Abstract
Background
Diarrheal diseases are responsible for high level of morbidity and mortality, particularly in children below 5 years. Salmonella and Shigella spp. are pathogenic microbes responsible for the major diarrheal associated mortality. The purpose of this study was to determine the prevalence, factors associated with Salmonella and Shigella isolates infections and their antimicrobial susceptibility patterns among diarrheic children aged below 5 years attending BRGH and GRH, Ethiopia.
Methods
A health institution based cross-sectional study was conducted from April to July 2016. One stool samples was collected from 422 diarrheic children under the ages of five and were cultured on to Hektoen Enteric (HE) and Salmonella-Shigella agar. Isolation identification of the Salmonella and Shigella isolates were conducted using standard bacteriological methods. Antibiotic susceptibility was done by Kirby–Bauer disk diffusion method. The isolates were defined as multidrug resistant if it was resistant to two or more antimicrobial agents. Descriptive statistics were employed and logistic regression models were constructed to determine factors associated with Shigella/Salmonella prevalence.
Results
The prevalence of Salmonella and Shigella isolates were 6.9 and 4.3%, respectively. Children aged between 1 to 3 years were significantly associated with Salmonella infection [AOR = 19.08, 95% CI (2.68–135.86)]. The odd of prevalence of Salmonella/Shigella isolates was significantly associated with absence of latrine, absence of hand washing after latrine, and in unimmunized children in adjusted odd ratio. Unimproved water sources and hand washing before meal had also higher odd of prevalence although the difference was not significant. All Salmonella and Shigella isolates were resistant to amoxicillin (100%). In addition, all Shigella isolates were completely resistant to chloramphenicol, and tetracycline, and were multidrug resistant. However, all Salmonella and Shigella isolates were susceptible to ciprofloxacin and ceftriaxone.
Conclusion
There was a relatively low prevalence of Salmonella and Shigella species in the study areas and were significantly associated with lack of personal hygiene and environmental sanitation. There were also higher drug resistance and multidrug resistant pattern. Personal hygiene and environmental sanitation, including access to latrine and supply of safe drinking water are suggested. Checking susceptibilities of Shigella and Salmonella isolates causing diarrhea is also suggested.
Similar content being viewed by others
Background
Diarrhea is a major cause of morbidity and mortality among children < 5 years old in sub-Saharan Africa [1]. It is the second cause of death (16%) after pneumonia in children under 5 years of age worldwide, with developing countries being the most affected [2]. The most common causes of infectious diarrhea include: viruses, bacteria, protozoa and unidentified and mixed infections [3]. Among bacterial pathogens, Salmonella and Shigella are of particular concern as causes of enteric fevers, food poisoning and gastroenteritis [3, 4].
Salmonella, with its more than 2500 different serotypes, is a leading cause of food-borne infections worldwide. Salmonella spp. causes self-limiting gastro-enteritis and the more severe forms of systemic typhoid fever. Typhoid fever is principally caused by Salmonella enteric serotype Typhi in humans. Salmonella outbreaks are related to unhygienic food preparation, cooking, reheating and storage practices that are contaminated with the pathogen [3, 4]. In addition, poor access to good latrine, poor sanitation and hygienic status, hand washing habit before and after meal and / or latrine, absence of proper sewage disposal system were responsible for typhodial type of salmonella infections [5,6,7,8,9]. Shigellosis is caused by Shigella spp. and it is a worldwide problem although more prevalent in developing countries [10,11,12]. Shigellosis is common in areas where living standards are very low and access to safe and adequate drinking water and proper waste disposal systems are often very limited, or even absent [6,7,8,9, 13,14,15,16,17]. Shigella spp. are limited to the intestinal tract of humans and cause bacillary dysentery leading to watery or bloody diarrhea.
Studies conducted in Ethiopia also revealed an increasing trend in the prevalence of Salmonella isolates and Shigella spp. [5,6,7,8,9, 14]. Antimicrobial resistance of Salmonella and Shigella are emerging global challenges, especially in developing countries where there is an increased misuse of antimicrobial agents in humans and animals [10, 11]. In Ethiopia in the past three decades, studies indicated that Salmonella and Shigella have developed varied rate of resistance against the first line antibiotics such as ampicillin, tetracycline, co-trimoxazole, chloramphenicol [5,6,7,8,9, 13, 14, 16,17,18,19,20,21,22], second generation fluoroquinolones such as norfloxacin and/or ciprofloxacin [7, 9, 13, 16, 19, 20, 22], the third generation cephalosporins (ceftriaxone) [7,8,9, 18, 20]. However, there is no published information on the magnitude of Salmonella and Shigella infection and their response to commonly prescribed drugs among diarrheic children aged below 5 years in Bale Robe General Hospital (BRGH) and Goba Referral Hospital (GRH), South Eastern Ethiopia. Understanding the magnitude and drug susceptibility pattern of Salmonella and Shigella are important in designing public health intervention measure in the area. The purpose of this study was to therefore determine the prevalence, associated factors and antimicrobial susceptibility patterns of Salmonella and Shigella isolates among children aged below 5 years with diarrhea in RGH and GRH, South Eastern Ethiopia from April 2016 up to July 2016.
