Abstract
Background
Diarrhea, particularly of enteric bacterial pathogen, remains a major cause of morbidity and mortality in Ethiopia. Despite the high prevalence of diarrheal disease among under-five children, antibiotic resistance of bacterial pathogens test is not part of routine childcare in the study area. This study aimed to investigate the prevalence and antimicrobial susceptibility status of Salmonella and Shigella species among diarrheic children attending public health institutions in Ambo town, west Showa, Ethiopia.
Methods
Institutional based, cross-sectional study was carried out from January to July 2014 among 239 diarrheic children below five years of age in Ambo town, Ethiopia. Information about patient demographics, signs, and symptoms was obtained from the parents/guardians of each child using a questionnaire. Stool samples from diarrheic children were collected and processed for isolation of Salmonella and Shigella using conventional microbiology procedures. Suspected Salmonella isolates were confirmed by genus-specific PCR and serotyped using a slide agglutination test. Susceptibility to 10 commonly used antimicrobials was assessed using the Kirby Bauer disc diffusion method.
Results
From the 239 children screened, only nine (3.8%) of them were positive for either Salmonella (n = 3) or Shigella (n = 6) and 19 (7.9%) positive for the intestinal parasite. Three species of Shigella were identified: Shigella flexinari (n = 3), Shigella boydii (n = 2), and Shigella sonnei (n = 1). The three Salmonella isolates were S. chicago, S. caracas, and S. saintpaul. Salmonella and Shigella isolates were resistant to ampicillin (88.9%), followed by tetracycline (66.7%), cotrimoxazole (55.6%), chloramphenicol (44.4%), amoxicillin (33.3%), nalidixic acid (11.1%) and cefotaxime (11.1%). All isolates were sensitive to amikacin, ciprofloxacin, and gentamycin.
Conclusion
In this study, either Salmonella or Shigella species were detected only in 3.8% of diarrheic children in Ambo town, suggesting the dominance of other causes of diarrhea in the study area. A further study targeting other causes of diarrhea should be conducted to establish the major causes of childhood diarrhea in the study area.
Similar content being viewed by others
Background
Infectious diarrhea, especially those due to enteric bacterial pathogens, remains a significant public health problem worldwide. While it primarily contributes to morbidity in developed countries, it accounts for significant mortality among children in low and middle-income countries (LMICs) [1]. Despite declining diarrhea-related death in the last 20 years [2], it still accounts for 21% of under-five child mortality, which translates to 2.5 million child deaths [3]. Africa and South Asia are still home to more than 80% of child deaths. Among 15 high burden countries that have three-quarters of all deaths from diarrhea, Ethiopia was ranked fifth [4], by having 27% diarrhea-related deaths [5].
The increasing antimicrobial resistance among enteric pathogens has become a contemporary global health threat. Notably, the Shigella, Vibrio cholerae, Enteropathogenic E. coli (EPEC), and Salmonella species are a critical concern of the developing world responsible for the high rate of diarrhea-related deaths. One reason for increasing antimicrobial resistance is the unrestricted use of over the counter drugs without medical supervision [6].
In Ethiopia, according to a study conducted in Jimma Health Center, 49.6% were positive for an intestinal parasite, Shigella, and Salmonella species. While Shigella species showed 100% resistance to ampicillin, amoxicillin, and cotrimoxazole, Salmonella isolates were resistant to amoxicillin. In contrast, all Shigella and Salmonella species were susceptible to ceftriaxone, ciprofloxacin, and gentamycin [7].
The magnitude of the Shigella species resistant to nalidixic acidis, an emerging problem in Ethiopia, ranges from 6.5% [8] to 16.7% [7] in Jimma, Southwestern Ethiopia; at 5.9% in Butajira [9] and 10% in Hawassa [10]. On the contrary, Shigella species isolated from diarrheic patients were susceptible to amikacin, ciprofloxacin, and gentamycin in Harar [11] and Jimma [9].
