Abstract
Background
The endoplasmic reticulum (ER) Ca2+ sensor, stromal interaction molecule1 (STIM1) activates the plasma membrane (PM) channel Orai1 in order to mediate store-operated Ca2+ entry (SOCE) in response to ER store depletion. Enhanced expression of STIM1 in cancer tissue has been associated with poor patient prognosis. Therefore, this study investigated the functional consequences of enhanced expression of STIM1 and Orai1 in a tumor-initiating subpopulation of Huh-7 hepatocellular carcinoma (HCC) cells that express epithelial cell adhesion molecule (EpCAM) and Prominin 1 (CD133).
Methods
We performed qRT-PCR, intracellular Ca2+ monitoring, protein analyses, and real-time cell proliferation assays on EpCAM(+)CD133(+) subpopulation of tumor-initiating Huh-7 HCC cells expressing high levels of STIM1 and/or Orai1. Statistical significance between the means of two groups was evaluated using unpaired Student’s t-test.
Results
Enhanced STIM1 expression significantly increased ER Ca2+ release and proliferation rate of EpCAM(+)CD133(+) cells.
Conclusion
STIM1 overexpression may facilitate cancer cell survival by increasing ER Ca2+-buffering capacity, which makes more Ca2+ available for the cytosolic events, on the other hand, possibly preventing Ca2+-dependent enzymatic activity in mitochondria whose Ca2+ uniporter requires much higher cytosolic Ca2+ levels.
Similar content being viewed by others
Background
Hepatocellular carcinoma (HCC) appears to be the third leading cause of cancer-related deaths worldwide [1,2,3,4,5,6,7,8,9,10,11]. The primary issue in HCC cases is the high recurrence rates [12] possibly due to the existence of chemotherapy-resistant tumor-initiating cell (TIC) subpopulations [13]. Tumor-initiating cells constitute 0.01–1% of tumor mass [14, 15]. These cells express certain cell surface antigens used for separating them from other cell types within the heterogeneous tumor cell lines [16]. Epithelial cell adhesion molecule (EpCAM) and Prominin 1 (CD133) are frequently used to identify Huh-7 human HCC TICs [17, 18] as NOD/SCID mice developed tumor after receiving Huh-7 cells expressing these two antigens [19].
SOCE, a major Ca2+ influx through Ca2+-release activated Ca2+ (CRAC) channels in non-excitable cells [20,21,22,23,24,25], has been shown to be operational both in normal hepatocytes and HCC [26]. SOCE components are the ER-resident Ca2+ sensor stromal interaction molecule 1 (STIM1) [27] and the PM Ca2+ channel Orai1 [28,29,30]. The roles of STIM1 and Orai1 in carcinogenesis, tumor initiation, proliferation and metastasis have recently attracted significant attention [27, 31]. Indeed, altered expression of STIM1 and Orai1 is a hallmark of many cancer types, suggesting their potential value as prognostic biomarkers in cancer [27, 32,33,34,35].
TICs appear to be responsible for high recurrence rates as well as for chemoresistance [36]. HCC cells are a non-excitable cell type, where SOCE plays a crucial role in Ca2+ homeostasis and signaling [37]. In many cancer types including HCC, enhanced expression of STIM1 and Orai1 have been shown to enhance carcinogenesis including proliferation, migration and invasion processes [26, 38, 39]. Previous studies have reported that STIM1 and Orai1 molecules mix at a specific ratio to encode functional CRAC channel assembly [40, 41]. Based on crystallographic and electrophysiological studies, STIM1 exists as a dimer under resting conditions, and binds to Orai1 in a nonlinear fashion such that all six Orai1 binding sites must be occupied for the activation of SOCE [42]. However, the structural basis of STIM1 interaction with Orai1 within the channel assembly is not known. Therefore, the purpose of this study is to investigate the functional impact of altered stoichiometry of STIM1 and/or Orai1 by employing overexpression plasmid vectors on intracellular Ca2+ dynamics as well as carcinogenic properties of Huh-7 EpCAM(+)CD133(+) cells.
Methodsn
Cell culture
Human HCC cell lines (Huh-7) were provided by Dr. Ozturk (IBG İzmir), originally from Dr. Jack Wands Laboratory (Massachusetts General Hospital, Boston, MA) as a gift, and tested for authenticity via DNA profiling (Applied Biosystem’s Identifier kit, PN 4322288) at DNA Sequencing & Analysis Shared Resource, University of Colorado Cancer Center. The authenticity was reconfirmed by Idexx Bioresearch Company (Germany) just before initiating our studies. In addition to these, the cells have been also checked regularly in our laboratory for mycoplasma contamination by using MycoAlert Mycoplasma Detection kit (Lonza). Parental Huh-7 HCC cells and the sorted cells after Fluorescence Activated Cell Sorting (FACS, FACSAria III, BD) were maintained in complete growth medium (Dulbecco’s modified Eagle medium, DMEM, Sigma) containing 10% heat-inactivated fetal bovine serum (FBS, Biowest), 2 mM L-glutamine (Sigma) and 0.1 mM non-essential amino acids (Sigma).
