Abstract
Background
Multi-drug resistance in microorganisms is a serious problem at national as well as at a global level. Many researches have suggested alternatives to antibiotics with minimal or no major side effects. LAB is one of the most human-friendly probiotic strains known to mankind from times immemorial. With the objective to deal with progressing antibiotic resistance among microorganisms, the present work demonstrates the inhibitory activity of LAB consortium against MDR clinical isolates.
Methods
Total of nine hospital isolates of staphylococci were obtained and distinguished as S.aureus and coagulase-negative Staphylococcus (CoNS) based on their ability to ferment mannitol and form clumping with citrated plasma. All the test organisms were tested for antibiotic sensitivity with HiMedia (India) Octadisc Combi 92. Sets of L .plantarum, L .acidophilus and L.casei var. rhamnosus were prepared and tested against a standard culture of S.aureus NCIM 2129 by agar well diffusion method. To identify the primary source of substances responsible for inhibitory action, whole broth, cell-free supernatant, and cell lysate was prepared from the above-mentioned set. These were tested for their inhibitory action initially against standard S.aureus NCIM 2127, followed by clinical isolates.
Results
The antibiotic sensitivity profile revealed that all clinical isolates were multi-drug resistant. The maximum inhibitory potential was seen in a combination of the three LAB in the ratio 1:1:1. Highest antagonistic activity was observed with whole broth and cell lysate of LAB consortium. In liquid broth assay, the cell lysate of LAB consortium astoundingly exhibited up to 85% inhibition of multi-drug resistant Staphylococcus isolates.
Conclusions
Our results suggest antagonistic role of LAB metabolites against methicillin resistant staphylococci.
Similar content being viewed by others
Background
Extensive use of antibiotics, even to cure a common cold, has led to the heightened development of resistance towards a variety of drugs amongst pathogens all around the globe [1]. Staphylococci are Gram-positive, clump-forming, salt tolerant and often hemolytic pathogens, responsible for numerous infections like skin lesions, abscesses, osteomyelitis, endocarditis, furunculosis, urinary tract infections, toxic shock syndrome, and food poisoning. They are the leading cause of nosocomially acquired infections like catheter-associated bacteremia and necrotizing pneumonia [2]. Staphylococcus species can be classified as coagulase-positive staphylococci (CoPS), and the coagulase-negative staphylococci (CoNS). CoPS mainly represent Staphylococcus aureus, an opportunistic microorganism with many potential virulence factors like surface proteins and other agents that promote colonization of host tissues, inhibit phagocytosis and damage host tissues causing disease symptoms. CoNS represent diverse species of staphylococci, like S. epidermis, S. haemolyticus, S. lugdunensis, and S. saprophyticus, with normally fewer virulence factors but an ability to form biofilms on implanted devices causing severe issues [3].
Staphylococci are well known for their ability to become resistant to antibiotics [4, 5], inclusive of second and third line drugs [6, 7]. Methicillin-Resistant Staphylococcus aureus (MRSA) is a perilous group of the bacterial pathogen which combines virulence, antibiotic/ drug-resistance, and a heavy rate of transfer. These infectious pathogens not only do increase treatment cost tremendously but also contribute to increased mortality and morbidity rates [8]. Limited established treatment options exist for such invasive infections [9]. Multiple drug resistance in Staphylococci is a major and growing problem and is now subdivided into hospital-acquired MRSA (HA-MRSA) and community-acquired MRSA (CA-MRSA) [10]. The epidemiology of MRSA is constantly changing, resulting in a variation in drug-resistance patterns throughout regions and countries [11]. Vancomycin once represented the paragon to treat such invasive infections. However, an increase in reports of in vitro resistance to vancomycin and clinical failures with such invasive infection accentuated the need to develop alternative therapies for treatment [10,11,12].
Similarly, the prevalence of multi-drug resistant (MDR) strains of common bacterial pathogens is increasing worldwide [13, 14]. Although antibiotics are available for the treatment of these infections, because of their numerous adverse effects and development of resistant strains, there is an urgent need to search for alternatives to synthetic antibiotics [15]. Therefore, various approaches have been adopted to deal with the progressing multi-drug resistance among such pathogenic species. Treatment with selected probiotic strains is one such solution that is comparatively safe and stable as they do not increase the risk of multi-drug resistance of these pathogens [16, 17]. Most Lactic Acid Bacteria (LAB), despite their origin, have the potential to inhibit the growth of pathogens, including problematic antibiotic-resistant isolates due to their ability to produce several antimicrobial metabolites [18]. Many researchers have proven the inhibitory activity of different probiotic strains against such infectious pathogens [19,20,21,22,23].