With regard to Salmonella, while there was a high level of resistance to ampicillin in Harar (100%) [11], Bahir Dar (93.9%) [12], Addis Ababa (82.3%) [13], Jimma (62.5%) [7], and Butajira (60%) [9], all isolates from Hawassa were susceptible to ampicillin [14]. This level difference might be related to the heavy reliance on empirical antibiotics treatment for infectious diseases in Ethiopia and irregularities in implementing the treatment protocol. Consequently, the problem increases the risk of the emergence of antibiotic-resistant bacteria strains [15].
The level of antibiotic resistance gauges the clinical and the community malpractices related to the use of antibiotics and the associated risk of emerging infections. Despite this, Ethiopia’s health facilities do not routinely perform the test for antibiotic resistance, at least among the most vulnerable segment of the population, the children affected by diarrhea. As a result, there is a poor understanding of antibiotic resistance on the most common etiologies of diarrhea, Salmonella, and Shigella species in central Ethiopia. Hence, this study intended to explore the magnitude and antimicrobial susceptibility of Salmonella and Shigella species isolated from diarrheic children in Ambo town.
Methods
Study design, area, and period
An institutional-based, cross-sectional study was carried out in Ambo Town Public Health Institutions (ATPHI)—at Ambo General Hospital, Ambo Health Center, and Awaro Health Center—from January to July 2014. These institutes provide health services for Ambo town and the surrounding districts. Ambo is one of the districts in the Western Shewa Zone, Oromia Region of Ethiopia. The 2007 national census of Ethiopia reported total populations for this district to be 108,406, of whom 54,186 were men and 54,220 were women [16].
Sample size
The sample size for the study was determined using a single population proportion formula. The prevalence and antimicrobial susceptibility of bacterial pathogens isolated from childhood diarrhea in Kenya were 17.7% [17]. It used as a reference and at a 95% level of confidence and a 10% non-response rate, the total of 250 children with diarrheal disease targeted to be included in the study as;
Where: n = sample size.
Zα/2 = level of confidence.
P = diarrhea prevalence from previous study = 17.7%.
q = 1-p.
d = margin of error (0.05).
The responded client size was 239 children less than five years of age presented with diarrhea to pediatric OPDs and Wards’. Diarrheic stool is defined as having loose or watery stools at least three times per day, or more frequently than usual for an individual (as per the WHO definition) [4]. A systematic random sampling method used to draw participants based pattern of previous patient flow. Accordingly, every other child whom the parents/guardians briefed on the aim of the study agreed and signed the consent to participate included in the study. Children who did not take an antibiotic for the current diarrheal attack included in the study.
Sample collection, handling, and transport
The collected clinical data includes body temperature, demographic data, and medical history. A single diarrheic stool specimen collected after a physical examination. At a hospital and health centers, the stool samples inoculated in Cary-Blair’s transport medium (CA, USA). The samples transported in a cool box to Ambo University Microbiology Laboratory within four hours of collection, and it processed on the same day.
Microscopic examination
Stool examination performed at health institution laboratories (Ambo General Hospital, Ambo Health Center, and Awaro Health Center). Macroscopic and microscopic examination of the safeguarded specimens (formalin 10%) completed then focuses on formalin-ether sedimentation for intestinal parasites, WBC, and RBC conducted immediately upon sample collection before being inoculated into Cary-Blair’s transport medium.
Culture and identification
All stool specimens cultured for isolation of Shigella and Salmonella species. The collected samples inoculated aerobically first in Selenite F broth (HIMEDIA, India) for the enrichment of Salmonella and Shigella species. Then, the samples inoculated to Xylose lysine desoxycholate agar (XLD) (OXOID, England) and incubated at 35–37 °C for 14–16 h. A loopful of the fecal suspension directly inoculated onto MacConkey agar (SRL, India), and Salmonella-Shigella (SRL, India) agar, and incubated at 37 °C for 18–24 h. MacConkey agar used to characterize most enteric bacteria toward their lactose utilization property, XLD (Shigella: red colonies, Salmonella: red with/without a black center), and SS agar used for the isolation of Shigella and Salmonella species. The presumptive colonies of each representative isolates, then characterized using standard biochemical tests.
