Abstract
Background
Previous clinical trials have demonstrated the potential efficacy of rechallenge with anti- epidermal growth factor receptor (EGFR) monoclonal antibodies (mAbs) for patients with RAS/BRAF V600E wild-type metastatic colorectal cancer (mCRC). Moreover, post hoc biomarker analyses of clinical trials has suggested that RAS status in circulating tumor DNA (ctDNA) has a high probability to select patients who could benefit from anti-EGFR mAb rechallenge.
Methods
This trial is composed of 2 phases: a monitoring phase (REMARRY) and a trial phase (PURSUIT). A monitoring phase, the REMARRY study, aims to evaluate the dynamics of plasma RAS status during the subsequent treatments after refractory to anti-EGFR therapy in patients with mCRC with RAS/BRAF V600E wild-type tumors who have progressed after a response to previous anti-EGFR therapy, using a highly sensitive digital polymerase chain reaction OncoBEAM RAS CRC kit in a central laboratory (Sysmex, Japan). A trial phase, the PURSUIT trial, is a multicenter, single-arm phase II trial to assess the efficacy and safety of rechallenge therapy with panitumumab plus irinotecan in patients without RAS mutations in ctDNA (plasma RAS negative) in the REMARRY study. Key eligibility criteria of the PURSUIT trial include RAS/BRAF V600E wild-type mCRC in tumor tissue refractory or intolerant to fluoropyrimidine, oxaliplatin, and irinotecan; progression after complete or partial response to previous anti-EGFR therapy; plasma RAS negative (defined as plasma mutant allele frequencies [MAF] of all RAS ≤ 0.1%) within 28 days prior to enrollment; 4 months or more between the last administration of previous anti-EGFR mAb and the start of protocol treatment; and Eastern Cooperative Oncology Group (ECOG) Performance Status (PS) ≤ 1. The primary endpoint is the confirmed objective response rate (ORR). The target sample size of the PURSUIT trial is 50 patients. Biomarker analyses will be performed in parallel using the OncoBEAM RAS CRC kit and a next-generation sequencing-based ctDNA analysis (Guardant360).
Discussion
Our trial aims to confirm the clinical benefit of anti-EGFR mAb rechallenge therapy in patients with plasma RAS negative. Moreover, through biomarker analyses, our trial will shed light on which patients would benefit from rechallenge in addition to being plasma RAS negative.
Trial registration
The REMARRY study: UMIN, UMIN000036424. Registered date: April 5, 2019. The PURSUIT trial: jRCT, jRCTs031190096. Registered date: October 1, 2019.
Similar content being viewed by others
Background
Anti-epidermal growth factor receptor (EGFR), monoclonal antibodies (mAbs), panitumumab, and cetuximab are key standard drugs for patients with metastatic colorectal cancer (mCRC) with RAS wild-type tumors [1,2,3,4], achieving a median overall survival (OS) of approximately 30 months [1, 2, 5, 6]. Recently, the potential efficacy of rechallenge with anti-EGFR mAbs in a later setting for patients who had benefited from previous anti-EGFR mAb therapy has been suggested in retrospective and prospective studies [7,8,9,10,11,12,13,14,15]. The CRICKET trial, a single-arm phase II trial of rechallenge with cetuximab in 28 patients with a response to previous anti-EGFR mAbs, demonstrated a promising objective response rate (ORR) of 21% [11], whereas the Japanese phase II JACCRO-CC-08 and -09 trials showed limited efficacy of rechallenging anti-EGFR mAbs, with an ORR of 2.9–8.3% [13].
Plasma RAS status in circulating tumor DNA (ctDNA) is gaining attention as a novel predictive biomarker for the efficacy of rechallenging anti-EGFR mAbs. In the CRICKET trial, an enhanced ORR of 30% and longer progression-free survival (PFS) was observed in patients without RAS mutations in ctDNA just before the rechallenge [11]. Moreover, in a combined analysis of the JACCRO-CC-08 and -09 trials, negative for RAS mutations in ctDNA was associated with improved PFS and OS in rechallenge therapy with anti-EGFR mAbs [13]. Although post hoc analyses in clinical trials have indicated that plasma RAS status potentially predicts the efficacy of rechallenge therapy with anti-EGFR mAbs, the utility of liquid biopsy has not been prospectively validated. Furthermore, the appropriate mutant allele frequency (MAF) cut-off level in RAS mutations has not been established because a different cut-off had been adopted in each post hoc analysis.