Selection of EpCAM(+)CD133(+) and EpCAM(−)CD133(−) Huh-7 cells with FACS
Huh-7 HCC cells were trypsinized, washed, and resuspended in FACS buffer (1XPBS, 1 mM EDTA, 25 mM HEPES, 1% FBS) and filtered through 0.2 μm filter. Cells were passed through cell strainers with pore diameters of 100 and 30 μm (Miltenyi) to eliminate cell aggregates. Cells (15 × 106) were centrifuged to obtain pellets, then, resuspended in 105 μl FACS buffer followed by reincubation with 30 μl FcR blocking reagent (Miltenyi), 15 μl EpCAM-FITC (Miltenyi) and 15 μl CD133-PE (Miltenyi) for 10 min on ice. After incubation, cells were washed with FACS buffer and sorted via a fluorescence-activated cell sorter (FACS Aria III, BD Biosciences). Cells with and without EpCAM and/or CD133 were separately collected inside FBS containing tubes. After sorting, purity percentages for EpCAM(+)CD133(+) were determined with FACSCalibur (BD Biosciences) on the fifth day.
Transfection of EpCAM(+)CD133(+) Huh-7 cells with STIM1 and Orai1 overexpression plasmids
Cells were seeded on 6 well-plate (105 cells/well) and transfection was performed after 24 h with X-tremeGENE HP DNA Transfection Reagent (Roche). Following removal of the cell media, serum-reduced media (Opti-MEM) were added and incubated for additional 1 h. 100 μl Opti-MEM, 1.5 μg plasmid DNA (MO70-STIM1-eYFP, pDEST501-Orai1-CFP and pCMV6 empty vector as a control) and 1 μl X-tremeGENE HP DNA Reagent-containing transfection mix was added to each well and incubated for 30 min at room temperature. Transfection mix was added on the cells dropwise and shaked gently. Plasmids were gently provided by Dr. M Trebak (Penn State University).
RNA isolation and cDNA synthesis
Cells were seeded on 6-well plate (15 × 104/well). Total RNA was isolated by using High Pure RNA Isolation (Roche) according to the manufacturer’s instructions. cDNA synthesis from the total RNA samples were performed by using Transcriptor First Strand cDNA Synthesis Kit (Roche) according to the manufacturer’s instructions.
Real-time quantitative RT-PCR (qRT-PCR)
FastStart DNA Master SYBR Green I kit was used in real-time qRT-PCR experiments performed (LightCycler 1.5, Roche Applied Science). Primer sequences are shown in Table 1. All expression levels were normalized to that of internal 18S rRNA ([Target gene]/[18S rRNA] × 100).
Protein isolation and Western blot
Protein isolation was performed on 15 × 104 cells seeded on 6-well plate by cOmplete Lysis-M, EDTA-free (Roche) according to the manufacturer’s instructions. Protein extracts, separated by SDS-PAGE were transferred onto PVDF membranes, then, incubated with antibodies targeted against STIM1 (3 μg/μl, Abcam), Orai1 (1:750, Abcam) and β-actin (1:5000, Sigma) overnight at 4 °C. Membranes were incubated with secondary antibodies (1:5000, anti-rabbit or anti-mouse, LI-COR) for 1 h via shaking at room temperature. Protein bands were visualized in an infrared imager (Odyssey, LI-COR) based on the appropriate channel properties (680RD or 800CW) of secondary antibodies.
Intracellular Ca2+
Cells seeded on circular coverslips were loaded with 5 μM Fura-2/AM (Molecular Probes) in HEPES-buffered saline. Changes in intracellular Ca2+ levels were monitored via a front-surface spectrofluorometer (PTI QM8/2005) as described earlier [43].
Real-time monitoring of proliferation by real-time cell analyzer (RTCA)
Real-time label-free impedance-based monitoring of cellular proliferation assay was performed by using xCELLigence MP (Roche Applied Science). Transfected cells were incubated in 6-well plates for 48 h. After the incubation period, 5000 cells/well were seeded in E-plate 96. Cell proliferation was monitored at every 15 min for 72 h. Changes in proliferation rate were expressed as “cell index” (RTCA software 1.2.1, Roche Applied Science).
Data analysis
Data expressed as mean ± standard error of the mean (S.E.M.). “n” denotes the number of samples. Statistical significance between the means of two groups was evaluated using Student’s t-test (unpaired data). Significance was accepted at 0.05 level of probability.