Biochemical tests performed to characterize the enteric gram-negative bacteria include gram stain morphology, pigment production, motility, urease, citrate, hydrogen sulfide utilization, oxidase, indole, lysine, and sugar fermentation. The media used were nutrient broth (CONDA, Spain), lysine iron agar (LIA) (OXIOD, England), MRVP, Simmons citrate agar (HIMEDIA, India), Kligler iron agar (KIA) (SRL, India), Sulfide-Indole-Motility (SIM), urea broth base (OXIOD, England), Motility Indole Ornithine Medium (MIO) (OXIOD, England). The 3% H2O2 was used to identify Salmonella, Shigella species, and other enteric bacteria as adopted from the Basic laboratory Procedures in Clinical bacteriology WHO (Vandepitteet al., 2nd ed. 2003).
Slide agglutination test was used to serotype the isolates of Salmonella and Shigella species using polyvalent/monovalent antisera. Presumptive Salmonella colonies were confirmed by genus-specific PCR [18]. The slide agglutination test for serotyping of Shigella species was carried out using antisera following the manufacturer instruction (Remel Europe Ltd). The Salmonella isolates were serotyped at the Public Health Agency of Canada, National Microbiology Laboratory at Guelph, OIE Salmonella Reference Laboratory, Guelph, Ontario [19].
Antimicrobial susceptibility
Antibacterial susceptibility test for the six Shigella and three Salmonella isolates was performed on Mueller-Hinton agar plates (SRL, India) using the Kirby-Bauer technique. The 0.5 McFarland standard used to prepare inoculum for the antimicrobial disk diffusion susceptibility test [20]. The antimicrobial susceptibility of bacterial isolates were screened for ten antibiotics amikacin (AK 30 μg), ampicillin (AM, 10 μg), amoxicillin (AX, 10 μg), cotrimoxazole (SXT, 25 μg), cefotaxime (CF, 30 μg), chloramphenicol (CH, 30 μg), ciprofloxacin (CP, 5 μg), gentamycin (GM, 10 μg), nalidixic acid (NA, 30 μg), and tetracycline (TTC, 30 μg).
The plates were incubated at 37 °C for 24 h, and the zone of inhibition diameters were measured with a ruler and, interpreted according to CLSI guidelines, and the results recorded as sensitive (S), resistant (R), or intermediate (I) based on CLSI [20]. The study declared multidrug resistance (MDR) if the isolates were resistant to more than two (> 2) of the antimicrobial agents belonging to different classes [21, 22]. Escherichia coli ATCC 25922 used as a quality control strain during the antimicrobial susceptibility test [20].
Statistical analysis
The data were entered into EpiData 3.02 and then transferred to SPSS version 17.0 statistical software for data processing and analysis. The descriptive statistics such as mean, standard deviation, and proportion used.
Results
A total of 239 children included in the study. Of these, more than three-quarters of them were from Ambo Hospital. Slightly more than half (52.3%) of the children were boys, and two-third of all the children were from urban areas. The children’s age ranged from 5 to 60 months, with a mean of 27.96 (SD ± 17.09) months (Tables 1, and 2).
As shown in Table, four out of five children were living in non-crowded housing conditions, which is < 1.5 persons-per-room [23]. One out of six households was using water from unsafe sources such as rivers and domestic wells. Two out of five households with diarrheic children had at least one domestic animal in their household (Table 1). However, no statistically significant difference was observed in the proportion of identified enteric bacterial pathogens across socio-demographics characters.
Clinical characteristics
Fifteen (6.3%) children took one of the four antibiotics: ceftriaxone, amoxicillin, metronidazole, or cotrimoxazole in the past four weeks—cotrimoxazole being the most commonly prescribed drug (Table 1). The most common clinical complaints recorded were fever (78.2%) followed by vomiting (66.1%), and mucoid diarrhea (41.8%) (Fig. 1).
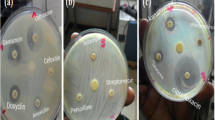
From seven positive stool samples, six Shigella and three Salmonella species were isolated. Of the isolated Shigella three (1.3%) were Shigella flexneri (3; 1.3%), two (0.8%) were Shigella boydii, and one was (0.4%) Shigella sonnei. The three Salmonella (3; 1.3%) isolates were S. chicago, S. caracas, and S. saintpaul (Fig. 2, Table 3). From identified positive stool samples, while five had a single infection of bacterial pathogens, two were infected by both bacterial pathogens (Table 2).