This trial is designed to prospectively monitor plasma RAS status in patients experiencing initial response, followed by disease progression with prior chemotherapy containing anti-EGFR mAbs, and to evaluate the efficacy of rechallenge therapy with panitumumab plus irinotecan in patients negative for RAS mutations in ctDNA.
Methods/design
Overall trial design
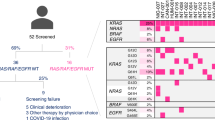
This trial is composed of 2 phases: a monitoring phase (REMARRY) and a trial phase (PURSUIT). The overall trial design is shown in Fig. 1.
Overall trial design. Liquid biopsies for OncoBEAM RAS CRC kit and/or Guardant360 will be performed in the PURSUIT trial at baseline, cycle 3, and after discontinuation of protocol treatment. C3: Cycle 3; G360: Guardant360; OncoBEAM: OncoBEAM RAS CRC kit; SOC: Standard of care. *Substitution of the result just before enrollment
Monitoring phase (REMARRY)
The REMARRY study prospectively monitors plasma RAS status after refractory to anti-EGFR therapy in mCRC patients with RAS/BRAF V600E wild-type tumors in a tumor tissue sample who have progressed after a complete or partial response to previous anti-EGFR mAb therapy, which aims to evaluate the dynamics of plasma RAS status. Plasma RAS status is measured at disease progression during subsequent therapies, using a highly sensitive digital polymerase chain reaction (PCR) OncoBEAM RAS CRC kit in a central laboratory (Sysmex, Japan).
Trial phase (PURSUIT)
The PURSUIT trial is a multicenter, single-arm phase II trial which assesses the efficacy and safety of rechallenge therapy with panitumumab plus irinotecan in patients with plasma RAS negative (defined as plasma MAF of all RAS ≤ 0.1%) in the REMARRY study.
Patient
Key eligibility criteria include RAS/BRAF V600E wild-type mCRC in tumor tissue refractory or intolerant to fluoropyrimidine, oxaliplatin, and irinotecan; progression after a complete or partial response to previous anti-EGFR mAb therapy; plasma RAS negative (MAF of all RAS ≤ 0.1%) within 28 days prior to enrollment; 4 months or more between the last administration of previous anti-EGFR mAbs and the start of protocol treatment; and Eastern Cooperative Oncology Group (ECOG) Performance Status (PS) ≤1. Details of the eligibility criteria are listed in Table 1.
Treatment
Patients will receive panitumumab 6 mg/kg plus irinotecan 150 mg/m2 biweekly until progressive disease, unacceptable toxicity, informed consent withdrawal, or patient’s death. The starting dose of irinotecan can be reduced to 120 mg/m2 or 100 mg/m2 according to adverse events during previous irinotecan therapy.
Outcomes and statistical considerations
The primary endpoint of the PURSUIT trial is the confirmed ORR, defined as the proportion of patients who achieve confirmation of complete or partial response by the investigator’s assessment with a minimum interval of 4 weeks. The secondary endpoints include PFS, time to treatment failure, duration of response, OS, disease control rate, and incidences of adverse events. Efficacy will be evaluated according to Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1, using computed tomography at 6 and 12 weeks after the start of treatment and every 8 weeks thereafter. The ORR threshold is set at 10%, based on the results of previous clinical rechallenge trials with anti-EGFR mAbs [11, 13,14,15]. The required sample size was calculated as 45, with an ORR of 25% deemed promising (one-sided α, 0.05; β, 0.15) [11]. Considering drop-outs and ineligible patients, the target sample size is 50 patients. The primary endpoint will be analyzed in a full analysis set (PURSUIT-FAS) of all patients enrolled in the PURSUIT trial, receiving at least one dose of protocol treatment and satisfying all the inclusion and exclusion criteria. All statistical analyses will be performed using SAS software, version 9.2 (SAS Institute).