Methods
Study area, design and period
Institution based cross-sectional study was conducted at BRGH and GRH from April 2016 up to July 2016 in order to determine the prevalence and antimicrobial susceptibility patterns of Salmonella and Shigella species in diarrheic children under the ages of five. BRGH and GRH are located are 430 Km and 445 Km, respectively away from Addis Ababa in South Eastern Ethiopia. The two hospitals provide health services for two major towns of the zones and to the surrounding districts.
Study population
The source population was all children under the ages of five and who lived either in Robe town, Goba town or in the adjacent rural villages during the study period. The study population was all diarrheic children under the ages of five in the selected hospitals who obtained medical service during the study period. Study population who had taken antibiotics 2 weeks before the study and with incomplete demographic information were excluded from the study.
Sample size determination and sampling techniques
The sample size (n) was determined though a single proportion formula by taking an estimated prevalence of 0.5 (for unknown prevalence) [23]. In addition, 10% of the sample size was added to the normal sample size to minimize errors [24]. A total of 422 study population was included in the study both for socio-demographic survey and stool sample collection. Simple random sampling methods were used to sample study population. Accordingly, 283 and 139 study participants were enrolled from Goba Referral Hospital and Robe General Hospital in the study, respectively.
Data and specimen collection
A structured questionnaire was used to collect data on socio-demographic characteristics (age, sex and residence) of the study population (children), and associated factors (status of drinking water, availability of latrine, hand washing habit before meal and after toilet, immunization status, availability of waste disposal system, presence of domestic animal and milk treatment status). Such data were retrieved from children parents or guardians by experienced physician or nurse (Table 1; Additional file 1). These factors were selected based on literature survey and considering personal hygiene habits and environmental sanitation of the people in the study areas. Children parents or guardians were also informed to bring freshly passed stool and rectal swab of the study subjects in a sterile stool cup using clean applicator stick. Accordingly, one stool sample was collected from 422 children under the age of five. The collected stool was placed immediately in Cary Blair transport (Oxoid, UK) and transported to laboratory for immediate processing. Instructions were given to children parents or guardians on how to collect stool samples from study children.
Culture and identification
Approximately, 1 g of stool specimen was suspended in Selenite-Cystine broth (Oxoid, UK) contained in sterile test tube for overnight. A loopful of the suspension of the specimen was streaked on to two different media namely, Hektoen Enteric (HE) Agar and Salmonella-Shigella (SS) agar, both from Oxoid, UK. These plates were incubated aerobically at 37 °C overnight to enhance the recovery of the two pathogens. The isolates were purified by subculturing on nutrient agar plates. A colorless colony with or without black center on SS agar media, and a blue green colony with or without black center on HE agar were presumably isolated as Salmonella-like isolates. A colorless colony on SS and a green, moist and raised colony on HE agar were presumably isolated as Shigella-like isolates [3, 25, 26]. Colonies exhibiting characteristic reactions of Salmonella and Shigella-like were further characterized by the pattern of biochemical reactions after inoculation on to Triple sugar iron agar, lysine iron agar, Simon’s citrate agar, and MIU test (motility test, Indole and Urease production) for final identification using the standard procedures [3, 25, 26].
Antibiotic susceptibility pattern
The antimicrobial susceptibility testing of Salmonella and Shigella isolates was done by Kirby- Bauer disc diffusion method according to Clinical and Laboratory Standards Institute [27] guidelines. Colonies from pure cultures of Salmonella or Shigella were taken and transferred to a tube containing 5 ml sterile distilled water. It is mixed gently until a homogenous suspension was formed. The suspension was incubated at 37 °C until the turbidity became adjusted to a 0.5 McFarland standard and then was uniformly inoculated on to Muller Hinton Agar (Oxoid, UK) by using sterile cotton swab under laminar hood. The inoculated plates were left at room temperature to dry for 3–5 min A sterilized forceps was then used to lightly press the antibiotic discs manually on the surface of a Muller-Hinton plate to make firm attachment. Accordingly, each isolate was subjected to 11 antibiotics discs on Muller Hinton agar (Oxoid, UK): amoxicillin (AML) (2 μg), ceftriaxone (CTR) (30 μg), ciprofloxacin (CIP) (5 μg), chloramphenicol (C) (30 μg), doxycycline (DO) (30 μg), and tetracycline (TE) (10 μg) (all from Oxoid, UK). The plates were then incubated at 37 °C for 24 h. The control disc was impregnated with sterile distilled water. Diameters of the zone of inhibition was measured to the nearest millimeter using a metallic caliper, and the isolates were classified as sensitive, intermediate and resistant according to the standardized table supplied by the CLSI [27]. Salmonella/Shigella isolate was defined as multidrug resistant if it was resistant to two or more antimicrobial agents tested [9, 18, 28].