Results
Selection of EpCAM(+)CD133(+) and EpCAM(−)CD133(−) Huh7 cells
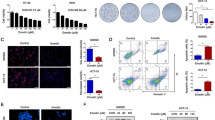
EpCAM(+)CD133(+) and EpCAM(−)CD133(−) Huh-7 cells were selected from a parental Huh-7 cell line via a FACS. Figure 1a and b show the percentages of EpCAM(+)CD133(+) and EpCAM(−)CD133(−) cells after sorting (Day 0) and on the 5th day (Day 5) Fig. 1c. On Day 5, as cells reach about 70% confluency in order to be ready for the transfection procedure, the EpCAM(+)CD133(+) cell population decreased from 96.6 to 64.3%.
EpCAM and CD133 antigen-expressing Huh-7 cell distribution after separation. a EpCAM(+)CD133(+) 96.6% in Day 0, P5 gate for EpCAM(+)CD133(+), (b) EpCAM(−)CD133(−) Huh-7 cells 99.5% in Day 0, P4 gate for EpCAM(−)CD133(−) and (c) EpCAM(+)CD133(+) in Day 5. EpCAM-FITC: fluorescein isothiocyanate conjugated EpCAM, CD133-PE: Phycoerythrin conjugated CD133
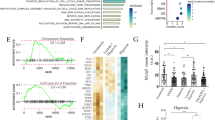
In addition to microscopic examinations, overexpression (OE) efficiency of STIM1 and Orai1 in all experimental conditions (STIM1-OE, Orai1-OE, STIM1 + Orai1-OE) on EpCAM(+)CD133(+) cells was confirmed via real time qRT-PCR. STIM1 and Orai1 expression levels were not significantly different between EpCAM(+)CD133(+) and EpCAM(−)CD133(−) cells (data not shown). In STIM1-OE and STIM1 + Orai1-OE EpCAM(+)CD133(+) cells (Fig. 2) STIM1 increased both in STIM1-OE (p < 0.05, Fig. 2a) and STIM1 + Orai1-OE cells (**p < 0.01, Fig. 2b) as expected.
Orai1 mRNA level increased in Orai1-OE (**p < 0.01, Student t-test, unpaired data n = 4, Fig. 3a) and STIM1 + Orai1-OE (**p < 0.01, Student t-test, unpaired data, n = 4, Fig. 3b) EpCAM(+)CD133(+) cells (Fig. 3) comparable to the control, which is similar to that of STIM1 mRNA expression levels revealed in previous data.
In STIM1-OE EpCAM(+)CD133(+) Huh-7 cells, STIM1 protein level was significantly higher (3 fold) than that of the control (**p < 0.01, Student t-test, unpaired data, Fig. 4a and b).
STIM1 protein expression in STIM1-OE EpCAM(+)CD133(+) Huh-7 cells. Shown are (a) STIM1 control (77 kDa) vs. STIM1-OE bands (STIM1 OE; STIM1 + eYFP ~ 103 kDa) and (b) cumulative data of STIM1 protein expression levels. STIM1 band intensities were normalized to β-actin’s (STIM1/β-actin; **p < 0.01, Student t-test, unpaired data, n = 4)
Although not statistically significant, the Orai1 protein level was lower in STIM1-OE samples comparable to that of the control (Fig. 5).
STIM1 protein levels decreased in STIM1 + Orai1-OE EpCAM(+)CD133(+) cells (p < 0.01, Fig. 6) possibly due to administration of STIM1 and Orai1 plasmids together.
STIM1 protein expression in STIM1 + Orai1-OE EpCAM(+)CD133(+) cells. Shown are (a) STIM1-OE (STIM1 + eYFP ≈103 kDa) vs. STIM1 (77 kDa) bands in WB analysis and (b) cumulative data of STIM1 protein expression levels. STIM1 band intensities were normalized to β-actin’s (STIM1/β-actin; **p < 0.01, Student t-test, unpaired data, n = 4)
Intracellular Ca2+
Intracellular basal Ca2+ levels were significantly higher in EpCAM(+)CD133(+) comparable to those of EpCAM(−)CD133(−) cells. Although Ca2+ elevation due to ER release was significantly higher in EpCAM(+)CD133(+) cells (*p < 0.05, Student t-test, unpaired data, n = 4–6), SOCE was not altered (Fig. 7).
Changes in ER Ca2+ release and SOCE in EpCAM(+)CD133(+) and EpCAM(−) CD133(−) cells. Shown are (a) EpCAM(+)CD133(+) vs. EpCAM(−)CD133(−) (99%, Fig. 1) cells (Mean ± S.E.M.) and (b) cumulative data of ER Ca2+ release and SOCE (*p < 0.05, Student t-test, unpaired data, n = 4–6). ΔF340/380: changes in intracellular Ca+ 2i levels
Although there was an apparent increase both in ER Ca2+ release and SOCE, the data did not reach statistical significance in STIM1-OE EpCAM(+)CD133(+) cells (Fig. 8). No significant change was observed in basal, ER Ca2+ release (expected) and SOCE, possibly due to increased coupling efficiency between depleted ER and Orai1. Although ER Ca2+ release and SOCE decreased in Orai1-OE EpCAM(+)CD133(+) cells, the data were not statistically significant (Fig. 9). SOCE increased significantly (p < 0.05, Fig. 10) in STIM1+ Orai1-OE cells without any change in ER Ca2+ release.