Ethidium bromide-stained 2% agarose gel showing the results of electrophoresis of products of the PCR reaction. A 496-bp band is seen in each lane with the product of the PCR for Salmonella species; bands are not seen in negative lanes. A 496-bp band is seen in lane 2, 3 and 4 with the product of the PCR for genus Salmonella. Lane 1 = ladder; Lane 2–5 = Clinical Isolate; Lane 6 = Positive control; Lane 7 = Negative control
Nineteen (7.9%) children were positive for the intestinal parasite. The most common identified intestinal protozoan was Giardia lamblia eight (3.3%), followed by Entamoeba cyst/trophozoite in five (2.1%) (Table 4). Fecal leukocytes and RBC were observed in 113 (55.6%) and 55 (23%) children, respectively. Despite the low proportion, mucoid diarrhea (2.1%) had a higher frequency of Salmonella and Shigella isolates (Table 5).
Antimicrobial susceptibility of isolated Salmonella and Shigella isolates
The antimicrobial profile for isolated Salmonella and Shigella species (n = 9) to 10 antimicrobials drugs determined by the disk diffusion method following the recommendations of the CLSI [20]. The isolated Salmonella and Shigella species had variable degrees of resistance to all antimicrobial agents tested. The susceptibility of 31.1% of the isolates was below standard resistance breakpoints for ampicillin, amoxicillin, cotrimoxazole, chloramphenicol, nalidixic acid, tetracycline, and cefotaxime. The highest enteropathogenic bacteria resistance was observed against ampicillin (88.9%) followed by tetracycline (55.6%) and cotrimoxazole (55.6%), chloramphenicol (44.4%), amoxicillin (33.3%), nalidixic acid (11.1%), and cefotaxime (11.1%). All isolates were sensitive to amikacin, ciprofloxacin, and gentamycin.
The resistance against tested ten antimicrobials was higher for Salmonella species (33.3%) than the Shigella species (30%). The highest rate of resistance observed was against ampicillin, which was 100% for Salmonella species and 83.3% for Shigella species. Conversely, the least antimicrobial resistance was to amikacin, ciprofloxacin, and gentamycin, where all isolates were sensitive. Shigella species showed no to a low level of resistance for cefotaxime and nalidixic acid (16.7%), respectively. Salmonella species showed no resistance against nalidixic acid. The resistance against the most commonly prescribed antibiotics, cotrimoxazole was 66.7% for Salmonella and 50% for Shigella isolates (Table 6).
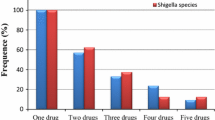
Regarding multidrug resistance, five Shigella species and two Salmonella species were multidrug-resistant. Out of the resistant species while, two (22.2%) were resistant to one antimicrobial two (22.2%) were resistant to five antibiotic agents (Table 6).
Discussion
This study intended to examine the prevalence, and antimicrobial susceptibility status of Salmonella and Shigella strains isolated from under-five children with diarrhea presented to Ambo town public health institutions. The overall prevalence of Salmonella and Shigellainfection was 3.8%, and there was a high rate of multidrug resistance, especially for Salmonella species. While the highest resistance observed against ampicillin, the least was for amikacin, ciprofloxacin, and gentamycin.
The overall prevalence of Salmonella and Shigella infection was less than the findings from Jimma, in Southwestern Ethiopia (8.4%) [7], Southern Ethiopia (22.5%) [14], and rural coastal India (11.2%) [24]. On the other hand, the magnitude of isolated Shigella species (2.5%) was comparable to findings from Jimma, Southwest Ethiopia (2.3%) [7], Nekemte (2.1%) [25], and Addis Ababa (3.2%) [13]. Conversely, the Shigella isolation rate was lower than the results from Jimma (8.8%) [13], Butajira, Central Ethiopia (4.5%) [9], and rural coastal India (4.2%) [24].
The lower Shigella and Salmonella isolation might be due in part to other potential enteric pathogen causes of diarrhea such as Rotavirus, Campylobacter species, Yersinia enterocolitica, Aeromonas species, and protozoans.