Data quality control
In order to generate quality and reliable data, all questions were prepared in a clear and precise way and translated into two local languages (Afan Oromo and Amharic). Completeness of the questionnaire was checked whether the necessary information was properly full filled or not. All the instruments used for sample processing were checked for sterility and proper functioning. For quality control, strains of E. coli ATCC 25922, Shigella flexneri ATCC 12021, and Salmonella Typhimurium ATCC 14028 were obtained from Ethiopian Public Health Institute. The sterility of prepared media was checked by incubating one of the prepared media for 24 h at 37 °C. Growth media that showed bacterial growth were discarded.
Data analysis
The data was analyzed using IBM SPSS Statistics for Windows version 20 (IBM Corp., Armonk, NY, USA). Prevalence/ isolation rate (the outcome or dependent variable) was expressed as percentage of stool samples of study subjects showing cultured-confirmed Salmonella or Shigella isolates divided by the total number of screened stool samples of study subjects. Socio-demographic factors and associated factors were independent factors from which predictors factors for the outcome variables were identified. Descriptive statistics was used to analyze the rate of isolation of Salmonella or Shigella isolates. Bivariate and multivariable logistic regression analyses were used to compute crude ratio (COR) and adjusted odds ratio (AOR). The independent variables were checked for the presence of little or no multicollinearity using Variance Inflation Factor (VIF) (VIF = 1, not correlated; between 1.0–5.0, moderately correlated and > 5.0 highly correlated). The goodness of fit of the employed model was evaluated using the Hosmer-Lemeshow test. A p-value > 0.05 indicated that the model fit well to run the logistic regression analysis. The logistic regression analysis was done by step-wise manner. First, each independent variable and outcome variable was first evaluated by bivariate logistic regression to calculate Crude odd ratio (COR). The COR of independent variables whose 95% confidence interval (CI) excluding one were fitted to multivariable logistic regression to calculate the adjusted odd ratio (AOR). In AOR, the 95% confidence interval (CI) of odds ratio (OR) excluding one was significantly associated with the corresponding dependent variable.
Ethical consideration
This study was approved by College of Natural and Computational Science of Madda Walabu University. Ethical clearance was obtained from Bale Zone Health Office. Permission to conduct this study was obtained from each hospital administration. Both oral assent and written informed consent were obtained from the parents or guardians of the study subjects before administration of the study. Information of the study subjects and that of the parents or guardians of the study subjects were kept confidential.
Results
Socio-demographic data
A total of 422 stool samples were collected from study participants in BRGH (n = 139) and GRH (n = 283) and the response rate was 100%. Among study subjects, 179 (42.4%) were male and 243 (57.6%) were female. Out of the total study subjects, 176 (41.7%) and 246 (58.3%) lived in urban and rural areas, respectively. The age of the study participants ranged from 3 months to 60 months (mean age 31 months ±4.7 months). Among them, 129 (30.6%) were infants, 86 (20.4%) were between 1 to 3 years and 207 (49%) were between 3 to 5 years. Most of the participants utilized an improved water (81.28%). Availability of latrine, handwashing before meal and after toilet, and child immunization were recorded from 59.24, 90.3, 81.04 and 73.7% of the respondents, respectively (Table 1).
Prevalence of Salmonella and Shigella isolates
The isolation rate of Salmonella and Shigella isolates are shown in Table 1. Salmonella and Shigella isolates were positive in 29 (6.9%) and in 18 (4.3%) study subjects, respectively. 22 (7.8%) of the Salmonella isolates were from GRH, and 7 (5.0%) were from BRGH. Of the 18 Shigella isolates, 15 (5.3%) were from GRH and 3 (2.2%) were from RGH. A higher isolation rate of Salmonella and Shigella isolates were recorded from study subjects in rural areas (9.3 and 5.3%) than urban areas (3.4 and 2.3%) (Table 1).
Associated factors for Salmonella and Shigella species infections
Associated factors for the prevalence of Salmonella and Shigella infection were shown in Tables 2 and 3, respectively. Children whose ages were between 1 to 3 years were 19.8 times [AOR = 19.8; 95% CI (2.68–135.86)] susceptible to Salmonella infection than children younger than 1 year. Absence of latrine and parents who didn’t wash their hands after latrine had 3.84 times [AOR = 3.84; 95% CI (1.31–11.2)], and 12.66 times [AOR = 12.66; 95% CI (4.54–35.3)], respectively higher odd of exposing their children to Salmonella infection. Children who drank from an unimproved water sources and parents who served foods to their children before washing their hands were more likely exposing their children to Salmonella infection although the difference was not significantly different from their counterparts. Absence of child immunization [AOR = 3.54; 95% CI (1.32–9.51)] was significantly associated with Salmonella infection) (Table 2).