Changes in ER Ca2+ release and SOCE in STIM1-OE EpCAM(+)CD133(+) cells. Shown are (a) control vs. STIM1-OE EpCAM(+)CD133(+) (64%, Fig.1) cells (Mean ± SEM) and (b) cumulative data of ER Ca2+ release and SOCE (Student t-test, unpaired data, n = 4–6). ΔF340/380: changes in intracellular Ca+ 2 levels
Changes in ER Ca2+ release and SOCE in Orai1-OE EpCAM(+)CD133(+) cells. Shown are data from (a) control vs. Orai1 OE EpCAM(+)CD133(+) (64%, Fig. 1) cells (Mean ± S.E.M.) and (b) cumulative data of ER Ca2+ release vs. SOCE in (Student t-test, unpaired data, n = 5). ΔF340/380: changes in intracellular Ca+ 2 levels
Changes in ER Ca2+ release and SOCE in STIM1 + Orai1 overexpressed EpCAM(+)CD133(+) cells. Shown are (a) control vs. STIM1 + Orai1-OE EpCAM(+)CD133(+) (64%, Fig. 1) (Mean ± S.E.M.) and (b) cumulative data of ER Ca2+ release and SOCE (*p < 0.05, Student t-test, unpaired data, n = 5). ΔF340/380: changes in intracellular Ca+ 2 levels
Cell proliferation patterns in EpCAM(+)CD133(+) and EpCAM(−)CD133(−), STIM1-OE and STIM1 + Orai1-OE EpCAM(+)CD133(+) cells
Elevations in impedance (cell index) in RTCA show an increased cellular proliferation rate in real-time. In this study, differences in the cell proliferation pattern were monitored in two groups [EpCAM(+)CD133(+) vs. EpCAM(−)CD133(−) cells and STIM1-OE and STIM1 + Orai1-OE EpCAM(+)CD133(+) cells]. The proliferation rate at 48th h was significantly higher in EpCAM(−)CD133(−) cells comparable to that of EpCAM(+)CD133(+) (**p < 0.01, Fig. 11).
Real-time proliferation patterns of EpCAM(+)CD133(+) vs. EpCAM(−)CD133(−) cells during 48 h. Seeding density was 5000 cells/well. Shown are (a) real-time proliferation pattern of EpCAM(+)CD133(+) vs. EpCAM(−)CD133(−) and (b) cumulative cell index data (**p < 0.01, Student t-test, unpaired data, n = 24)
We also monitored the effects of STIM1 and STIM1 + Orai1 overexpression on cell proliferation in EpCAM(+)CD133(+) Huh-7 cells. Comparable to the control, STIM1-OE cells at 72nd h showed the highest proliferation rate (p < 0.01, Fig. 12); higher than that of STIM1 + Orai1-OE.
Real-time proliferation patterns of control, STIM1-OE and STIM1 + Orai1-OE EpCAM(+)CD133(+) Huh-7 cells. Cell seeding density was, 5000 cells/well. Shown are (a) real-time proliferation pattern of STIM1-OE vs. STIM1 + Orai1-OE EpCAM(+)CD133(+) during 72 h and (b) cumulative cell index data (***p < 0.001, ###p < 0.001, ##p < 0.05; *control vs. STIM1-OE, #control vs. STIM1 + Orai1-OE, Student t-test, unpaired data, n = 32)
The difference in multidrug resistance gene (MDR1) expression between tumor-initiating cells and tumor cell lines as well as the effects of STIM1 and Orai1 overexpression on MDR1 transcription in a number of experimental settings were investigated as increases in SOCE appeared to be associated with chemoresistance [44]. MDR1 mRNA levels were significantly higher in EpCAM(+)CD133(+) cells that in EpCAM(−)CD133(−) cells (**p < 0.01, Student t-test, unpaired data, n = 4, Fig. 13). Elevation of MDR1 in EpCAM(+)CD133(+) was potentiated by inducing STIM1 or an Orai1 expression and drastically increased (6-fold) by STIM1 + Orai1 overexpression (*p < 0.05, Student t-test, unpaired data, n = 4, data not shown).
Discussion
In addition to being involved in intracellular Ca2+ homeostasis mechanism of non-excitable cells, SOCE appears to be operational in hepatocellular carcinogenesis [26]. In this study, the role of SOCE components, STIM1 and Orai1, reportedly involved in intracellular Ca2+ regulation was investigated on Huh-7 TICs expressing cell surface antigens EpCAM and CD133 through monitoring intracellular Ca2+ dynamics (ER Ca2+ release and SOCE), proliferation and MDR1 expression responsible partly for drug resistance. High intracellular Ca2+ concentration comprises toxic and proapoptotic conditions for cells. Excessive Ca2+ is buffered by certain proteins (e.g., calsequestrin and calreticulin) inside ER and by mitochondria. ER Ca2+ release and SOCE are significantly higher in EpCAM(+)CD133(+) cells comparable to that of EpCAM(−) CD133(−).