Among Shigella isolates, Shigella flexneri (12.5%) was the most dominant, followed by Shigella boydii (8.3%) and Shigella sonnei (1.4%), which is comparable with a study done in Northern India [26]. On the contrary, study findings in Jimma, Southwest Ethiopia, and Salvador, Bahia, Brazil showed that the isolates were not comparable [8, 27], respectively. The discrepancy might be due to a difference in the study population, study time, and illness due to other enteric pathogens.
The rate of identification of Salmonella species(1.3%) in this study was comparable to other studies done in Ethiopia, 1% from Hossana [28] and elsewhere 1.6% from North a [26]. However, it was lower than other studies in Ethiopia 7.8% from Bahir Dar town [12], but lower than a study done in, Ethiopia (6.2% from Jimma, [7], and other studies focused on multidrug-resistant Salmonella concord among children in Jimma (2.5%) and Addis Ababa (6.7%) [13], Hawassa (2.5%) [14], and Butajira(10.5%) [9]. The reason might be the difference in susceptibility methods breakpoints used.
There was a high rate of resistance to ampicillin for both Salmonella and Shigella species. Ampicillin is among the top dispensed drugs in Ethiopia for the last several years. The majority of such antibiotics (85%) are prescribed empirically [29]. That has made ampicillin familiar, popular, and accessible among the people, hence leading to a high rate of self-prescription. According to Mihrate et al., (2014), Ampicillin (11.1%) is the third most common self-prescribed drug only next to amoxicillin (61.1%) and, cotrimoxazole (27.8%). The self-prescription such antibiotics are common for gastrointestinal disorders [30, 31]. Similarly, high levels of resistance were reported in previous studies [32,33,34]. The public health implication of such high resistance could be associated with the suboptimal water and sanitation conditions and inadequate sewage disposal systems. This could be further complicated by the hand hygiene practice of caregivers and/or mothers.
All Salmonella and Shigella isolates in this study displayed resistance to one or more antimicrobial, including ampicillin, tetracycline, amoxicillin, cotrimoxazole, nalidixic acid, and cefotaxime. Among the isolates, there were no resistances for amikacin, ciprofloxacin and, gentamycin—except 1 and 2 intermediate for ciprofloxacin and gentamycin, respectively, which is comparable with other studies [11, 14, 24, 27, 35]. The highest antibiotic resistance of Shigella against ampicillin (83.5%) observed was comparable with a study done on Shigella isolates in Awassa (93%) [10], Jimma (70.1%) [8], Gondar (79.9%) [36], Harar (100%) [11], Jimma (100%) [7], and Southwestern Nigeria (90.5%) [37]. However, it is higher than a study done in Hawassa (63.6%) [14] and Butajira (47.1%) [9]. The differences may be due to the different susceptibility methods breakpoints used [38].
The antibiotic resistance of Shigella species to tetracycline (66.7%) was comparable with a study done in Jimma (63.6%) [8] and in Harar (70.6%) [11], but lower than a study done in other parts of Ethiopia; in Butajira (82.4%) [9], in Gondar University teaching hospital (86%) [39], in Awassa (90%) [10], and Gondar (86%) [36]. The difference might be due to strains that are moderately susceptible to tetracycline in some areas of the country. Cotrimoxazole showed 50% resistance against Shigella which is comparable with a study done in Awassa (56.0%) [10] and Addis Ababa (45.7%), [40]. Fifty percent of Shigella spp. showed resistance to amoxicillin, which was not comparable with studies done in Hawassa [14], Harar [11], Jimma [7] which count 100%, and Southwestern Nigeria which counts 81% [37]. The difference might be attributed to the difference in laboratory techniques used for the susceptibility test [38].
Antibiotic resistance against chloramphenicol 33.3% in this study is comparable with a study done in Harar (29.5%) [11], in Butajira (29.4%) [9] and Egypt [41], but was lower than a study done in Gondar University Hospital, Northwest Ethiopia [36], Awassa (63.3%) [10], and Jimma (40.3%) [8]. This result is also not comparable to a study done in Southwestern Nigeria (85.7%) [37]. The differences in the findings might be due to a biophysical environment and antibiotic resistance [42].