The present study aimed to evaluate the in vitro antibacterial activity of an effective consortium of lactobacilli against MDR staphylococci.
Methods
Strains and culture conditions
Three Lactobacillus species, Lactobacillus plantarum NCIM 2374 (NCIB 6376), Lactobacillus acidophilus NCIM 2660 (ATCC 11975) and Lactobacillus casei var. rhamnosus NCIM 2364 (ATCC 7469), were collected from the National Collection of Industrial Microorganisms(NCIM) at the National Chemical Laboratory (NCL), Pune.
A standard strain of Staphylococcus aureus NCIM 2127 was collected from NCIM, NCL, Pune. Nine random clinical isolates of Staphylococcus species (labeled as A to I) were collected from the Microbiology Laboratory at the Bharati Hospital, Katraj, Pune, India.
For revival and maintenance of LAB, de Mann-Rogosa-Sharpe (MRS) medium (HiMedia, India) was used, while the clinical isolates were enriched with Brain Heart Infusion (BHI) agar (HiMedia, India). All these cultures were incubated overnight (about 18 h) at 37 °C at still and at shaking (120 rpm) conditions respectively.
Colony morphology and Gram reaction of the cultures were tested. Staphylococci strains were tested for their reactivity with mannitol and citrated plasma, as stated by Turner and Schwartz [24], to distinguish S. aureus from other Staphylococcus species.
Substantiation of the multi-drug resistance
A standard Staphylococcus aureus NCIM 2127 (ATCC 9144) and clinically obtained Staphylococcus A to I were tested for their sensitivity towards methicillin and other antibiotics. The cultures were allowed to grow overnight in BHI broth at 37 °C. The overnight cultures of staphylococci were adjusted to 1.0 at OD600 and swabbed on Muller-Hilton agar plates. An octa-disc ring (OCTADISCS COMBI 92 HiMedia, India) containing the following antibiotics: Amikacin 30 μg (AK), Ciprofloxacin 5 μg (CIP), Gentamicin 10 μg (GEN), Ceftazidime 30 μg (CAZ), Cefepime 30 μg (CPM), Cefoxitin 30 μg (CX), Cefoxatime 30 μg (CTX) and Ceftriaxone 30 μg (CTR) was placed on each swabbed plate. The plates were incubated at 37 °C and were observed the next day for the zone of clearance. Zone diameters were measured according to Barry A. et al. [25]. and were compared with the standard measures given by the CLSI Performance Standards for Antimicrobial Susceptibility Testing [26].
Antimicrobial activity
Antimicrobial activity of Lactobacillus strains was assayed to evaluate their ability to inhibit the clinical isolates of MDR microorganisms.
Seven sets of whole broth (WB) of L. plantarum, L. acidophilus and L. casei var. rhamnosus, individually as well as in combinations, were prepared as described as below to test their antimicrobial activity against the standard culture S. aureus NCIM 2127.
-
I: L. plantarum
-
II: L. acidophilus
-
III: L. casei
-
IV: L. plantarum + L. acidophilus (1:1)
-
V: L. acidophilus + L. casei (1:1)
-
VI: L. plantarum + L. casei (1:1)
-
VII: L. plantarum + L. acidophilus + L. casei (1:1:1)
The activity was determined using the agar well diffusion method as described by Barbara et al., [27] with slight modification. For the preparation of each set, the culture broth of 0.5 O.D (McFarland Standard) for each culture was used. An overnight incubated culture of S. aureus was swabbed on 0.7% soft agar medium containing BHI+ MRS (1:1) (with 2% agar-agar base). Wells of 5 mm diameter were then punched on these pre-swabbed plates and 50 μl of each set was added in the wells accordingly. Inoculated sets were allowed for diffusion for 1 h at room temperature. The plates were then incubated overnight at 37 °C. Zone diameters were measured according to Barry A. et al. [25]. Inhibition was scored positive if the width of the clear zone around the well was observed.
The set giving the best result was used further to check the activity of WB, cell-free broth (CFB), and cell lysate (CL).