Biomarker analysis
Liquid biopsies will be performed in the PURSUIT trial at baseline, cycle 3, and after discontinuation of protocol treatment. The ctDNA will be analyzed using a highly sensitive digital PCR method, OncoBEAM RAS CRC kit, and a targeted next-generation sequencing, Guardant360. Figure 1 shows at which point each analysis is performed. OncoBEAM RAS CRC kit, which uses beads, emulsion, amplification, magnetics (BEAMing) digital PCR technology, detects 34 mutations in KRAS/NRAS codons 12, 13, 59, 61, 117, and 146 in plasma [16]. This test is an in vitro diagnostic test, CE-marked in Europe and approved by the Pharmaceuticals and Medical Devices Agency in Japan to detect RAS mutations in ctDNA derived from mCRC. Several prospective and retrospective studies comparing RAS status as determined by BEAMing in plasma and the tissue reference method have reported high concordance rates, from 86.4 to 93.3% [17,18,19,20]. Guardant360 is a hybrid capture-based next-generation sequencing panel of ctDNA by Guardant Health, which is a Clinical Laboratory Improvement Amendments-certified, College of American Pathologists-accredited, New York State Department of Health-approved laboratory, as previously described [21]. Briefly, Guardant360 detects 74 gene alterations, including single nucleotide variants, indels, amplifications, and fusions, with a reportable range of ≥0.04, ≥0.02, ≥0.04%, and ≥ 2.12 copies, respectively.
Integrated analysis
Data on baseline characteristics and clinical outcomes will be collected on patients enrolled in the REMARRY study receiving rechallenge with anti-EGFR mAb in clinical practice from the PURSUIT trial (clinical practice set [plasma MAF of all RAS > 0.1%]). An integrated analysis, including PURSUIT-FAS (MAF ≤0.1%) and the clinical practice set (MAF > 0.1%), will be performed to determine a clinically significant plasma RAS MAF cut-off value.
Trial organization
This trial is supported by a nationwide cancer biomarker screening project, SCRUM-Japan [22]. Participating institutions include 28 core centers in Japan.
Discussion
Post hoc analyses of clinical trials have indicated the clinical significance of plasma RAS status at baseline as a predictive biomarker for the efficacy of rechallenge with anti-EGFR mAbs in patients with mCRC. Beyond these data, our trial will reveal some important points to select patients who benefit from rechallenge with anti-EGFR mAbs.
First, our trial’s findings will enable us to estimate the optimal cut-off value for RAS MAF in ctDNA associated with the efficacy of rechallenge with anti-EGFR mAbs. Given the cut-off values have varied in previous reports, the optimal value remains unclear. Although the absolute cut-off value is defined as 0.1% in the PURSUIT trial based on the previous retrospective or post-hoc analyses [14, 23], integrated analysis of rechallenge with anti-EGFR mAbs in PURSUIT-FAS (MAF ≤0.1%) and the clinical practice set (MAF > 0.1%) will be performed to determine the optimal cut-off value of plasma RAS.
Second, our trial could shed more light on the relationship of temporal-spatial tumor heterogeneity and rechallenge efficacy. Previous reports have focused mainly on plasma RAS status just before rechallenge; the role of plasma RAS status just after refractory to previous anti-EGFR therapy as a biomarker for rechallenge remains unknown. Moreover, it is unclear whether acquired alterations other than RAS mutations, including BRAF, EGFR, HER2, MET, and PIK3CA, affect the efficacy of rechallenge with anti-EGFR mAbs [24,25,26,27,28]. Our trial monitors serial ctDNA status from just after refractory to the previous anti-EGFR therapy using OncoBEAM RAS CRC kit and a plasma-targeted next-generation sequencing panel (Guardant360), allowing us to reveal how the dynamics of RAS mutations and other acquired alterations influence rechallenge efficacy.
Third, our trial could also clarify the significance of clinical factors in a plasma RAS-negative population. Although clinical factors, including the anti-EGFR mAb-free interval and PFS for previous anti-EGFR therapy, have been assessed in patients without a plasma RAS test, it is unknown whether clinical factors still predict the efficacy of rechallenge with anti-EGFR mAbs in patients with plasma RAS negative. Our trial will help patient selection by using clinical factors and molecular markers to enhance the efficacy of rechallenge with anti-EGFR mAbs in patients with mCRC.
Availability of data and materials
Not applicable.
Abbreviations
- ctDNA:
-
Circulating tumor DNA
- ECOG:
-
Eastern Cooperative Oncology Group
- EGFR:
-
Epidermal growth factor receptor
- mAb:
-
Monoclonal antibody
- MAF:
-
Mutant allele frequency
- mCRC:
-
Metastatic colorectal cancer
- ORR:
-
Objective response rate
- OS:
-
Overall survival
- PFS:
-
Progression-free survival
References
Douillard J-Y, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab–FOLFOX4 treatment and RAS mutations in colorectal Cancer. New Engl J Med. 2013;369(11):1023–34. https://doi.org/10.1056/NEJMoa1305275.