The odd of prevalence of Shigella infection were significantly higher in the absence of latrine [AOR = 4.58; 95% CI (1.1–18.9)], in children whose parents didn’t wash their hand after latrine [AOR = 3.31; 95% CI (1.02–10.90)]. Children who drank from unimproved water sources and that was served by parents who didn’t wash their hands before meal were also more likely exposed to Shigella infection although the difference was not significantly varied from their counterparts. Unimmunized children had about 3.63 times higher infection risk than those who were immunized [AOR = 3.63; 95% CI (1.04–12.65)] (Table 3).
Antimicrobial susceptibility test
The Salmonella and Shigella isolates displayed different rate of susceptibility to the evaluted antibiotics (Fig. 1; Table 4). All Salmonella isolates (29) and Shigella isolates (18) were 100% susceptible to ciprofloxacin, and ceftriaxone (Table 4). However, Salmonella isolates showed high rate of resistance against amoxicillin (100%) and tetracycline (79.3%), while low resistance was observed against chloramphenicol (24.1%). Salmonella isolates showed 100% intermediate susceptibility against doxycycline. The Shigella isolates showed high rate of resistance against chloramphenicol, tetracycline, and amoxicillin (100% each) and to doxycycline (88.9%) (Table 4).
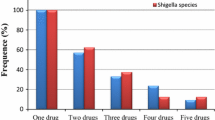
The results of multiple drug resistant (MDR) patterns of Salmonella and Shigella are presented in Table 5. Most of the Salmonella isolates (n = 23, 79.31%) and Shigella isolates (n = 18,100%) were found to be multiple drug resistant (resistant to two and above antimicrobial drugs). 14 (48.3%) and 9 (31.03%) of Salmonella isolates displayed MDR against two and three drugs, respectively. On the other hand, 2 (11.1%) and 16 (88.9%) Shigella isolates were resistant to three and four drugs, respectively (Table 5).
Discussion
This study was the first report on the prevalence and antimicrobial susceptibility pattern of Salmonella and Shigella isolates in RGH and GRH. The isolation rate of Salmonella isolates (6.9%) in our study (Table 1) was comparable with the study in Jimma, South west Ethiopia (6.2%) [6], but higher than Hawassa town, South Ethiopia (2.5%) [7], Kenya (3.5%) [29] and Botswana (3.0%) [30], and was lower than Bahir Dar, North Ethiopia (7.8%) [31] and Arba Minch, South Ethiopia (12.6%) [9]. Shigella isolates had the isolation rate of 4.3% (Table 1) in conformity with the results from Arba Minch (4.8%) [9] and Nepal (4.6%) [32], but it was lower than that results from Bahir Dar (9.5%) [31], South Ethiopia (8.3%) [33], and Botswana, Gaborone (21%) [30]. This might be due to variations in personal hygiene and environmental sanitation, and food handling practices of the community and due to climatic differences which affect the viability of infective pathogens.
In our study, children within the age range of 1–3 years were more susceptible to diarrhea caused by Salmonella (12.8%) and Shigella (7%) isolates. This finding was in agreement with several studies [6, 9, 33, 34]. Children at this age are naturally taking contaminated soils, food and water into their mouth and may acquire disease causing microbes including pathogenic Salmonella and Shigella spp. from the environment easily. Children from rural areas were more susceptible to diarrhea due to Salmonella and Shigella infection than children from urban areas, a result in conformity with study of Vargas et al. [35]. The reason might be because less awareness on personal hygiene and environmental sanitation and a reduced provision of health facilitates in rural areas compared to urban areas.
Our study revealed that study subjects who used water from unimproved sources, and had no access to latrine had higher odd of prevalence of Salmonella and Shigella isolates than their counterparts although the difference was significant for the former factor (Tables 2 and 3). It was indicated that the consumption of contaminated food and/ or water is responsible for diarrheal diseases caused by Salmonella and Shigella isolates [3, 4]. Children whose parents did not wash their hands before serving food to them and after toilet had higher odd of prevalence of Salmonella and Shigella spp. in concordance with several studies [8, 9, 15,16,17]. This was a result of fecal-oral transmission of these bacteria from the parents to their children during feeding and/or handling [3, 4]. Children whose parents didn’t possess waste disposal facility and reared domestic animals had higher odd of prevalence of Salmonella and Shigella isolates although the difference was not significant as compared to their counter parts (Tables 2 and 3). This was in concordance with studies in Arba Minch [9], in Thailand [15] and in Guinea-Bissau [36]. It is revealed that unhygienic living circumstances and close relations between humans and animals may substantially contribute to the occurrence of salmonellosis and shigellosis [11, 37].