To prepare CFB, the set was centrifuged at 8000 rpm for 15 min at 4 °C. The supernatant was collected and passed through a 0.2-μm syringe filter (BioEra, India) to remove any remaining bacterial cells and cell debris. The cell lysate (CL) was prepared as per the protocol by Kang et al (2012). Cells separated from the above centrifugation were washed twice with saline and resuspended in lysis buffer (10 mM Tris HCl, pH 8.0; 1 mM EDTA; 0.1% (w/v) SDS). The sample was sonicated for 5–15 min until it appears milky. The sonicated sample was then centrifuged at 8000 rpm for 15 min at 4 °C and the supernatant was used for the experiment [28]. These samples ie. WB, CFB, and CL were used to investigate their activity against the clinical isolates by agar well diffusion method as described above.
To understand the antimicrobial activity of CL better, the liquid medium containing a mixture of LAB-CL with clinically obtained microorganisms was studied according to Barbara et al (2010) with slight modification. A 96-well ELISA plate was used for this purpose. 20 μl of each test organism (OD600=0.8) were mixed with 50 μl of CL. Each well was added with 130 μl of nutrient broth. Test microorganisms and CL individually were considered as controls. Optical Density at 595 nm was obtained with the help of Epoch microplate spectrophotometer (Biotek, USA).
All the experiments were performed in triplicates independently.
Results
Determination of coagulase positive and coagulase negative staphylococci
According to the MSA slant color change (reactivity with mannitol) and coagulase test, Staphylococcus isolates were distinguished to CoPS and CoNS. It was observed that Staphylococcus isolates A, B, C, D, E, and I were able to ferment mannitol, hence changing the MSA slant colour to yellow and showed agglutination when mixed with citrated plasma, hence may belong to S.aureus species; whereas Staphylococcus isolates F, G, and H neither showed any colour change on MSA nor any agglutination or clumping with the citrated plasma. It was also observed that S.aureus NCIM 2127, tested as a reference, was coagulase positive and was able to ferment mannitol.
Substantiation of multi-drug resistance
The overnight incubated plates with antibiotic discs were observed and the zone diameters were measured. These zone diameters were compared with the CLSI standards [26] to obtain inferences (Table 1).
As a result of the antibiotic susceptibility test for the clinical isolates, all were observed to be resistant against gentamicin (GEN) and cefoxitin (CX). Among the other six antibiotics, amikacin (CTX) possessed the maximum inhibitory potential against the nine Staphylococcus strains. All the clinical isolates were MDR as they exhibited resistance towards at least three antibiotics belonging to different classes. S. aureus NCIM 2127 was observed to be sensitive towards all the antibiotics.
Synergistic activity of Lactobacillus cultures
Agar well diffusion procedure was performed with seven sets as described above. The overnight incubated plates were observed and the zone diameters were measured (Table 2).
It was observed that all the three strains of Lactobacillus sp. exhibited antimicrobial activity against S.aureus. The zone of inhibition was observed to be maximum with set VII ie. a consortium of all three LAB cultures with zone diameter of 20.3 ± 0.47 mm. This was followed by set IV ie. L.plantarum and L. acidophilus with a zone diameter of 14.3 ± 0.30.
Inhibitory effect of selected LAB consortium
As discussed above, set VII showed the best activity in the agar well diffusion assay against S. aureus NCIM 2127. Hence, it was further used as WB, CFB and CL for comparative evaluation initially against S. aureus NCIM 2127 and then against all the clinical isolates. Data represented in Table 3 reveals the inhibitory action of actively growing cells, acid supernatant and cell lysate of the mixture of Lactobacillus.
It was observed that WB, CFB as well as CL, all three exhibits antibacterial activity against S.aureus, MRSt as well as on MDR isolates.
The whole broth of set VII showed the maximum inhibitory effect on MRSt A to I and S.aureus NCIM 2127. Supernatant exhibited the least inhibitory effect on all the clinical isolates. On comparing with CFB, it was observed that CL had higher inhibitory activity against all the clinical isolates.
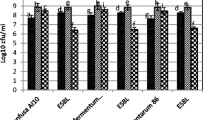
Quantitative assay using LAB cell lysate
To understand the action of cell lysate of LAB against MDR organisms better, cultures were treated with cell lysate in liquid broth and checked for the change in optical density.
It was observed (Fig. 1) that the standard culture of S.aureus NCIM 2127 showed the maximum ie. 96.6% inhibition. Amongst the MRSt, CoNS isolates (F, G, and H) showed inhibition in the range of 57.4–65%. The cell lysate was able to inhibit the MDR/MRSA isolates (A, B, C, D, E, and I) by an average of 83.1%.
