes was proportional to the degree centrality obtained from topology analysis. The results indicated that TP53, VEGFA, PIK3CA, AKT1, SRC, STAT3, INS, IGF1, HRAS, CTNNB1, IL6, EGFR, APP and PTEN were important target genes of AD (degree > 40).
DMSCO target-AD target network
In order to determine the association between DMSCO potential targets and AD disease related targets, the Venn online tool (https://bioinfogp.cnb.csic.es/tools/venny/index.html) was used to obtain 37 shared targets of DMSCO and AD (Fig. 3a). The interaction network between these genes was established and 35 targets indicated high interaction (confidence score ≥ 0.7) in the PPI diagram generated from the STRING database (Fig. 3b). The network consists of 35 nodes and 189 edges and the size of nodes is proportional to degree centrality obtained from topology analysis. Topology analysis indicated that TP53, VEGFA, AKT1, MMP9, IL6, STAT3, TNF, CASP3, CCND1 and IL1B were the top 10 shared targets from the perspective of degree centrality. Subsequently, we established the interaction network of DMSCO target-AD targets (Fig. 3c), which was composed of 331 nodes and 3226 edges. The node size was proportional to degree centrality obtained from topology analysis.
Shared targets between AD targets and DMSCO targets. a Shared targets between AD targets and DMSCO targets were identified by Venny 2.1. b Network of the shared targets was generated by Cytoscape 3.6.0 software. Pink round nodes represent the shared targets, and the size of nodes is proportional to degree centrality by topology analysis. c DMSCO target-AD targets network was generated by Cytoscape 3.6.0 software. Yellow hexagon nodes stand for DMSCO targets, pink eclipse nodes represent AD-related targets, and light blue round rectangle nodes represent the shared targets of DMSCO and AD. The size of nodes is proportional to degree centrality by topology analysis
Key targets of the DMSCO-AD interaction network
In the DMSCO target-AD target interaction network, 43 key targets and 814 interactions were obtained through central network evaluation (Fig. 4). The information of key targets of degree centrality ≥ 50 was listed in Table 2. GO enrichment analysis indicated that these targets mainly existed in the nucleus, cytoplasm, plasma membrane and other regions of cell and were involved in transcriptional regulation, apoptosis, signal transduction, drug response, cell proliferation, gene expression, protein phosphorylation and other biological processes. They exhibited various molecular functions, such as binding proteins, enzymes, ATP, DNA and transcription factors (Fig. 5a). KEGG pathway enrichment analysis indicated that 119 pathways were affected by the active component of DMSCO (P < 0.05). The top pathways (count number ≥ 15) included cancer, PI3K-AKT, MAPK, TNF, focal adhesion, prolactin, HIF-1, thyroid hormone, FoxO, Rap1 and Ras signaling pathways (Fig. 5b). The distribution of key targets in the PI3K-AKT and MAPK pathways is shown in Fig. 6.
Network of key targets. Key targets were identified by central network evaluation and its network was generated by Cytoscape 3.6.0 software. Pink hexagon nodes stand for DMSCO targets, yellow round rectangle nodes represent AD-related targets, and lavender eclipse nodes represent the shared targets of DMSCO and AD. The size of nodes is proportional to degree centrality by topology analysis
The key targets related to each compound were listed in Table 3. Ursolic acid, anethole and β-sitosterol were related to the largest number of key targets, including 17, 7 and 5 key targets, respectively. These results suggested that ursolic acid, anethole and β-sitosterol were the main compounds in DMSCO that were most closely associated with the treatment of AD.
It is worth noting that the results of enrichment and pathway analysis of drug targets of DMSCO and key targets of the DMSCO-AD interaction network are not completely consistent. By comparison, it was found that the major biological processes of DMSCO drug targets are mRNA transcription regulation, drug response and signal transduction, while the major biological processes of key targets are mRNA transcription regulation, DNA-templated and signal transduction. The cellular components of DMSCO drug targets are mainly plasma membrane, cytosol and cytocomponents, while the cellular components of key targets are mainly nucleus, cytoplasm and cytosol. The molecular functions of DMSCO drug targets are mainly protein binding, enzyme binding and protein homodimerization activity, while the molecular functions of key targets are protein binding, enzyme binding and protein-like binding. These results suggested that drug targets may have a wider range of action than key targets. Key targets are the parts of the DMSCO drug targets that specifically treats AD. In other words, key targets have a more specific treating direction. As for the pathway, key targets mainly focused on PI3K-Akt and MAPK signaling pathway, while the drug targets are mainly enriched in hepatitis B and neuroactive ligand-receptor interaction in addition to these two pathways. These results suggested that in addition to AD, DMSCO may has effects on other diseases, such as hepatitis B, which is worth further study.