The antimicrobial resistance of enteric pathogens such as Salmonella and Shigella have been increased all over the world as a result of indiscriminate use of antimicrobial agents [18, 32, 34, 35, 38]. High level of resistance of Salmonella isolates to amoxicillin (100%) revealed in this study was comparable with studies in different areas of Ethiopia (100%) [6, 9, 21]. The resistance of Salmonella isolates against chloramphenicol (24.1%) was comparable with study in Addis Ababa (21.7%) [8], but lower than earlier studies in Harar (62.3%) [21] and in Addis Ababa (83.7%) [18]. The current use of this drug is limited in RGH and GRH. The rate of resistance of Salmonella isolates to tetracycline in our study (79.3%) was higher than in Harar (71.4%) [21], in Hawassa town (0%) [7] and in Mozambique 15% [28]. It was revealed that antibiotic resistance of Salmonella isolates from diarrheic children in Southeastern Africa was conferred by tem-like β-lactamases for ampicillin, floR genes and CAT activity for chloramphenicol, tetA genes for tetracycline, and dfrA1 genes for trimethoprim-sulfamethoxazole/ co-trimoxazole [28]. The susceptibility of Salmonella isolates in our study to ciprofloxacin and ceftriaxone (100%) was higher and consistent with some studies in Ethiopia and elsewhere [6, 8, 28], However, ciprofloxacin resistant (6.67%) Salmonella isolates was reported from Nekemte Hospital [20]. It was reported earlier that ciprofloxacin was effective and well tolerated for treatment of typhoid fever in children [39, 40]. In addition ceftraxione resistant isolates were reported from Hawassa town (75%) [7] and Nakemete Hospital (3.33%) [20]. Ceftriaxone is safe to use, including in children, is slowly bactericidal against Salmonella serovar Typhi in vitro, and is able to penetrate and kill intracellular bacteria [41].
Our study showed that Shigella isolates were 100% resistant to three antibiotics (amoxicillin, chloramphenicol, and tetracycline). The resistance to amoxicillin in our study (100%) was very high and in conformity with previous studies in Ethiopia [6, 7]. The resistance of Shigella isolates to tetracycline in our study (100%) was also higher than earlier studies in Addis Ababa (97.3%) [18], in Bahir Dar (93.8%) [19], in Gonder (89.7%) [17] and in Mozambique 66% [28]. Similarly, the resistance of Shigella isolates to chloramphenicol (100%) was higher than studies in Arba Minch (62.5%) [9], and in Gonder (67.8%) [14]. The resistance of Shigella isolates to many drugs is due to the widespread use of antibiotics in medicine, veterinary medicine, and agriculture [3, 42]. Shigella isolates produce R plasmids that code for several resistance genes and can confer multiple antibiotic resistance [3]. Similarly, analyses of antibiotics resistance genes of Shigella isolates causing diarrhea in children under age of 5 years in South eastern Africa revealed the presence of oxa-1-like β-lactamases for ampicillin, dfrA1 genes for trimethoprim-sulfamethoxazole/co-trimoxazole, tetB genes for tetracycline and Chl acetyltransferase (CAT) activity for chloramphenicol [28]. All Shigella isolates in our study exhibited 100% susceptibility to ciprofloxacin and ceftriaxone, a result consistent with studies in Ethiopia [6, 19, 20] and in Mozambique [28]. A relatively higher rate of resistance of Shigella isolates to ciprofloxacin than our study was reported from Mekelle Hospital (6.7%) [16] and Gonder Hospital (9.2%) [22], and to ceftraxione was reported from Hawassa town (54.5%) [7] and Addis Ababa (4.3%) [8].
In our study, Salmonella isolates (79.3%) and Shigella isolates (100%) exhibited multidrug resistant pattern (Table 5). The multidrug resistant pattern of Salmonella isolates in our study were lesser than reported from Haramaya, Eastern Ethiopia (85.7%) [43], but higher than Mozambique (23%) [28]. However, the Shigella isolates in our study exhibited higher multidrug resistant pattern than those isolated from Haramaya (85.7%) [43], from Mozambique (65%) [28], from Mekele Hospital (80%) [16] and from Gonder University Teaching Hospital (79%) [13]. There are also several reports on multiple antimicrobial resistance among strains of Shigellia and Salmonella species in Ethiopia [5,6,7,8,9, 31, 33, 34].
The treatment of diarrheal diseases caused by Salmonella and Shigellla species should consider the age restrictions associated with usage of certain antimicrobials in children. Empiric therapy may be started with oral co-trimoxazole or metronidazole, but in severe cases parenteral treatment with ceftriaxone or fluoroquinolones (ciprofloxacin) might be considered [39,40,41, 44]. But, the use of fluoroquinolones and tetracyclines in the empirical treatment of diarrhea in small children is constrained by its adverse effects on musculoskeletal events and its adverse effect on the teeth younger than 8 years of age, respectively [45]. Third generation cephalosporins such as ceftraxione is the drug of choice for treatment of severe acute diarrheal caused by pathogens such as Salmonella and Shigella spp. and they have fewer adverse effects on children and effective against fluoroquinolone resistant strains of these bacterial species [45].