Discussion
The study revealed that from the nine clinical isolates of Staphylococcus species, six were coagulase positive and had an ability to ferment mannitol, hence belong to S.aureus species; whereas three were coagulase negative and were unable to ferment mannitol. Coagulase protein is an important virulent agent of S. aureus which can clot plasma into the gel [29]. However, there are reports available showing agglutination organisms other than Staphylococcus species [30].
A further confirmatory test is to find the ability of S.aureus to ferment mannitol. S.aureus ferments mannitol sugar and produce acid as an end product. Hence on inoculation with MSA, the color turns from pink to yellow [31]. CoNS do not ferment mannitol and hence the color of the slant remains pink [3]. CoNS include species other than S. aureus, like S. epidermis, S. haemolyticus, S. lugdunensis, and S. saprophyticus [32].
The antibiotic susceptibility assay revealed that the Staphylococcus isolates were resistant towards cefoxitin. Methicillin is a semisynthetic derivative of penicillin to treat penicillin-resistant Staphylococcus infections. Methicillin resistance is a marker of resistance to β lactam antibiotics (i.e., the penicillin and most cephalosporin antibiotics), which are some of the most commonly used antibiotics globally. Further, MRSA can easily develop resistance towards many other steroidal and non-steroidal antibiotics [33]. This means that even though methicillin is not used much these days, the resistance of bacteria towards it indicates drug resistance against multiple antimicrobial agents.
It is a well-known fact that microbes acquire multi-drug resistance due to various factors. Two most common mechanisms are (i) genetic mutations within the microorganism and (ii) triggering of mobile genetic elements that secure drug resistance genes [34]. Aminoglycoside (AG) antibiotics like amikacin and gentamicin are used to treat many Gram-negative and some Gram-positive infections. Gentamicin is the most used AG antibiotic while amikacin (semisynthetic AG) is usually prescribed less to prevent resistance development. Amikacin is persuasive towards most AG-resistant pathogens since it has a refracting nature towards most aminoglycoside-modifying enzymes. However, the upsurging resistance to amikacin has led to limitations of drugs for the treatment of infections in neonates [35, 36]. Research by Wenchang et al., [37] revealed that resistance development towards amikacin is associated with the thickening of the bacterial cell wall. However, resistance to AG drugs does not always mean resistance to other antibiotics. The strains used by the researcher showed susceptibility to antibiotics like ceftizoxime, chloramphenicol, ciprofloxacin, gentamicin, rifampicin, tetracycline, teicoplanin, linezolid, and vancomycin. The current work partially relates to these observations as the clinical isolates of MRSA are resistant towards amikacin and gentamicin. All of these clinical isolates can be considered as MDR as they are resistant towards antibiotics belonging to three different classes (aminoglycosides, quinolone and, cephalosporins) [38, 39].
The strains used in the study are Lactobacillus plantarum NCIM 2374 (NCIB 6376), Lactobacillus acidophilus NCIM 2660 (ATCC 11975) and Lactobacillus casei var. rhamnosus NCIM 2364 (ATCC 7469). All of these are industrially used strains and were selected with reference to the study concluded by Sikorska and Smoragiewicz [2013]. Their study concluded that most active strains against MRSA were Lactobacillus reuteri, Lactobacillus rhamnosus GG, Propionibacterium freudenreichii, Propionibacterium acnes, Lactobacillus paracasei, L. acidophilus, L. casei, Lactobacillus plantarum, Lactobacillus bulgaricus, Lactobacillus fermentum and Lactococcus lactis. Their effects were mediated both by direct cell competitive exclusion as well as the production of acids or bacteriocin-like inhibitors [40]. Similarly, many studies have been conducted using these LAB species to test their antimicrobial activity against MRSA [21, 27, 41].
Set VII showed maximum inhibition against S.aureus. These results demonstrate the synergistic effect of the combination of L.plantarum, L.casei and L. acidophilus (Set VII) in ratio 1:1:1, exhibiting highest antimicrobial activity as compared to the other sets.