Cutsem EV, Lenz H-J, Köhne C-H, Heinemann V, Tejpar S, Melezínek I, et al. Fluorouracil, Leucovorin, and irinotecan plus Cetuximab treatment and RAS mutations in colorectal Cancer. J Clin Oncol. 2015;33(7):692–700. https://doi.org/10.1200/JCO.2014.59.4812.
Sobrero AF, Maurel J, Fehrenbacher L, Scheithauer W, Abubakr YA, Lutz MP, et al. EPIC: phase III trial of Cetuximab plus irinotecan after Fluoropyrimidine and Oxaliplatin failure in patients with metastatic colorectal Cancer. J Clin Oncol. 2008;26(14):2311–9. https://doi.org/10.1200/JCO.2007.13.1193.
Seymour MT, Brown SR, Middleton G, Maughan T, Richman S, Gwyther S, et al. Panitumumab and irinotecan versus irinotecan alone for patients with KRAS wild-type, fluorouracil-resistant advanced colorectal cancer (PICCOLO): a prospectively stratified randomised trial. Lancet Oncol. 2013;14(8):749–59. https://doi.org/10.1016/s1470-2045(13)70163-3.
Venook AP, Niedzwiecki D, Lenz H-J, Innocenti F, Fruth B, Meyerhardt JA, et al. Effect of first-line chemotherapy combined with Cetuximab or bevacizumab on overall survival in patients with KRASWild-type advanced or metastatic colorectal Cancer. JAMA. 2017;317(23):2392–401. https://doi.org/10.1001/jama.2017.7105.
Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran S-E, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15(10):1065–75. https://doi.org/10.1016/S1470-2045(14)70330-4.
Saif MW, Kaley K, Chu E, Copur MS. Safety and efficacy of Panitumumab therapy after progression with Cetuximab: experience at two institutions. Clin Colorect Cancer. 2010;9(5):315–8. https://doi.org/10.3816/CCC.2010.n.046.
Wadlow RC, Hezel AF, Abrams TA, Blaszkowsky LS, Fuchs CS, Kulke MH, et al. Panitumumab in patients with KRAS wild-type colorectal Cancer after progression on Cetuximab. Oncol. 2012;17:14–e34.
Pietrantonio F, Perrone F, Biondani P, Maggi C, Lampis A, Bertan C, et al. Single agent panitumumab in KRAS wild-type metastatic colorectal cancer patients following cetuximab-based regimens. Cancer Biol Ther. 2013;14(12):1098–103. https://doi.org/10.4161/cbt.26343.
Liu X, George GC, Tsimberidou AM, Naing A, Wheler JJ, Kopetz S, et al. Retreatment with anti-EGFR based therapies in metastatic colorectal cancer: impact of intervening time interval and prior anti-EGFR response. BMC Cancer. 2015;15(1):713. https://doi.org/10.1186/s12885-015-1701-3.
Cremolini C, Rossini D, Dell’Aquila E, Lonardi S, Conca E, Re MD, et al. Rechallenge for patients with RAS and BRAF wild-type metastatic colorectal Cancer with acquired resistance to first-line Cetuximab and irinotecan. JAMA Oncol. 2018;5(3):343–50. https://doi.org/10.1001/jamaoncol.2018.5080.
Rossini D, Germani MM, Pagani F, Pellino A, Dell’Aquila E, Bensi M, et al. Re-treatment with anti-EGFR antibodies in metastatic colorectal Cancer patients: a multi-institutional analysis. Clin Colorect Cancer. 2020;19(3):191–199.e6. https://doi.org/10.1016/j.clcc.2020.03.009.
Sunakawa Y, Nakamura M, Ishizaki M, Kataoka M, Satake H, Kitazono M, et al. RAS Mutations in Circulating Tumor DNA and Clinical Outcomes of Rechallenge Treatment With Anti-EGFR Antibodies in Patients With Metastatic Colorectal Cancer. Jco Precis Oncol. 2020;4:898–911.