Conclusions
The overall prevalence of Salmonella and Shigella isolates was 6.9 and 4.3%, respectively. Children in the ages of 1 to 3 years had relatively higher isolation rate of both Salmonella and Shigella isolates compared to other ages. Drinking from an unimproved water sources, absence of latrine, absence of hand washing habit before meal and after toilet by parents, and unimmunized children resulted in higher odd of prevalence of Salmonella and Shigella infection in children. The Salmonella and Shigella isolates displayed high rate of resistance to commonly used drugs such as ampicillin,tetracycline, chloramphenicol and doxycycline. This indicates that these drugs have a reduced efficacy in the treatment of diarrhea caused by Salmonella and Shigella isolates. But, all Salmonella and Shigella isolates were highly susceptible to ciprofloxacin and ceftriaxone drugs of choice recommended for diarrheal diseases caused by these pathogens in the two hospitals. Multiple antimicrobial resistances were high among Salmonella and Shigella isolates. The limitations of this study were failure of identification of Serogroup of Salmonella and Shigella species as a results of financial constraints and lack of laboratory facilities in the study areas. Periodic awareness on personal hygiene and environmental sanitation, frequent microbiological analyses of food and water are suggested to reduce diarrhea caused by Salmonella and Shigella spp. The choice of the drugs for the treatment of diarrhea caused by Salmonella/Shigella isolates should be supported by an in vitro susceptibility studies of individual drugs.
Availability of data and materials
The original data for this study is available from the corresponding author.
Abbreviations
- (C):
-
Chloramphenicol
- (DO):
-
Doxycycline
- AML:
-
Amoxicillin
- AOR:
-
Adjusted Odds Ratio
- BRGH:
-
Bale Robe General Hospital (BRGH)
- CI:
-
Confidence Interval
- CIP:
-
Ciprofloxacin
- CLSI:
-
Clinical Laboratory Standards Institute
- COR:
-
Crude Odds Ratio
- CTR:
-
Ceftriaxone
- GRH:
-
Goba Referral Hospital
- HE:
-
Hektoen Enteric Agar
- MDR:
-
Multi-Drug Resistance
- SPSS:
-
Statistical Package for Social Sciences
- SS Agar:
-
Salmonella-Shigella Agar
- TE:
-
Tetracycline
- UK:
-
United Kingdom
- VIF:
-
Variance Inflation Factor
- WHO:
-
World Health Organization
References
Boschi-Pinto C, Young M, Black RE. The Child Health Epidemiology Reference Group reviews of the effectiveness of interventions to reduce maternal, neonatal and child mortality. Int J Epidemiol. 2010;39(Suppl 1):i3–6.
UNICEF/WHO. Diarrhea: Why children are still dying and what can be done; 2009, Accessed 15 Mar 2019.
Madigan MT, Martinko JM, Stahl DA, Clark DP. Brock biology of microorganisms. 13th ed. San Francisco: Pearson Education, Inc., Publishing; 2012.
Jay JJ, Loessener MJ, Golden DA. Modern food microbiology. 7th ed. USA: Springer Science+ Business Media, Inc.; 2005.
Beyene G, Nair S, Asrat D, Mengistu Y, Engers H, Wain J. Multidrug resistant Salmonella Concord is a major cause of salmonellosis in children in Ethiopia. J Infect Dev Ctries. 2011;5(1):023–33.
Beyene G, Tasew H. Prevalence of intestinal parasite, Shigella and Salmonella species among diarrheal children in Jimma health center, Jimma southwest Ethiopia: a cross sectional study. Ann Clin Microbiol Antimicrob. 2014;13:10. https://doi.org/10.1186/1476-0711-13-10.
Mulatu G, Beyene G, Zeynudin A. Prevalence of Shigella, Salmonella and Campylobacter species and their susceptibility pattern among under five children with diarrhea in Hawassa town, South Ethiopia. Ethiop J Health Sci. 2014;24(2):101–8.
Mamuye Y, Metaferia G, Birhanu A, Desta K, Fantaw S. Isolation and antibiotic susceptibility patterns of Shigella and Salmonella among under 5 children with acute diarrhea: a cross-sectional study at selected public health facilities in Addis Ababa, Ethiopia. Clin Microbiol. 2015;4:186.
Ameya G, Tsalla T, Getu F, Getu E. Antimicrobial susceptibility pattern, and associated factors of Salmonella and Shigella infections among under five children in Arba Minch, South Ethiopia. Ann Clin Microbiol Antimicrob. 2018;17:1. https://doi.org/10.1186/s12941-018-0253-1.
Yan H, Li L, Alam MJ, Shinoda S, Miyoshi S. Prevalence and antimicrobial resistance of Salmonella in retail foods in northern China. Int J Food Microbiol. 2010;143(3):230–4.