Overexpression of STIM1 and Orai1 is shown in many cancer types like prostate cancer, breast cancer, glioblastoma and hepatocellular carcinoma [33]. More specifically, STIM1 overexpression is commonly seen in HCC [26, 39]. Among the three overexpression groups of EpCAM(+)CD133(+) Huh-7 cell subpopulation in our study, STIM1-OE showed the highest ER Ca2+ release. As STIM1 has Ca2+ binding EF hand domains located on the intracellular part of ER [45], its overexpression may buffer more Ca2+, leading to more Ca2+ available to be released from ER following SERCA blockade by CPA. STIM1 is the key initiating molecule in SOCE. After ER depletion, as a sensor of ER Ca2+ content, STIM1 accumulated in ER membrane closely located to PM with Orai1. At this ER and PM junctions, STIM1 interacts with Orai1 as a result SOCE is activated [46]. Lower ER release and SOCE in Orai1 OE EpCAM(+)CD133(+) Huh-7 cells comparable to the control cells could be due to changes in coupling stoichiometry between STIM1-Orai1 for SOCE [47]. Higher levels of the PM channel subunit (Orai1) might decrease effective coupling of two molecules (STIM1 and Orai1) yielding SOCE inhibition. Increases of ER release and SOCE in STIM1 + Orai1-OE EpCAM(+)CD133(+) cells, show presence of appropriate coupling stoichiometry between STIM1 and Orai1 for SOCE as both molecules are freely available for random interaction [32]. Similar SOCE elevations were also seen in STIM1 + Orai1-OE and only STIM1-OE DU145 (prostate cancer cell line) and HEK (human embryonic kidney) cells, respectively [40, 48, 49]. Overexpression of Orai1 in DU145 and HEK cells also inhibited SOCE, as observed in EpCAM(+)CD133(+) cells in our study [40, 47, 48].
TICs tended to remain in a quiescence state [50]. These EpCAM(+)CD133(+) cells have slow proliferation rates comparable to that of EpCAM(−)CD133(−) [49, 51] as was also observed in the present study. This may support their survival strategy in a cytotoxic environment [34, 52, 53]. The higher proliferation rate of STIM1-OE cells, comparable to that of STIM1 + Orai1-OE cells, showed that upregulation of these two genes (STIM1 and Orai1) suppresses the cell division/proliferation possibly through attenuated Ca2+ buffer capacity of ER. Again, the significantly higher proliferation rate observed with STIM1-OE cells over that of EpCAM(+)CD133(+) cells overexpressing both STIM1 and Orai1 (present data) confirms the poor prognosis of several cancer types with overexpressed STIM1 [54,55,56].
Cancer cells show resistance to chemotherapeutic treatments. This may result from drug inactivation, changing drug targets, DNA damage repair, and efflux of drug from cells by ABC transporters [57]. Because of the upregulated ABC transporters, cancer cells can pump chemotherapeutics out of the cell [58]. The “slow and steady” feature might also be maintained by higher MDR1 (an ABC transporter family member) expression. Upregulated MDR1 in EpCAM(+)CD133(+) Huh-7 cells in the present study is also in accordance with the increased MDR1 gene expression in lung cancer [59], ovary cancer [60], osteosarcoma [61] and glioblastoma’s [62] cancer stem cells. The signaling pathways (JAK/STAT, PI3K/AKT, MAPK/ERK), which take place in drug resistance, are regulated by Ca2+/calmodulin dependent protein kinase II (CaMKII), suggesting an interaction between Ca2+ and MDR mechanisms in liver cancer [38].
Conclusions
Based on the higher proliferation rates observed in STIM1-overexpressing EpCAM(+)CD133(+) Huh7 cells compared to that of STIM1 + Orai1-OE constructs, one may conclude that HCC stem cells might undergo a phenotypical switch process from a quiescent to proliferative stage by increasing ER Ca2+ buffering capacity due to higher levels of Ca2+-binding protein, STIM1. Furthermore, one may also speculate that increased ER Ca2+ buffering prevents Ca2+- dependent processes in mitochondria localized within the ER microenvironment by inhibiting Ca2+ uptake via low affinity/high capacity Ca2+ uniporter of mitochondria.
Availability of data and materials
The datasets used and/or analyzed in the present study are available from the corresponding author.