Osawa H, Shinozaki E, Nakamura M, Ohhara Y, Shindo Y, Shiozawa M, et al. 481P Phase II study of cetuximab rechallenge in patients with ras wild-type metastatic colorectal cancer: E-rechallenge trial. Ann Oncol. 2018;29(suppl_8):viii161. https://doi.org/10.1093/annonc/mdy281.029.
Santini D, Vincenzi B, Addeo R, Garufi C, Masi G, Scartozzi M, et al. Cetuximab rechallenge in metastatic colorectal cancer patients: how to come away from acquired resistance? Ann Oncol. 2012;23(9):2313–8. https://doi.org/10.1093/annonc/mdr623.
Diehl F, Schmidt K, Durkee KH, Moore KJ, Goodman SN, Shuber AP, et al. Analysis of mutations in DNA isolated from plasma and stool of colorectal cancer patients. Gastroenterology. 2008;135(2):489–98. https://doi.org/10.1053/j.gastro.2008.05.039.
Vidal J, Muinelo L, Dalmases A, Jones F, Edelstein D, Iglesias M, et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol. 2017;28(6):1325–32. https://doi.org/10.1093/annonc/mdx125.
Grasselli J, Elez E, Caratù G, Matito J, Santos C, Macarulla T, et al. Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol. 2017;28(6):1294–301. https://doi.org/10.1093/annonc/mdx112.
García-Foncillas J, Alba E, Aranda E, Diaz-Rubio E, López-López R, Tabernero J, et al. Incorporating BEAMing technology as a liquid biopsy into clinical practice for the management of colorectal cancer patients: an expert taskforce review. Ann Oncol. 2017;28(12):2943–9. https://doi.org/10.1093/annonc/mdx501.
Bando H, Kagawa Y, Kato T, Akagi K, Denda T, Nishina T, et al. A multicentre, prospective study of plasma circulating tumour DNA test for detecting RAS mutation in patients with metastatic colorectal cancer. Brit J Cancer. 2019;120(10):982–6. https://doi.org/10.1038/s41416-019-0457-y.
Odegaard JI, Vincent JJ, Mortimer S, Vowles JV, Ulrich BC, Banks KC, et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res. 2018;24:3539–49 clincanres.3831.2017.
Bando H. The current status and problems confronted in delivering precision medicine in Japan and Europe. Curr Prob Cancer. 2017;41(3):166–75. https://doi.org/10.1016/j.currproblcancer.2017.02.003.
Peeters M, Price T, Boedigheimer M, Kim TW, Ruff P, Gibbs P, et al. Evaluation of Emergent Mutations in Circulating Cell-Free DNA and Clinical Outcomes in Patients With Metastatic Colorectal Cancer Treated With Panitumumab in the ASPECCT Study. Clin Cancer Res. 2019;25:clincanres.2072.2018. https://doi.org/10.1158/1078-0432.ccr-18-2072.
Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21(7):795–801. https://doi.org/10.1038/nm.3870.
Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532–6. https://doi.org/10.1038/nature11156.
Montagut C, Dalmases A, Bellosillo B, Crespo M, Pairet S, Iglesias M, et al. Identification of a mutation in the extracellular domain of the epidermal growth factor receptor conferring cetuximab resistance in colorectal cancer. Nat Med. 2012;18(2):221–3. https://doi.org/10.1038/nm.2609.
Diaz LA, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nat Publ Group. 2012;486:537–40.
Morelli MP, Overman MJ, Dasari A, Kazmi SMA, Mazard T, Vilar E, et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol. 2015;26(4):731–6. https://doi.org/10.1093/annonc/mdv005.
Acknowledgements
We would like to thank all the patients and their families who participated in this trial; all the co-investigators and site personnel. We also thank Independent Data Monitoring Committee (Hiroshi Osawa, Yoshihiro Kakeji, and Ayumu Hosokawa) and EPS Corporation (Aiko Toya, Hinako Yanagiya, Tomoko Nagasawa, Hideaki Takada, and Takako Hinohara).