Kasper DL, Braunwald E, Fauci AS, Hauser SL, Longo DL, Jameson JL. Harrison’s Principles of Internal Medicine: McGraw-Hill Medical Publishing Division; 2005.
Choi SY, Jeon YS, Lee JH, Choi B, Moon SH. Multilocus sequence typing analysis of Shigella flexneri isolates collected in Asian countries. J Med Microbiol. 2007;56(Pt 11):1460–6.
Tiruneh M. Serodiversity and antimicrobial resistance pattern of Shigella isolates at Gondar University teaching hospital, Northwest Ethiopia. Jpn J Infect Dis. 2009;62(2):93–7.
Yismaw G, Negeri C, Kassu A. A five-year antimicrobial resistance pattern observed in Shigella species isolated from stool samples in Gondar University hospital Northwest Ethiopia. Ethiop J Health Dev. 2006;20(3):194–8.
Chompook P, Todd J, Wheeler JG, von Seidlein L, Clemens J, Chaicumpa W. Risk factors for shigellosis in Thailand. Int J Infect Dis. 2006;10(6):425–33.
Gebrekidan A, Dejene TA, Kahsay G, Wasihun AG. Prevalence and antimicrobial susceptibility patterns of Shigella among acute diarrheal outpatients in Mekelle hospital, Northern Ethiopia. BMC Res Notes. 2015;8:611.
Alemu A, Geta M, Taye S, Eshetie S, Engda T. Prevalence, associated risk factors and antimicrobial susceptibility patterns of Shigella infections among diarrheic pediatric population attending at Gondar town healthcare institutions, Northwest Ethiopia. Trop Dis Travel Med Vaccines. 2019;5:7.
Asrat D. Shigella and Salmonella serogroups and their antibiotic susceptibility patterns in Ethiopia. East Mediterr Health J. 2008;14(4):760–7.
Debas G, Kibret M, Biadglegne F, Abera B. Prevalence and antimicrobial susceptibility patterns of Shigella species at Felege Hiwot referral hospital, Northwest Ethiopia. Ethiop Med J. 2011;4(3):249–56.
Terfassa A, Jida M. Prevalence and antibiotics susceptibility pattern of Salmonella and Shigella species among diarrheal patients attending Nekemte referral hospital, Oromia, Ethiopia. Int J Microbiol. 2018;6:9214689.
Reda AA, Seyoum B, Yimam J, Andualem G, Fiseha S, Vandeweerd J-M. Antibiotic susceptibility patterns of Salmonella and Shigella isolates in Harar, eastern, Ethiopia. J Infect Dis Immun. 2011;3(8):134–9. https://academicjournals.org/journal/JIDI/article-abstract/E5B84D93856.
Huruy K, Kassu A, Mulu A, Gebretsadik S, Andargie G, Tadesse T, et al. High level of antimicrobial resistance in Shigella species isolate from diarrheal patients in University of Gondar Teaching Hospital, Gondar, Ethiopia. Pharmacologyonline. 2008;2:328–40.
Daniel WW. Biostatistics: A Foundation for Analysis in the Health Sciences: Wiley; 1999.
Naing L, Winn T, Rusli BN. Practical issues in calculating the sample size for prevalence practical issues in calculating the sample size for prevalence studies. Arch Orofac Sci. 2006;1:9–14.
Vandepitte J, Verhaegen J, Engbaek K, Rohner P, Piot P, Heuck CC. Basic laboratory procedures in clinical bacteriology: World Health Organization; 2003.
Cheesebrough M. District laboratory practice in tropical countries: Cambridge University Press; 2006.
CLSI, editor. Performance standards for antimicrobial susceptibility testing. 27th ed. CLSI supplement M100. Clinical and Laboratory Standards Institute: Wayne; 2017.
Mandomando I, Jaintilal D, Pons MJ, Vallès X, Espasa M, Mensa L, et al. Antimicrobial susceptibility and mechanisms of resistance in Shigella and Salmonella isolates from children under five years of age with diarrhea in rural Mozambique. Antimicrob Agents Chemother. 2009;53(6):2450–4.
Sang W, Oundo V, Schnabel D. Prevalence and antibiotic resistance of bacterial pathogens isolated from childhood diarrhea in four provinces of Kenya. J Infect Dev Ctries. 2012;6(7):572–8.
Urio EM, Collison EK, Gashe BA, Sebunya TK, Mpuchane S. Shigella and Salmonella strains isolated from children under 5 years in Gaborone, Botswana, and their antibiotic susceptibility patterns. Trop Med and Int Health. 2001;6(1):55–9.
Admassu M, Yemane G, Kibret M, Abera B, Nibret E, Adal M. Prevalence and antibiogram of Shigella and Salmonella spp. from under five children with acute diarrhea in Bahir Dar town, Ethiopia. Ethiop J Health Sci Technol. 2015;8(1):27–35.