Discussion
In the present study, network pharmacology was used to explore the key compounds and potential targets of DMSCO for the treatment of AD, providing a basis for the development of a feasible alternative therapy for AD. The results indicated that ursolic acid, polysitosterol, cornuside, sweroside, morroniside, loganin, garlic acid, stigmasterol, retinol, tetrahydroalstonine, oleanolic acid, anethole, methyleugenol and other compounds were the possible effective components of DMSCO responsible for its anti-AD effect. Ursolic acid, anethole and β-sitosterol were the key compounds in DMSCO that could be used for the treatment of AD.
Ursolic acid inhibits the extracellular signaling regulation kinases, p38 and JNK, leading to the inactivation of NF-κB and to the downregulation of COX-2 and iNOS, which are induced by Aβ1–42 peptides. This in turn leads to the production of anti-inflammatory and neuroprotective effects [22]. β-sitosterol exhibits potent anticholinesterase and antioxidant activities, which can improve memory and learning impairment in APP/PS1 double-transgenic AD mice [23, 24]. Anethole can reduce hypoxia-glucose deprivation-reoxygenation-induced nerve cell death and its mechanism is associated with antioxidant activity, antiexcitatory toxicity and mitochondrial protection [25]. Chainy et al. demonstrated that anethole could block the early and late cellular responses mediated by TNF-α, while also affecting the NF-κB, AP-1, JNK and MAPK pathways and inhibiting the H2O2-induced activation of NF-κB [26].
In addition to these three key compounds, other components also possess anti-AD effects. Loganin exhibits anti-inflammatory and memory improvement effects and can inhibit Aβ1–42-induced microglial activation and inflammatory response by inhibiting TLR4/TRAF6/NF-κB signal transduction [10]. Morroniside plays a neuroprotective role by increasing the activity of PP2A and inhibiting phosphorylation of tau protein at multiple sites including Thr217 [27]. Oleanolic acid inhibits the secretion of Aβ-activated inflammatory cytokines, such as IL-6, TNF-β and IL-1β and alleviates the neuronal apoptosis caused by Aβ, thus improving the cognitive deficits in rats with AD [28].
In the present study, 43 key targets of DMSCO were identified for AD treatment and the top 10 targets were TP53, MAPK1, AKT1, VEGFA, INS, IL6, STAT3, PIK3CA, MAPK8 and MAPK3. TP53, also known as p53, is a transcription factor that can initiate apoptosis. The expression and activity of p53 increase rapidly following hypoxia, DNA damage, oncogene activation, microtubule destruction and oxidative damage conditions [29], while p53-mediated apoptosis is associated with chronic neuronal degeneration. Neuregulin-1 can significantly reduce CoCl2-induced SH-SY5Y cell death by regulating HIF-1α and p53 [30]. It has been proposed that the detection of the Aβ42 peptide, tau, p16 and p53 protein expression levels is a promising method for diagnosing AD and evaluating the aging rate during the development of this disease [31]. VEGFR-2 plays a key role in angiogenesis and is involved in the development of the central nervous system. Aβ1–42 participates in the pathogenesis of AD by inhibiting VEGF-induced endothelial cell migration and VEGFR-2 activity [32]. AD is also associated with the JAK-STAT pathway [33]. Curcumin can regulate the inflammatory response of microglial cells through JAK/STAT/SOCS signaling, thus regulating the neuroinflammatory response [34].