It can be hypothesized that a combination of probiotic strains may complement each other’s effects or improve benefits or properties [42,43,44]. The objective of this study was, therefore, to determine if the chosen probiotics in the combinations tested may increase or enhance each other’s beneficial properties and their potential applications in favoring maximum inhibition against S.aureus and MDR staphylococci. The work can be co-related with research work by Karska-Wysocki [27] where the activity of L. acidophilus and L. casei in equal proportion (1:1) was observed to be higher when used in a combination.
To understand the primary action source of inhibition, the action of whole broth, cell-free supernatant and cell lysate were compared with each other. Generally, the LAB are the most implicated of the probiotic organisms, particularly those of the genera Lactobacillus and Bifidobacterium, which protects their territory by secreting acids like lactic, acetic, formic, succinic, glutamic, citric, and butyric acids; thereby creating an environment which is inhospitable to disease-causing bacteria. Lactobacilli are known to be microaerophilic by nature. They produce metabolites which can cause alterations in the oxidation-reduction potential, hence making the environment unfavorable for aerobic organisms. This action contributes to the overall inhibiting effect of these probiotic bacteria [45, 46]. Different Lactobacillus species have shown significant activities against classical antibiotic-resistant bacteria, MRSA and other emerging antibiotic resistant microorganisms. Voravuthikunchaia [47] stated that the antibacterial activity of the lactobacilli might cause growth inhibition and cell death with respect to the pathogen it is dealing with. Different mechanisms to exert antimicrobial effect are shown by the LAB, but the cell envelope is generally the target. Metabolic by-products such as bacteriocins, hydrogen peroxide (H2O2), and organic acids, produced by the lactobacilli during growth, contribute to their antibacterial activity. Other mechanisms proposed for their microbial antagonism are competition for nutrition, adhesion inhibition of pathogens to surfaces and stimulation of the immune system [47]. Studies have revealed that L.reuteri, isolated from a healthy vaginal ecosystem, [47] and L.fermentum [48]; can appreciably inhibit pathogens like MRSA. It has been observed that L. casei can displace and kill S. aureus adhering to human intestine mucus by 39 to 44% [49]. Charlier et al. (2008) reported that L.lactis had a specific antimicrobial activity against S.aureus [50]. Later, Koji (2005) stated the beneficial effects conferred by lactobacilli, including inhibition of Gram-negative and positive pathogenic bacteria [51].
Earlier, researchers have observed that the bacteriocins produced by LAB get adsorbed on the cell surface at specific pH [52,53,54,55,56]. Yang et al. [57], further discovered a technique for the purification of bacteriocins, responsible for the antimicrobial activity. In view of this, cell lysate of selected LAB consortium (present work) was prepared and studied further for their inhibitory action against MDR clinical isolates. The pH of 6.0–5.5 was observed after an overnight incubation, which is an ideal pH at which the proteins get adsorbed on the cells. Hence, the cell lysate contains both adsorbed proteins and intracellular proteins. This study revealed the potential of cell lysate of the combination of the LAB to inhibit MDR organisms. To our knowledge, antimicrobial activity by cell lysate is very less researched and present study gives an indication that the inhibitory activity of bacteriocins and some intracellular proteins of LAB can be of great importance.
Conclusions
From all the in vitro testing, it can be clearly concluded that S. aureus was vulnerable to the metabolites produced by the LAB cultures selected, individually as well as in combinations. However, a combination of all the three strains used together exhibited the best result. This study showed the potential of the whole broth and cell lysate of the combination of L.acidophillus, L. plantarum, and L. casei var. rhamnosus as a better inhibitor towards MDR clinical isolates.
Abbreviations
- CFB:
-
Cell-free broth(supernatant)
- CL:
-
Cell lysate
- LAB:
-
Lactic Acid Bacteria
- MDR:
-
Multi drug resistant
- MRSA:
-
Methicillin-Resistant Staphylococcus aureus
- MRSt:
-
Multi-drug resistant Staphylococcus
- WB:
-
Whole broth
References
Pfaller MA, Jones RN, Marshall SA, Coffman SL, Hollis RJ, Edmond MB, et al. Inducible amp C beta-lactamase producing gram-negative bacilli from blood stream infections: frequency, antimicrobial susceptibility, and molecular epidemiology in a national surveillance program (SCOPE). Diagn Microbiol Infect Dis. 1997;28:211–9.
Reddy PN, Srirama K, Dirisala VR. An update on clinical burden, diagnostic tools, and therapeutic options of Staphylococcus aureus. Infectious Diseases. 2017;10:1179916117703999.