The study suggested that resistance shadowed a ‘selection density’. In the assumption, ecology was the basis; i.e., in a particular geographic area as the more antibiotic used for individual persons, animals, or plants, there is a high probability of bacteria to develop antibiotic resistance. Antibiotic residues from human excreta stool can have an impact on the geographical ecosystem. They can easily pass to water, and soil thought manure and sewage. So, antibiotic-resistant bacterial strains may have been found in natural water [43].
This study has a small sample size and included only children with diarrhea and restricted to a public health facility, to this end, it will be difficult to generalize beyond the study settings. Besides, restricting the finding to Shigella and Salmonella species hindered the isolation of other common causes of diarrhea. Thus, a comprehensive study should be conducted to determine the common enteric pathogens that cause diarrhea along with sensitivity tests.
Conclusion
The low prevalence of Salmonella and Shigella species was identified from the diarrhea stool of children under five years in the study area. Among those isolates, antibiotic resistance was at a high rate against ampicillin. It was followed by cotrimoxazole and tetracycline while the isolates were sensitive to amikacin, ciprofloxacin, and gentamycin.
Availability of data and materials
The datasets used and analyzed in the study are available from the corresponding author on reasonable request.
Abbreviations
- ATCC:
-
American Type Culture Collection
- ATPHI:
-
Ambo Town Public Health Institutions
- AUML:
-
Ambo University Microbiology Laboratory
- CLSI:
-
Clinical and Laboratory Standards Institute
- CSA:
-
Central Statistical Agency
- EBR:
-
Ethiopian Birr
- EPEC:
-
Enteropathogenic Escherichia coli
- EPHI:
-
Ethiopian Public Health Institution
- OPD:
-
Outpatient Departments
- Spp.:
-
Species
- SPS:
-
Statistical Package for Social Science
- SS:
-
Salmonella Shigella Medium
- USA:
-
United States of America
- WHO:
-
World Health Organization
- XLD:
-
Xylose lysine desoxycholate agar
References
Cheng AC, McDonald JR, Thielman NM. Infectious diarrhea in developed and developing countries. J Clin Gastroenterol. 2005;39(9):757–73.
Black R. Epidemiology of diarrheal diseases. Johns Hopkins University. 2007.
Boschi-Pinto C, Lanata CF, Black RE. The global burden of childhood diarrhea. World Health Organization. 2009:225–43.
UNICEF/WHO. Why children are still dying and what can be done. UNICEF/WHO. 2009:1–44.
2011 Whs: World health statistics 2011. 2011.
Hoge CW, Gambel JM, Srijan A, Pitarangsi C, Echeverria P. Trends in antibiotic resistance among diarrheal pathogens isolated in Thailand over 15 years. Clin Infect Dis. 1998;26(2):341–5.
Beyene G, Tasew H. Prevalence of intestinal parasite, Shigella and Salmonella species among diarrheal children in Jimma health center, Jimma southwest Ethiopia: a cross sectional study. Ann Clin Microbiol Antimicrob. 2014;13:10.
Mache A. Antibiotic resistance and sero-groups of shigella among paediatric out-patients in Southwest Ethiopia. East Afr Med J. 2001;78(6):296–9.
Mengistu G, Mulugeta G, Lema T, Aseffa A. Prevalence and Antimicrobial Susceptibility Patterns of Salmonella serovars and Shigella species. J Microb Biochem Technol. 2014;s2(01):1–7.
Roma B, Worku S, Mariam ST, Langeland N. Antimicrobial susceptibility pattern of Shigella isolates in Awassa. Eth J Heth Dev. 2000;14(2):149–54.
Reda AA, Seyoum B, Yimam J, Andualem G, Sisay Fiseha S, Vandeweerd J. M: antibiotic susceptibility patterns of Salmonella and Shigella isolates in Harar, Eastern Ethiopia. J Infect Dis Immun. 2011;3(8):134–9.
Yemane G, Mulaw G, Gaim T. Prevalence and antimicrobial susceptibility of Salmonella species in diarrheal children under five-years of age in Bahir Dar town, Ethiopia2278 - 1145. Int J Int sci Inn Tech Sec A. 2014;13(2):12–7.