The pathological of AD is closely associated with oxidative stress, the inflammatory response and Aβ deposition. The present study demonstrated that DMSCO regulated AD-related pathways, such as the PI3K/AKT and MAPK pathways. The PI3K-AKT pathway is an important pathway that mediates neuronal survival. It can lead to autophagy disruption and slow down AD progression [35]. The PI3K/AKT/mTOR signals inhibit apoptosis and autophagy, regulate oxidative stress and play a neuroprotective role in SH-SY5Y cells treated with Aβ1–42 [36]. Deuterium-depleted water can inhibit H2O2-induced oxidative stress by upregulating the PI3K/AKT signaling [37]. The MAPK pathway mediates the inflammatory response induced by Aβ1–42 [38]. EGb761 suppresses the neuroinflammatory response induced by Aβ1–42 in virtue of inhibiting the phosphorylation of MAPK [39]. It has also been shown that miR330 inhibits oxidative stress damage in AD mice and alleviates mitochondrial dysfunction by regulating MAPK signal transduction, which reduces the production of Aβ [40].
Conclusions
In summary, the present study demonstrated that ursolic acid, anethole and β-sitosterol may be the main active ingredients of DMSCO in the treatment of AD. DMSCO can treat AD by regulating amyloid deposition, apoptosis, autophagy, inflammatory response and oxidative stress via the PI3K-AKT, MAPK and other signaling pathways. The present study provides a basis for the treatment of AD by DMSCO from the perspective of network pharmacology. We identified the main components, targets and pathways of DMSCO in the treatment of anti-AD, these findings have guiding significance for application of DMSCO, and provide new clues for the future study and development of anti-AD drugs.
Availability of data and materials
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Abbreviations
- AD:
-
Alzheimer’s disease
- Aβ:
-
Amyloid β
- ADME:
-
Absorption, distribution, metabolism and drug excretion
- BACE1:
-
Beta-site amyloid precursor protein cleaving enzyme 1
- BC:
-
Betweenness centrality
- CC:
-
Closeness centrality
- COX-2:
-
Cyclooxygenase-2
- DC:
-
Degree centrality
- DL:
-
Drug-likeness
- DMSCO:
-
The dried mature sarcocarp of Cornus officinalis Sieb. et Zucc.
- ETCM:
-
Encyclopedia of traditional Chinese medicine
- FC:
-
Fold change
- GEO:
-
Gene expression omnibus
- GO:
-
Gene ontology
- iNOS:
-
Inducible nitric oxide synthase
- JNK:
-
c-Jun N-terminal kinase
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- MAPK:
-
Mitogen-activated protein kinase
- NF-κB:
-
Nuclear factor kappa B
- OB:
-
Oral bioavailability
- OMIM:
-
Online Mendelian inheritance in man
- PI3K:
-
Phosphoinositide 3-kinase
- PPI:
-
Protein-protein network
- TCMSP:
-
Traditional Chinese medicine system pharmacology
- TNF-α:
-
Tumor Necrosis Factor α
- VEGFR:
-
Vascular endothelial growth factor receptor
References
Haines JL. Alzheimer disease: perspectives from epidemiology and genetics. J Law Med Ethics. 2018;46(3):694–8.
Alzheimer’s Association. Alzheimer’s disease facts and figures. Alzheimers Dement. 2015;11(3):332–84.
Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148(6):1204–22.
Graham WV, Bonito-Oliva A, Sakmar TP. Update on Alzheimer’s disease therapy and prevention strategies. Annu Rev Med. 2017;68:413–30.
Colin M, Dujardin S, Schraen-Maschke S, Meno-Tetang G, Duyckaerts C, Courade JP, et al. From the prion-like propagation hypothesis to therapeutic strategies of anti-tau immunotherapy. Acta Neuropathol. 2020;139(1):3–25.
Xie F, Xie N, Yu S, Yu N. Research progress of Di-Huang-yin-Zi in the treatment of senile dementia. J Liaoning U Tradit Chin Med. 2020;22(01):106–9 Chinese.
Qing ZQ. Effect of Liu-Wei-Di-Huang pills on delaying Alzheimer’s disease in 50 cases. Chin J Tradit Med Sci Tech. 2007;16(5):373–4 Chinese.
Ma D, Luo Y, Huang R, Zhao Z, Wang Q, Li L, et al. Cornel iridoid glycoside suppresses tau hyperphosphorylation and aggregation in a mouse model of tauopathy through increasing activity of PP2A. Curr Alzheimer Res. 2019;16(14):1316–31.