Becker K, Heilmann C, Peters G. Coagulase-negative Staphylococci. Clin Microbiol Rev. 2014;27(4):870–926.
Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Investig. 2003;111(9):1265–73.
Chambers HF, DeLeo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nature reviews. Microbiology. 2009;7(9):629–41.
Fair RJ, Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect Medicin Chem. 2014;6:25–64.
Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109(7):309–18.
Raygada JL, Levine DP. Methicillin-resistant Staphylococcus aureus: a growing risk in the hospital and in the community. Am Health Drug Benefits. 2009;2(2):86–95.
Drew RH. Emerging options for treatment of invasive, multidrug-resistant Staphylococcus aureus infections. Pharmacotherapy. 2007;27:227–49.
Choo EJ. Community-associated methicillin-resistant Staphylococcus aureus in nosocomial infections. Infect Chemother. 2017;49(2):158–9.
Rodríguez-Noriega E, Seas C, Guzmán-Blanco M, Mejía C, et al. Evolution of methicillin-resistant Staphylococcus aureus clones in Latin America. Int J Infect Dis. 2010;14:560–6.
Chaudhary AS. A review of global initiatives to fight antibiotic resistance and recent antibiotics discovery. Acta Pharm Sin B. 2016;6(6):552–6.
Allocati N, Masulli M, Alexeyev MF, Di Ilio C. Escherichia coli in Europe: An Overview. Int J Environ Res Public Health. 2013;10(12):6235–54.
Oli AN, Eze DE, Gugu TH, Ezeobi I, Maduagwu UN, Ihekwereme CP. Multi-antibiotic resistant extended-spectrum beta-lactamase producing bacteria pose a challenge to the effective treatment of wound and skin infections. Pan Afr Med J. 2017;27:66.
Maxton A, Benjamin JC, Ram GD, Bailey SB, Ramteke PW. Antibacterial activity of isolated human intestinal microbiota lactobacillus strains against methicillin resistant and susceptible Staphylococcus aureus. Afr J Microbiol Res. 2013;7:1802–8.
Tagg JR, Dierksen KP. Bacterial replacement therapy: adapting “germ warfare” to infection prevention. Trends Biotechnol. 2003;21:217–23.
Roghmann MC, McGrail L. Novel ways of preventing antibiotic-resistant infections: what might the future hold? Am J Infect Control. 2006;34:469–75.
Petrova M, Georgieva R, Dojchinovska L, Kirilov N, Iliev I, Antonova S, Hadjieva N, Ivanova I, Danova S. Lactic Acid Bacteria Against Pathogenic Microbes. Trakia J Sci. 2009;7(2):33–9.
Figueroa-González I, Rodríguez-Serrano G, Hernández-Sánchez H, Jiménez-Guzmán J, Gómez-Ruiz L, García-Garibay M, Cruz-Guerrero A. Effect of prebiotics on the growth of probiotics. Department of Biotechnology, Universidad Autónoma Metropolitana-Iztapalapa. XIII National Congress of Biotechnology and Bioengineering. 2009. Acapulco: Fairmont Acapulco Princess Hotel Oral work OIII-18; 2009.
Aminnezhad S, Kermanshahi RK, Ranjbar R. Evaluation of synergistic interactions between cell-free supernatant of Lactobacillus strains and amikacin and Genetamicin against Pseudomonas aeruginosa. Jundishapur J Microbiol. 2015;8(4):e16592.
Eggers S, Barker AK, Valentine S, Hess T, Duster M, Safdar N. Effect of Lactobacillus rhamnosus HN001 on carriage of Staphylococcus aureus: results of the impact of probiotics for reducing infections in veterans (IMPROVE) study. BMC Infec Dis. 2018;18:129.
Allen HK. Alternatives to antibiotics: Why and how. In: NAM Perspectives. Washington, DC: Discussion Paper, National Academy of Medicine; 2017. https://doi.org/10.31478/201707g.
Allen H, Trachsel J, Looft T, Casey T. Finding alternatives to antibiotics. Ann N Y Acad Sci. 2014;1323(1):91–100.
Turner FJ, Schwartz BS. The use of a lyophilized human plasma standardized for blood coagulation factors in the coagulase and fibrinolytic tests. J Lab Clin Med. 1958;52:888–94.
Barry A, Coyle M, Thornsberry C, Gerlach H, Hawkinson R. Methods of measuring zones of inhibition with the BauerKirby disk susceptibility test. J Clin Microbiol. 1979;10(6):885–9.
Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 26th ed. CLSI supplement M100S, 2016.
Karska-Wysocki B, Bazo M, Smoragiewicz W. Antibacterial activity of Lactobacillus acidophilus and Lactobacillus casei against methicillin-resistant Staphylococcus aureus (MRSA). Microbiol Res. 2010;165(8):674–86 ISSN 0944-5013.
Kang MS, Oh JS, Lee SW, et al. Effect of Lactobacillus reuteri on the proliferation of Propionibacterium acnes and Staphylococcus epidermidis. J Microbiol. 2012;50:137–42.
Sperber WH, Tatini SR. Interpretation of the tube coagulase test for identification of Staphylococcus aureus. Appl Microbiol. 1975;29(4):502-5.
Bayliss BG, Hall ER. Plasma coagulation by organisms other than Staphylococcus aureus. J Bacteriol. 1965;89(1):101–5.
D’Souza HA, Baron EJ. BBL CHROMagar Staphylococcus aureus is superior to mannitol salt for detection of Staphylococcus aureus in complex mixed infections. Am J Clin Pathol. 2005;123(6):806–8.
Freeman J, Platt R, Sidebottom DG, Leclair JM, Epstein MF, Goldmann DA. Coagulase-negative staphylococcal bacteremia in the changing neonatal intensive care unit PopulationIs there an epidemic? JAMA. 1987;258(18):2548–52. https://doi.org/10.1001/jama.1987.03400180082031.
Naimi TS, LeDell KH, Como-Sabetti K, et al. Compariso of community- and health care-associated methicillin resistant Staphylococcus aureus infection. JAMA. 2003;290:2976–84.
Khadri H, Alzohairy M. Prevalence and antibiotic susceptibility pattern of methicillin-resistant and coagulase-negative staphylococci in a tertiary care hospital in India. Int J Med Med Sci. 2010;2(4):116–20.
Woloj M, Tolmasky ME, Roberts MC, Crosa JH. Plasmid-encoded amikacin resistance in multidrug resistant strains of Klebsiella pneumoniae isolated from neonates with meningitis. Antimicrob Agents Chemother. 1986;29:315–9.
Gonzalez LS, Spencer JP. Aminoglycosides: a practical review. Am Fam Phys. 1998;58:1811–20.
Yuan W, Hu Q, Cheng H, Shang W, Liu N, Hua Z, Zhu J, Hu Z, Yuan J, Zhang X, Li S, Chen Z, Hu X, Jianfeng F, Rao X. Cell wall thickening is associated with adaptive resistance to amikacin in methicillin-resistant Staphylococcus aureus clinical isolates. J Antimicrob Chemother. 2013;68(5):1089–96.
Reddy P, Chadaga S, Noskin G. Antibiotic considerations in the treatment of multidrug-resistant (MDR) pathogens: A case-based review. Journal of Hospital Medicine. 2009; 4(6): E8-E15.
Mandal S, Mandal MD, Pal NK. Evaluation of combination effect of ciprofloxacin and cefazolin against Salmonella enterica serovar typhi isolates by in vitro methods. Calicut Med J. 2004;2(2):22–6.
Sikorska H, Smoragiewicz W. Role of probiotics in the prevention and treatment of meticillin-resistant Staphylococcus aureus infections. Int J Antimicrob Agents. 2013;42(6):475–81.
Hala F, Radeef M, Al-Mathkhury H. Capability of Lactobacillus acidophilus supernatant to inhibit production of lipase from methicillin-resistant Staphylococcus aureus. J. of university of anbar for pure science. 2011;5.
Myllyluoma E, Veijola L, Ahlroos T, Tynkkynen S, Kankuri E, Vapaatalo H, Rautelin H, Korpela R. Probiotic supplementation improves tolerance to helicobacter pylori eradication therapy - a placebo-controlled, double-blind randomized pilot study. Aliment Pharmacol Ther. 2005;21(10):1263–72.
Kajander K, Hatakka K, Poussa T, Farkkila M, Korpela R. A probiotic mixture alleviates symptoms in irritable bowel syndrome patients: a controlled 6-month intervention. Aliment Pharmacol Ther. 2005;22(5):387–94.
Myllyluoma E, Ahonen A-M, Korpela R, Vapaatalo H, Kankuri E. Effects of multispecies probiotic combination on Helicobacter pylori infection in vitro. Clin Vaccine Immunol. 2008;15(9):1472–82.