Beyene G, Nair S, Asrat D, Mengistu Y, Engers H, Wain J. Multidrug resistant Salmonella Concord is a major cause of salmonellosis in children in Ethiopia. J Infect Dev Ctries. 2011;5(1):23–33.
Mulatu G, Beyene G, Zeynudin A. Prevalence of Shigella, Salmonella and Cmpylobacter species and their susceptibility patters among under five children with diarrhea in Hawassa town, South Ethiopia. Ethiop J Health Sci. 2014;24(2):101–8.
Ten Hove RJ, Tesfaye M, Ten Hove WF, Nigussie M. Profiling of antibiotic resistance of bacterial species recovered from routine clinical isolates in Ethiopia. Ann Clin Microbiol Antimicrob. 2017;16(1):46.
Zekaria S. Central statistical agency, 2010–07, English [eng], Ethiopia [eth] Publisher(s); 2007.
Sang WK, Oundo V, Schnabel D. Prevalence and antibiotic resistance of bacterial pathogens isolated from childhood diarrhoea in four provinces of Kenya. J Infect Dev Ctries. 2012;6(7):572–8.
Cohen ND, Neibergs HL, McGruder ED, Whitford HW, Behle RW, Ray PM, Hargis BM. Genus-specific detection of salmonellae using the polymerase chain reaction (PCR). J Vet Diagn Investig. 1993;5(3):368–71.
P G, F-X. W: Antigenic formulas of Salmonella Serotypes. 9th ed. WHO Collaborating Centre for Reference and Research on Salmonella: France. 2007.
CLSI: Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Seventeenth Informational Supplement. CLSI document M100-S17 [ISBN 1–56238–625-5]. Clinical and Laboratory Standards Institute, 940 West Valley Road, Suite 1400, Wayne, Pennsylvania 19087–1898 USA, 2007. 2007, 27.
Bartoloni A, Pallecchi L, Benedetti M, Fernandez C, Vallejos Y, Guzman E, Villagran AL, Mantella A, Lucchetti C, Bartalesi F, et al. Multidrug-resistant commensal Escherichia coli in children, Peru and Bolivia. Emerg Infect Dis. 2006;12(6):907–13.
Wright SW, Wrenn KD, Haynes M, Haas DW. Prevalence and risk factors for multidrug resistant uropathogens in ED patients. Am J Emerg Med. 2000;18(2):143–6.
Blake KS, Kellerson RL, Simic A.: Measuring Overcrowding in Housing. 2007:38.
Ballal MDSM, Chakraborty R, Shetty V. Emerging trends in the etiology and antimicrobial susceptibility pattern of enteric pathogens in rural coastal India. Int J Clin Med. 2014;5:425–32.
Terfassa A JM: Prevalence and Antibiotics Susceptibility Pattern of Salmonella and Shigella Species among Diarrheal Patients Attending Nekemte Referral Hospital, Oromia, Ethiopia. Hindawi Int J Microbiol. https://doiorg/101155/2018/9214689 2018, 2018:6.
Taneja N, Mohan B, Khurana S, Sharma M. Antimicrobial resistance in selected bacterial enteropathogens in North India. Indian J Med Res. 2004;120(1):39–43.
Santos DRD, Santana JS, Barretto JR, Andrade MGM, Silva LR. Epidemiological and microbiological aspects of acute bacterial diarrhea in children from Salvador, Bahia, Brazil. BJID. 2005;9(1):77–83.
Abebe W, Earsido A, Taye S, Assefa M, Eyasu A, Godebo G. Prevalence and antibiotic susceptibility patterns of Shigella and Salmonella among children aged below five years with Diarrhoea attending Nigist Eleni Mohammed memorial hospital, South Ethiopia. BMC Pediatr. 2018;18(1):241.
Fekadu G, Sado E, Chennupati SV, GJJoAC B. Evaluation of antibiotics use and prescribing error among hospitalized pediatric patients in Nekemte referral hospital, Ethiopia. 2017;6(4):607–19.
Mihretie TMJAAUAA, Ethiopia: self-medication practices with antibiotics among urban dwellers of Bahir Dar town, north West Ethiopia. 2014.