Funding
The REMARRY study is supported by Sysmex Corporation, and the PURSUIT trial is supported by Takeda Pharmaceutical Company Limited, respectively. The funding sources provide financial support for study costs. This protocol has undergone peer-review by the funding sources. The funding sources had no role in the design, conduct, or analysis of the trial or the decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Contributions
HN and DK contributed equally to this article. HN, DK, TO, HT, and YK, as a task manager, participated in the entire coordinating of this trial, design and writing of the protocol, data collection, data analysis, data interpretation, and writing of the manuscript. YN contributed biomarker analysis using the next-generation sequencing-based ctDNA analysis. HB, TK, EO, ES, YS, KY, SY, TY1, and TY2 as the protocol preparation committee, participated in all phases of this trial, including design and writing of the protocol, data collection, data analysis, data interpretation, and preparation of the manuscript. TY1, as the chief of statistical analysis, participated in the statistical setting of trial, design, and data analysis. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The REMARRY and PURSUIT trials are conducted in accordance with the Declaration of Helsinki, the Japanese Ethical Guidelines for Medical and Health Research Involving Human Subjects, and the Clinical Trial Acts in Japan. Each trial has been approved by the institutional review board of each participating institution and National Cancer Center Hospital East Certified Review Board, respectively. Written informed consent shall be obtained from all patients before enrollment.
Consent for publication
Not applicable.
Competing interests
DK reports honoraria from Takeda, Chugai, Lilly, Merck Serono, Taiho, Ono, Bristol-Myers Squibb, and Sysmex. HB reports research funding from AstraZeneca and Sysmex and honoraria from Taiho and Lilly. TK reports honoria from Bayer, Chugai, Yakult Honsha, Sanofi, Lilly, Taiho, Takeda, Merck. EO reports honoraria from Taiho, Yakult Honsha, Merck Biopharma, Bayer, Lilly, Ono, Takeda, and Chugai. YN reports research funding from Taiho, Guardant Health, Genomedia, and Chugai. ES reports honoraria from Taiho, Chugai, Takeda, Merck Biopharma, Lilly, Sanofi, and Daiichi-Sankyo. YS reports research funding from Taiho, Takeda, Chugai, Lilly, Daiichi-Sankyo, Merck Serono, and Sanofi and honoraria from Taiho, Chugai, Takeda, Yakult Honsha, Sanofi, Bayer, Bristol Myers Squib, Merck Biopharma, Lilly, Nippon Kayaku, and Kyowa Hakko Kirin and a consulting or advisory role for Takeda and Daiichi-Sankyo. KY reports research funding from Taiho and honoraria from Daiichi-Sankyo, Lilly, Yakult Honsha, Merck Serono, Bristol Myers Squibb, Ono, MSD, Sanofi, Chugai, Takeda, Bayer, and Taiho. SY reports honoraria from Taiho, Ono, Sanofi, Bayer, Lilly, Bristol-Myers Squibb, Merck Biopharma, Chugai, Yakult Honsha, and MSD. TY1 reports research funding from Chugai, Takeda, Taiho, Daiichi Sankyo, Ono, Boehringer Ingelheim, Bayer, Merck Serono, Astellas, and Lilly and honoraria from Chugai, Takeda, Taiho, Boehringer Ingelheim, Bayer, and Pfizer; fees for consultancy from Gilead Sciences, Daiichi-Sankyo, HUYA Biosciences, and Sysmex. TY2 reports research funding from Taiho, Ono, Amgen, Parexel International, MSD, Chugai, Daiichi-Sankyo, Sumitomo Dainippon, and Sanofi and honoraria from Taiho, Takeda, Chugai, Lilly, Bayer, and Merck Biopharma. HT reports research funding from Daiichi-Sankyo, Takeda, Dainippon Sumitomo Pharma, Array BioPharma, Sysmex, MSD Oncology, Ono, and Novartis and honoraria from Mitsubishi Tanabe Pharma, Nippon Kayaku, Takeda, Taiho, Yakult Honsha, Bayer, Merck Serono, Bristol-Myers Squibb, Daiichi-Sankyo, Chugai, Sanofi, MSD, Lilly, Medical & Biological Laboratories Co Ltd., and Novartis.
YK reports research funding from Takeda and honoraria from Bayer, Chugai, Yakult Honsha, Sanofi, Lilly, Taiho, Takeda, Merck and Sysmex. The remaining authors have declared no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Nakajima, H., Kotani, D., Bando, H. et al. REMARRY and PURSUIT trials: liquid biopsy-guided rechallenge with anti-epidermal growth factor receptor (EGFR) therapy with panitumumab plus irinotecan for patients with plasma RAS wild-type metastatic colorectal cancer. BMC Cancer 21, 674 (2021). https://doi.org/10.1186/s12885-021-08395-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-021-08395-2