Zalán Z, Hudáček J, Štětina J, et al. Eur Food Res Technol. 2010;230:395. https://doi.org/10.1007/s00217-009-1179-9.
Olanrewaju O. Antagonistic effect of Lactobacillus isolates from Kunnu and cow milk on selcted pathogenic microorganisms. Int J Food Safety. 2007;9:63–6.
Voravuthikunchai SP, Bilasoi S, Supamala O. Antagonistic activity against pathogenic bacteria by human vaginal lactobacilli. Anaerobe. 2006;12:221–6. https://doi.org/10.1016/j.anaerobe.2006.06.003.
Nawaz M, Wang J, Zhou A, et al. Curr Microbiol. 2011;62:1081. https://doi.org/10.1007/s00284-010-9856-2.
Vesterlund S, Karp M, Salminen S, Ouwehand AC. Staphylococcus aureus adheres to human intestinal mucus but can be displaced by certain lactic acid bacteria. Microbiology. 2006;152:1819–26.
Charlier C, Cretenet M, Even S, Le Loir Y. Interactions between Staphylococcus aureus and lactic acid bacteria: an old story with new perspectives. Int J Food Microbiol. 2008;131:30–9.
Koji N. Prevention of infection by probiotics. J Biosci Bioeng. 2005;100:583–92.
Bhunia AK, Johnson MC, Ray B, Kalchayanand N. Mode of action of pediocin AcH from Pediococcus acidilactici H on sensitive bacteria strains. J Appl Bacteriol. 1991;70:25–30.
Gonzalez CF, Kunka BS. Plasmid-associated bacteriocin production and sucrose fermentation in Pediococcus acidilactici. Appl Environ Microbiol. 1987;53:2534–8.
Klaenhammer TR. Bacteriocin from lactic acid bacteria. Biochimie. 1988;70:337–49.
Ray B. Bacteriocins of starter culture bacteria as biopreservatives: an overview. In: Ray B, Daeschel MA, editors. Food biopreservatives of microbial origin. Boca Raton: CRC Press, Inc; 1992. p. 177–205.
Tagg HG, Dajani AS, Wannamaker LW. Bacteriocin of gram-positive bacteria. Bacteriol Rev. 1976;40:722–56.
Yang R, Johnson MC, Ray B. Novel method to extract large amount of bacteriocins from lactic acid bacteria. Appl Environ Microbiol. 1992;58(10):3335–59.
Acknowledgements
We gratefully aknowledge the support from Bharati Vidyapeeth (Deemed to be) University to undertake this work. We are also thankful to Microbiology Department of Bharati Hospital for providing clinical pathogens.
Funding
The work was not supported by any external funding.
Availability of data and materials
The standard cultures of Lactobacillus and S.aureus were procured from the National Collection of Industrial Microorganisms(NCIM) at the National Chemical Laboratory (NCL), Pune, India.
The clinical isolates and datasets, used during the current study, were obtained from the Bharti Hospital, Katraj, Pune 411,046, India; and can be availed from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
JB carried out the experimental work and wrote the manuscript. RB helped out in planning of the experiments and data analysis, and revised the manuscript. Both authors have read and approved the manuscript, and ensure that this is the case.
Corresponding author
Ethics declarations
Authors’ information
Jinal Bhola: Post graduate student of Biotechnology at Rajiv Gandhi Institue of IT and Biotechnology, Bharati Vidyapeeth (Deemed to be) University, Pune, India.
Dr. Rama Bhadekar: Associate Professor, Department of Microbial Biotechnology, Rajiv Gandhi Institue of IT and Biotechnology, Bharati Vidyapeeth (Deemed to be) University, Pune, India.
Ethics approval and consent to participate
The clinical isolates were obtained from the Microbiology Department at the Bharati Hospital, Katraj, Pune 411,046, India. Ethical permission to use the clicinal isolates for this study was obtained from the Institutional Ethics Committee (DCGI Reg. No. ECR 518) and Approval Letter Reference: BVDU/MC/E52 Dated: 16/03/2017.
Consent for publication
Not Applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Bhola, J., Bhadekar, R. Invitro synergistic activity of lactic acid bacteria against multi-drug resistant staphylococci. BMC Complement Altern Med 19, 70 (2019). https://doi.org/10.1186/s12906-019-2470-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12906-019-2470-3