Abbreviations
- ABC:
-
ATP-Binding Cassette
- BSA:
-
Bovine Serum Albumin
- CD133:
-
Clustering Domain 133 (Prominin 1)
- CRAC:
-
Calcium release-activated calcium
- DMEM:
-
Dulbecco’s modified eagle medium
- EpCAM:
-
Epithelial cell adhesion molecule
- ER:
-
Endoplasmic reticulum
- FACS:
-
Fluorescence-activated cell sorting
- FITC:
-
Fluorescein isothiocyanate
- HBS:
-
HEPES-buffered saline
- HCC:
-
Hepatocellular carcinoma
- MDR:
-
Multidrug resistance
- OE:
-
Overexpressed
- PE:
-
Phytoerythrin
- PM:
-
Plasma membrane
- RTCA:
-
Real-time cell analyzer
- SDS-PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SOCE:
-
Store-operated calcium entry
- STIM:
-
Stromal interaction molecule
- TIC:
-
Tumor-initiating cells
References
Zhao YJ, Ju Q, Li GC. Tumor markers for hepatocellular carcinoma. Mol Clin Oncol. 2013;1(4):593–8.
Yang SY, Zhang JJL, Huang XY. Orai1 and STIM1 are critical for breast tumor cell migration and metastasis. Cancer Cell. 2009;15(2):124–34.
Li Y, Farmer RW, Yang Y, Martin RC. Epithelial cell adhesion molecule in human hepatocellular carcinoma cell lines: a target of chemoresistence. BMC Cancer. 2016;16(16):228.
Tam K. The roles of doxorubicin in hepatocellular carcinoma. ADMET & DMPK. 2013;1(3):29–44.
Hoffmann K, Franz C, Xiao Z, Mohr E, Serba S, Buchler MW, et al. Sorafenib modulates the gene expression of multi-drug resistance mediating ATP-binding cassette proteins in experimental hepatocellular carcinoma. Anticancer Res. 2010;30(11):4503–8.
Callegari E, Gramantieri L, Negrini M, Sabbioni S. Emerging role of microRNAs in the treatment of hepatocellular carcinoma. Gastrointest Cancer. 2015;5:89–102.
D'Anzeo M, Faloppi L, Scartozzi M, Giampieri R, Bianconi M, Del Prete M, et al. The role of micro-RNAs in hepatocellular carcinoma: from molecular biology to treatment. Molecules. 2014;19(5):6393–406.
Negahdary M, Eftekhari A, Mirzaei S, Basirizadeh M, Ghobadzadeh S. Tumor markers and hepatocellular carcinoma. J Biol Today's World. 2015;4(6):124–31.
Behne T, Copur MS. Biomarkers for hepatocellular carcinoma. Int J Hepatol. 2012;859076.
Sugimachi K, Matsumura T, Hirata H, Uchi R, Ueda M, Ueo H, et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer. 2015;112(3):532–8.
Zhu AX. Systemic therapy of advanced hepatocellular carcinoma: how hopeful should we be? Oncologist. 2006;11:790–800.
Sun JH, Luo Q, Liu LL, Song GB. Liver cancer stem cell markers: progression and therapeutic implications. World J Gastroenterol. 2016;22(13):3547–57.
Malik B, Nie D. Cancer stem cells and resistance to chemo and radio therapy. Front Biosci. 2012;4:2142–9.
Chiba T, Iwama A, Yokosuka O. Cancer stem cells in hepatocellular carcinoma: therapeutic implications based on stem cell biology. Hepatol Res. 2016;46(1):50–7.
Schwarz-Cruz-y-Celis Á, Meléndez-Zajgla J. Cancer stem cells. Rev Invest Clínica. 2011;63(2):179–86.
Kim WT, Ryu CJ. Cancer stem cell surface markers on normal stem cells. BMB Rep. 2017;50(6):285–98.
Lee TK, Cheung VC, Ng IO. Liver tumor-initiating cells as a therapeutic target for hepatocellular carcinoma. Cancer Lett. 2013;338(1):101–9.
Liu R, Shen Y, Nan KJ, Mi BB, Wu T, Guo JY, et al. Association between expression of cancer stem cell markers and poor differentiation of hepatocellular carcinoma. Medicine. 2015;94(31).
Chen Y, Yu D, Zhang H, He H, Zhang C, Zhao W, et al. CD133(+)EpCAM(+) phenotype possesses more characteristics of tumor initiating cells in hepatocellular carcinoma Huh7 cells. Int J Biol Sci. 2012;8(7):992–1004.
Tojyo Y, Morita T, Nezu A, Tanimura A. Key components of store-operated Ca2+ entry in non-excitable cells. J Pharmacol Sci. 2014;125(4):340–6.
Hogan PG, Rao A. Store-operated calcium entry: mechanisms and modulation. Biochem Biophys Res Commun. 2015;460(1):40–9.
Smyth JT, Hwang SY, Tomita T, DeHaven WI, Mercer JC, Putney JW. Activation and regulation of store-operated calcium entry. J Cell Mol Med. 2010;14(10):2337–49.
Putney JW. The physiological function of store-operated calcium entry. Neurochem Res. 2011;36(7):1157–65.
Zhan ZY, Zhong LX, Feng M, Wang JF, Liu DB, Xiong JP. Over-expression of Orai1 mediates cell proliferation and associates with poor prognosis in human non-small cell lung carcinoma. Int J Clin Exp Patho. 2015;8(5):5080–8.