Ansari S, Sherchand JB, Parajjuli K, Mishra SK, Dahal RK, Shrestha S, et al. Bacterial etiology of acute diarrhea in children under five years of age. Nepal J Health Resource Council. 2012;10(22):218–23.
Abebe W, Earsido A, Taye S, Assefa M, Eyasu A, Godebo G. Prevalence and antibiotic susceptibility patterns of Shigella and Salmonella among children aged below five years with diarrhea attending Nigist Eleni Mohammed memorial hospital, South Ethiopia. BMC Pediatrics. 2018;18:241.
Kibet SC, Matiru VN, Kibaba PW, Mucheke A, Louis H. Prevalence and antibiotic susceptibility patterns of Shigella and Salmonella causing Diarrhoea in children below 5 years at Thika level five district hospital. WNOFNS. 2017;11:28–36.
Vargas M, Gascón J, Casals C, Schellenberg D, Urassa H, Kahigwa E, et al. Etiology of diarrhea in children less than five years of age in Ifakara, Tanzania. Am J Trop Med Hyg. 2004;70(5):536–9.
Im J, Nichols C, Bjerregaard-Andersen M, Sow AG, Løfberg S, Tall A, et al. Prevalence of Salmonella Excretion in Stool: A Community Survey in 2 Sites, Guinea-Bissau and Senegal. Clin Infect Dis. 2016;62(Suppl 1):S50–5.
Birhaneselassie M, Williams D. A study of Salmonella carriage among asymptomatic food-handlers in southern Ethiopia. Inter J Nutr Food Sci. 2013;2(5):243–5.
Basnyat B. Typhoid fever in the United States and antibiotic choice. JAMA. 2010;303(1):34.
Thomsen LL, Paerregaard A. Treatment with ciprofloxacin in children with typhoid fever. Scand J Infect Dis. 1998;30(4):355–7.
Dutta P, Rasaily R, Saha MR, Mitra U, Bhattacharya SK, Bhattacharya MK, et al. Ciprofloxacin for treatment of severe typhoid fever in children. Antimicrob Agents Chemother. 1993;37(5):1197–9.
Acharya G, Crevoisier C, Butler T, Ho M, Tiwari M, Stoeckel K, et al. Pharmacokinetics of ceftriaxone in patients with typhoid fever. Antimicrob Agents Chemother. 1994;38(10):2415–8.
Ecker L, Olarte L, Vilchez G, Ochoa TJ, Amemiya I, Gil AI, et al. Physicians’ responsibility for antibiotic use in infants from periurban Lima, Peru. Rev Panam Salud Publica. 2011;30(6):574–9.
Marami D, Hailu K, Tolera M. Prevalence and antimicrobial Susceptibility pattern of Salmonella and Shigella species among asymptomatic food handlers working in Haramaya University cafeterias, Eastern Ethiopia. BMC Res Notes. 2018;11:74.
Bruzzese E, Giannattasio A, Guarino A. Antibiotic treatment of acute gastroenteritis in children. [version 1; referees: 2 approved]. F1000Research. 2018;7(F1000 Faculty Rev):193. https://doi.org/10.12688/f1000research.12328.1.
Diniz-Santos DR, Silva RL, Silva N. Antibiotics for the empirical treatment of acute infectious diarrhea in children. Braz J Infect Dis. 2006;10(3):217–27.
Acknowledgements
We would like to acknowledge Madda Walabu University, Department of Biology, Department of Chemistry, and Department of Laboratory Technology Department for facilitating laboratory work. All physicians and laboratory staff member BRGH and GRH are hereby acknowledged for their support in collection of study specimens. The Ethiopian Public Health Institute was acknowledged for providing us standardized bacteria used for quality control.
Funding
This study was funded by Madda Walabu University for the second author. The funds were used in the data collection, analysis, and report writing only.
Author information
Authors and Affiliations
Contributions
MG participated in the design of the study, conducted data collection, analyzed the data and drafted the paper. AA participated in the conception and design of the study, analysis and interpretation of the data and revised subsequent drafts of the paper. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by College of Natural and Computational Science of Madda Walabu University. Ethical clearance was obtained from Bale Zone Health Office. Permission was obtained from each hospital administration. Both oral assent and written informed consent were obtained from the parents or guardians of the study subjects before administration of the study. Information of the study subjects and that of the parents or guardians of the study subjects were kept confidential.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
Consent form and socio-demographic data and clinical morbidities of children aged below five years with diarrhea attending Robe General Hospital and Goba Referral Hospital, South East Ethiopia.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Assefa, A., Girma, M. Prevalence and antimicrobial susceptibility patterns of Salmonella and Shigella isolates among children aged below five years with diarrhea attending Robe General Hospital and Goba Referral Hospital, South East Ethiopia. Trop Dis Travel Med Vaccines 5, 19 (2019). https://doi.org/10.1186/s40794-019-0096-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40794-019-0096-6