Chen K, Lu Y, Liu C, Zhang L, Fang Z, Yu G. Morroniside prevents H2O2 or Abeta1-42-induced apoptosis via attenuating JNK and p38 MAPK phosphorylation. Eur J Pharmacol. 2018;834:295–304.
Cui Y, Wang Y, Zhao D, Feng X, Zhang L, Liu C. Loganin prevents BV-2 microglia cells from Abeta1-42 -induced inflammation via regulating TLR4/TRAF6/NF-kappaB axis. Cell Biol Int. 2018;42(12):1632–42.
Bhakta HK, Park CH, Yokozawa T, Tanaka T, Jung HA, Choi JS. Potential anti-cholinesterase and beta-site amyloid precursor protein cleaving enzyme 1 inhibitory activities of cornuside and gallotannins from Cornus officinalis fruits. Arch Pharm Res. 2017;40(7):836–53.
Li S, Zhang B. Traditional Chinese medicine network pharmacology. Theory, methodology and application. Chin J Nat Med. 2013;11(2):110–20.
Ru J, Li P, Wang J, Zhou W, Li B, Huang C, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Aust J Chem. 2014;6:13.
Xu HY, Zhang YQ, Liu ZM, Chen T, Lv CY, Tang SH, et al. ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic Acids Res. 2019;47(D1):D976–82.
Huang C, Yang Y, Chen X, et al. Large-scale cross-species chemogenomic platform proposes a new drug discovery strategy of veterinary drug from herbal medicines. PLoS One. 2017;12(9):e0184880.
Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016;44(D1):D380–4.
Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–64.
Amberger JS, Bocchini CA, Scott AF, Hamosh A. OMIM.Org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019;47(D1):D1038–43.
Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833–9.
Safran M, Dalah I, Alexander J, Rosen N, Iny ST, Shmoish M, et al. GeneCards version 3: the human gene integrator. Database (Oxford). 2010;2010:q20.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57.
Yoon JH, Youn K, Ho CT, Karwe MV, Jeong WS, Jun M. P-Coumaric acid and ursolic acid from Corni fructus attenuated beta-amyloid (25-35)-induced toxicity through regulation of the NF-kappaB signaling pathway in PC12 cells. J Agric Food Chem. 2014;62(21):4911–6.
Ayaz M, Junaid M, Ullah F, Subhan F, Sadiq A, Ali G, et al. Anti-Alzheimer’s studies on beta-sitosterol isolated from polygonum hydropiper L. Front Pharmacol. 2017;8:697.
Ye JY, Li L, Hao QM, Qin Y, Ma CS. Beta-Sitosterol treatment attenuates cognitive deficits and prevents amyloid plaque deposition in amyloid protein precursor/presenilin 1 mice. Korean J Physiol Pharmacol. 2020;24(1):39–46.
Ryu S, Seol GH, Park H, Choi IY. Trans-anethole protects cortical neuronal cells against oxygen-glucose deprivation/reoxygenation. Neurol Sci. 2014;35(10):1541–7.
Chainy GB, Manna SK, Chaturvedi MM, Aggarwal BB. Anethole blocks both early and late cellular responses transduced by tumor necrosis factor: effect on NF-kappaB, AP-1, JNK. MAPKK and apoptosis Oncogene. 2000;19(25):2943–50.
Yang CC, Kuai XX, Gao WB, Yu JC, Wang Q, Li L, et al. Morroniside-induced PP2A activation antagonizes tau hyperphosphorylation in a cellular model of neurodegeneration. J Alzheimers Dis. 2016;51(1):33–44.
Wang K, Sun W, Zhang L, Guo W, Xu J, Liu S, et al. Oleanolic acid ameliorates Abeta25-35 injection-induced memory deficit in Alzheimer's disease model rats by maintaining synaptic plasticity. CNS Neurol Disord Drug Targets. 2018;17(5):389–99.
Mendrysa SM, Ghassemifar S, Malek R. p53 in the CNS: perspectives on development, stem cells, and cancer. Genes Cancer. 2011;2(4):431–42.
Yoo SY, Yoo JY, Kim HB, Baik TK, Lee JH, Woo RS. Neuregulin-1 protects neuronal cells against damage due to CoCl2-induced hypoxia by suppressing hypoxia-inducible factor-1alpha and P53 in SH-SY5Y cells. Int Neurourol J. 2019;23(Suppl 2):S111–8.