Wu ZS, Qing JJ, Xia YX, Wang K, Zhang F. Suppression of stromal interaction molecule 1 inhibits SMMC7721 hepatocellular carcinoma cell proliferation by inducing cell cycle arrest. Biotechnol Appl Bioc. 2015;62(1):107–11.
Yang N, Tang Y, Wang F, Zhang H, Xu D, Shen Y, et al. Blockade of store-operated ca(2+) entry inhibits hepatocarcinoma cell migration and invasion by regulating focal adhesion turnover. Cancer Lett. 2013;330(2):163–9.
Xie J, Pan H, Yao J, Zhou Y, Han W. SOCE and cancer: recent progress and new perspectives. Int J Cancer. 2016;138(9):2067–77.
Xia JL, Wang HQ, Huang HX, Sun L, Dong ST, Huang N, et al. Elevated Orai1 and STIM1 expressions upregulate MACC1 expression to promote tumor cell proliferation, metabolism, migration, and invasion in human gastric cancer. Cancer Lett. 2016;381(1):31–40.
Motiani RK, Hyzinski-Garcia MC, Zhang X, Henkel MM, Abdullaev IF, Kuo YH, et al. STIM1 and Orai1 mediate CRAC channel activity and are essential for human glioblastoma invasion. Pflug Archiv Eur J Phy. 2013;465(9):1249–60.
Chen YF, Chiu WT, Chen YT, Lin PY, Huang HJ, Chou CY, et al. Calcium store sensor stromal-interaction molecule 1-dependent signaling plays an important role in cervical cancer growth, migration, and angiogenesis. Proc Natl Acad Sci USA. 2011;108(37):15225–30.
Venkatachalam K, van Rossum DB, Patterson RL, Ma HT, Gill DL. The cellular and molecular basis of store-operated calcium entry. Nat Cell Biol. 2002;4(11):E263–72.
Prevarskaya N, Skryma R, Shuba Y. Ion channels in Cancer: are Cancer hallmarks Oncochannelopathies? Physiol Rev. 2018;98(2):559–621.
Vashisht A, Trebak M, Motiani RK. STIM and Orai proteins as novel targets for cancer therapy. A review in the theme: cell and molecular processes in Cancer metastasis. Am J Physiol Cell Physiol. 2015;309:C457–C69.
Chen W, Dong J, Haiech J, Kilhoffer MC, Zeniou M. Cancer stem cell quiescence and plasticity as major challenges in Cancer therapy. Stem Cells Int. 2016;2016:1740936.
Zui P, JianJie MA. Open sesame: treasure in store-operated calcium entry pathway for cancer therapy. Sci China Life Sci. 2015;58(1):48–53.
Xu XL, Xing BC, Han HB, Zhao W, Hu MH, Xu ZL, Li JY, Xie Y, Gu J, Wang Y, Zhang ZQ. The properties of tumor-initiating cells from a hepatocellular carcinoma patient's primary and recurrent tumor. Carcinogenesis. 2010;31(2):167–74.
El Boustany C, Bidaux G, Enfissi A, Delcourt P, Prevarskaya N, Capiod T. Capacitative calcium entry and transient receptor potential canonical 6 expression control human hepatoma cell proliferation. Hepatology. 2008;47(6):2068–77.
Wen L, Liang C, Chen E, Chen W, Liang F, Zhi X, et al. Regulation of multi-drug resistance in hepatocellular carcinoma cells is TRPC6/calcium dependent. Sci Rep. 2016;6:23269.
Jardin I, Rosado JA. STIM and calcium channel complexes in cancer. Biochim Biophys Acta. 2016;1863(6 Pt B):1418–26.
Soboloff J, Spassova MA, Tang XD, Hewavitharana T, Xu W, Gill DL. Orai1 and STIM reconstitute store-operated calcium channel function. J Biol Chem. 2006;281(30):20661–5.
Scrimgeour N, Litjens T, Ma L, Barritt GJ, Rychkov GY. Properties of Orai1 mediated store-operated current depend on the expression levels of STIM1 and Orai1 proteins. J Physiol. 2009;587(Pt 12):2903–18.
Yen M, Lewis RS. Numbers count: how STIM and Orai stoichiometry affect store-operated calcium entry. Cell Calcium. 2019;12(79):35–43.
Selli C, Erac Y, Tosun M. Simultaneous measurement of cytosolic and mitochondrial calcium levels: observations in TRPC1-silenced hepatocellular carcinoma cells. J Pharmacol Toxicol Methods. 2015;72:29–34.
Tang BD, Xia X, Lv XF, Yu BX, Yuan JN, Mai XY, et al. Inhibition of Orai1-mediated ca(2+) entry enhances chemosensitivity of HepG2 hepatocarcinoma cells to 5-fluorouracil. J Cell Mol Med. 2017;21(5):904–15.