Sisay M, Mengistu G, Edessa DJBP, Toxicology: Epidemiology of self-medication in Ethiopia: a systematic review and meta-analysis of observational studies 2018, 19(1):56.
Aseffa A, Gedlu E, TJEAmj A. Antibiotic resistance of prevalent Salmonella and Shigella strains in northwest Ethiopia. 1997;74(11):708–13.
Asrat D. Shigella and Salmonella serogroups and their antibiotic susceptibility patterns in Ethiopia; 2008.
SJEmj G-S. Antimicrobial resistance patterns of clinical bacterial isolates in southwestern Ethiopia. 2007;45(4):363–70.
Karambu S, Matiru V, Kiptoo M, Oundo J. Characterization and factors associated with diarrhoeal diseases caused by enteric bacterial pathogens among children aged five years and below attending Igembe District Hospital, Kenya. Panafr Med J. 2013;16(37):2947.
Yismaw G, Negeri C, Kassun A. A five-year antimicrobial resistance pattern observed in Shigella species isolated from stool samples in Gondar University hospital, Northwest Ethiopia. Ethiop J Health Dev. 2006;20(3):195–8.
Efuntoye MO, Adenuga A SHIGELLA SEROTYPES AMONG NURSERY AND PRIMARY SCHOOL CHILDREN WITH DIARRHOEA IN AGO-IWOYE AND IJEBU-IGBO, SOUTHWESTERN NIGERIA. JPCS 2011, 2:28–32.
Sharp S. Verification of antimicrobial susceptibility testing methods “a practical approach”. Clin Microbiol. 2013;61.
Tiruneh M. Serodiversity and antimicrobial resistance pattern of Shigella isolates at Gondar University teaching hospital, Northwest Ethiopia. Jpn J Infect Dis. 2009;62(2):93–7.
Daniel A. Shigella and Salmonella serogroups and their antibiotic susceptibility patterns in Ethiopia. East Med He J. 2008;14(4):760–7.
Wasfy MO, Frenck R, Ismail TF, Mansour H, Malone JL, Mahoney FJ. Trends of multiple-drug resistance among Salmonella serotype Typhi isolates during a 14-year period in Egypt. Clin Infect Dis. 2002;35(10):1265–8.
Sahoo KC, Tamhankar AJ, Sahoo S, Sahu PS, Klintz SR, Lundborg CS. Geographical variation in antibiotic-resistant Escherichia coli isolates from stool, cow-dung and drinking water. Int J Environ Res Public Health. 2012;9(3):746–59.
Barbosa TM, Levy SB. The impact of antibiotic use on resistance development and persistence. Drug Resist Updat. 2000;3(5):303–11.
Acknowledgments
We would like to appreciate the staff of the microbiology laboratory at Ambo University and Addis Ababa University, Institute of Pathobiology, for their technical support of sample processing. We gratefully appreciate all parents/guardians who volunteered to provide data and guided their child for stool sample collection. We extend our thanks to Rasha Darghawth and Lauren Eby for the English language edition.
Funding
Addis Ababa University’s financial funding support was for data collection, but not for study design, interpretation, and writing manuscript. Ambo University had support reagents, material, and laboratory facilities. Supporting institutions did not participate in the writing of this paper, nor did it read or approve it.
Author information
Authors and Affiliations
Contributions
WT, AM, and TA designed the research. WT & TE performed experiments. WT, AM, AA, TE, and TA contributed to data analysis and interpretation. WT and TA wrote the manuscript. WT, AM, AA, TE, and TA critically reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical clearance was obtained from the Ethics Review Committee of Department of Microbiology, Immunology, and Parasitology, College of Health Sciences, Addis Ababa University. Formal permission were letters obtained from the District Health Offices. Informed written consent obtained from the parents/guardians of every child before taking the stool samples. Positive cases were reported to a physician or health professional. No fee was charged.
Consent for publication
Not applicable.
Competing interests
The author(s) declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Tosisa, W., Mihret, A., Ararsa, A. et al. Prevalence and antimicrobial susceptibility of Salmonella and Shigella species isolated from diarrheic children in Ambo town. BMC Pediatr 20, 91 (2020). https://doi.org/10.1186/s12887-020-1970-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12887-020-1970-0