Zuev VA, Dyatlova AS, Lin'Kova NS, Kvetnaya TV. Expression of Abeta42, tau-protein, p16, p53 in buccal epithelium: prospects for use in the diagnostics of Alzheimer's disease and rate of aging. Bull Exp Biol Med. 2019;166(5):676–9.
Patel NS, Mathura VS, Bachmeier C, Beaulieu-Abdelahad D, Laporte V, Weeks O, et al. Alzheimer's beta-amyloid peptide blocks vascular endothelial growth factor mediated signaling via direct interaction with VEGFR-2. J Neurochem. 2010;112(1):66–76.
Nevado-Holgado AJ, Ribe E, Thei L, Furlong L, Mayer MA, Quan J, et al. Genetic and real-world clinical data, combined with empirical validation, nominate Jak-Stat signaling as a target for Alzheimer's disease therapeutic development. Cells. 2019;8(5):E425. https://doi.org/10.3390/cells8050425.
Porro C, Cianciulli A, Trotta T, Lofrumento DD, Panaro MA. Curcumin regulates anti-inflammatory responses by JAK/STAT/SOCS signaling pathway in BV-2 microglial cells. Biology (Basel). 2019;8(3):E51. https://doi.org/10.3390/biology8030051.
Rai SN, Dilnashin H, Birla H, Singh SS, Zahra W, Rathore AS, et al. The role of PI3K/Akt and ERK in neurodegenerative disorders. Neurotox Res. 2019;35(3):775–95.
Song L, Yao L, Zhang L, Piao Z, Lu Y. Schizandrol a protects against Abeta1-42-induced autophagy via activation of PI3K/AKT/mTOR pathway in SH-SY5Y cells and primary hippocampal neurons. Naunyn Schmiedeberg's Arch Pharmacol. 2020. https://doi.org/10.1007/s00210-019-01792-2.
Wu Y, Qin D, Yang H, Wang W, Xiao J, Zhou L, et al. Neuroprotective effects of deuterium-depleted water (DDW) against H2O2-induced oxidative stress in differentiated PC12 cells through the PI3K/Akt signaling pathway. Neurochem Res. 2020. https://doi.org/10.1007/s11064-020-02978-4.
Chang KW, Zong HF, Ma KG, Zhai WY, Yang WN, Hu XD, et al. Activation of alpha7 nicotinic acetylcholine receptor alleviates Abeta1-42-induced neurotoxicity via downregulation of p38 and JNK MAPK signaling pathways. Neurochem Int. 2018;120:238–50.
Meng M, Ai D, Sun L, Xu X, Cao X. EGb 761 inhibits Abeta1-42-induced neuroinflammatory response by suppressing P38 MAPK signaling pathway in BV-2 microglial cells. Neuroreport. 2019;30(6):434–40.
Zhou Y, Wang ZF, Li W, Hong H, Chen J, Tian Y, et al. Protective effects of microRNA-330 on amyloid beta-protein production, oxidative stress, and mitochondrial dysfunction in Alzheimer's disease by targeting VAV1 via the MAPK signaling pathway. J Cell Biochem. 2018;119(7):5437–48.
Acknowledgements
Not applicable.
Funding
This study was supported by National Natural Science Foundation of China (grant number 81873246), Natural Science Foundation of Shanghai Municipality (grant number 18ZR1439900), The Fourth Batch of National Research and Training Program for Outstanding Talents in Traditional Chinese Medicine from State Administration of Traditional Chinese Medicine (grant number 2017–24) and Scientific and Technological Innovation Projects of Longhua Hospital (grant number CX201766). These funds provide financial support for the study of network pharmacology, English language editing and article publishing fees. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
HMA and BH designed the study, coordinated technical support and funding; BH revised the manuscript; YJQ performed the study and drafted the manuscript; RRZ, LMZ, CG, LC and XP participated the study. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not Applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Qu, YJ., Zhen, RR., Zhang, LM. et al. Uncovering the active compounds and effective mechanisms of the dried mature sarcocarp of Cornus officinalis Sieb. Et Zucc. For the treatment of Alzheimer’s disease through a network pharmacology approach. BMC Complement Med Ther 20, 157 (2020). https://doi.org/10.1186/s12906-020-02951-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12906-020-02951-2