Huang Y, Zhou Y, Wong HC, Chen Y, Chen Y, Wang S, et al. A single EF-hand isolated from STIM1 forms dimer in the absence and presence of Ca2+. FEBS J. 2009;276(19):5589–97.
Chen YF, Chen YT, Chiu WT, Shen MR. Remodeling of calcium signaling in tumor progression. J Biomed Sci. 2013;20:23.
Soboloff J, Spassova MA, Dziadek MA, Gill DL. Calcium signals mediated by STIM and Orai proteins--a new paradigm in inter-organelle communication. Biochim Biophys Acta. 2006;1763(11):1161–8.
Xu Y, Zhang S, Niu H, Ye Y, Hu F, Chen S, et al. STIM1 accelerates cell senescence in a remodeled microenvironment but enhances the epithelial-to-mesenchymal transition in prostate cancer. Sci Rep. 2015;5:11754.
Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M, et al. Targeting notch, hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol. 2015;12(8):445–64.
Li L, Bhatia R. Stem cell quiescence. Clin Cancer Res. 2011;17(15):4936–41.
Moore N, Lyle S. Quiescent, slow-cycling stem cell populations in cancer: a review of the evidence and discussion of significance. J Oncol. 2011;2011:396076.
Vaidya A. The quintessential quiescence of cancer stem cells: a struggle towards better treatment. JCMT. 2016;2:242–4.
Takeishi S, Nakayama KI. To wake up cancer stem cells, or to let them sleep, that is the question. Cancer Sci. 2016;107(7):875–81.
Wang J, Shen J, Zhao K, Hu J, Dong J, Sun J. STIM1 overexpression in hypoxia microenvironment contributes to pancreatic carcinoma progression. Cancer Bio Med. 2019;17(1):100–8.
Wang JY, Sun J, Huang MY, Wang YS, Hou MF, Sun Y, et al. STIM1 overexpression promotes colorectal cancer progression, cell motility and COX-2 expression. Oncogene. 2015;34(33):4358–67.
Yang YJZ, Wang B, Chang L, Liu J, Zhang L, Gu L. Expression of STIM1 is associated with tumor aggressiveness and poor prognosis in breast cancer. Pathol Res Prac. 2017;9(213):1043–7.
Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N, et al. Drug resistance in cancer: an overview. Cancers. 2014;6(3):1769–92.
Fletcher JI, Haber M, Henderson MJ, Norris MD. ABC transporters in cancer: more than just drug efflux pumps. Nat Rev Cancer. 2010;10(2):147–56.
Wang B, Yang H, Huang YZ, Yan RH, Liu FJ, Zhang JN. Biologic characteristics of the side population of human small cell lung cancer cell line H446. Chin J Cancer. 2010;29(3):254–60.
Hu L, McArthur C, Jaffe RB. Ovarian cancer stem-like side-population cells are tumourigenic and chemoresistant. Br J Cancer. 2010;102(8):1276–83.
Adhikari AS, Agarwal N, Wood BM, Porretta C, Ruiz B, Pochampally RR, et al. CD117 and Stro-1 identify osteosarcoma tumor-initiating cells associated with metastasis and drug resistance. Cancer Res. 2010;70(11):4602–12.
Jin Y, Bin ZQ, Qiang H, Liang C, Hua C, Jun D, et al. ABCG2 is related with the grade of glioma and resistance to mitoxantone, a chemotherapeutic drug for glioma. J Cancer Res Clin Oncol. 2009;135(10):1369–76.
Acknowledgements
Authors acknowledge Dr. Xiaozhou Hu (Izmir Biomedicine and Genome Center, Dokuz Eylul University, Izmir, Turkey) for an excellent technical support in flow cytometry, Dr. Mehmet Ozturk (Izmir Biomedicine and Genome Center, Dokuz Eylul University, Izmir, Turkey) and Dr. Mohamed Trebak (Dept. of Cellular and Molecular Physiology, Penn State Cancer Institute, Hershey, PA, USA) for providing Huh-7 cells and plasmid vectors, respectively. Authors also thank Dr. Trebak and Dr. Donald Staub (School of Foreign Languages at Izmir University of Economics, Izmir, Turkey) for their critical comments on the manuscript.
Funding
This work was supported by the Scientific and Technological Research Council of Turkey (TUBITAK 113S399 to MT).
Author information
Authors and Affiliations
Contributions
Project proposal: YE, MT; Recipient of the project grant: MT; Experimental design: YE, MT; Experimental work: BK, YE; Analysis and interpretation: BK, YE, MT; Manuscript preparation: BK, YE, MT. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Karacicek, B., Erac, Y. & Tosun, M. Functional consequences of enhanced expression of STIM1 and Orai1 in Huh-7 hepatocellular carcinoma tumor-initiating cells. BMC Cancer 19, 751 (2019). https://doi.org/10.1186/s12885-019-5947-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-019-5947-z