Abstract
Tumor hypoxia is a common microenvironmental factor in breast cancers, resulting in stabilization of Hypoxia-Inducible Factor 1 (HIF-1), the master regulator of hypoxic response in cells. Metabolic adaptation by HIF-1 results in inhibition of citric acid cycle, causing accumulation of lactate in large concentrations in hypoxic cancers. Lactate can therefore serve as a secondary microenvironmental factor influencing cellular response to hypoxia. Presence of lactate can alter the hypoxic response of breast cancers in many ways, sometimes in opposite manners. Lactate stabilizes HIF-1 in oxidative condition, as well as destabilizes HIF-1 in hypoxia, increases cellular acidification, and mitigates HIF-1-driven inhibition of cellular respiration. We therefore tested the effect of lactate in MDA-MB-231 under hypoxia, finding that lactate can activate pathways associated with DNA replication, and cell cycling, as well as tissue morphogenesis associated with invasive processes. Using a bioengineered nano-patterned stromal invasion assay, we also confirmed that high lactate and induced HIF-1α gene overexpression can synergistically promote MDA-MB-231 dissemination and stromal trespass. Furthermore, using The Cancer Genome Atlas, we also surprisingly found that lactate in hypoxia promotes gene expression signatures prognosticating low survival in breast cancer patients. Our work documents that lactate accumulation contributes to increased heterogeneity in breast cancer gene expression promoting cancer growth and reducing patient survival.




Similar content being viewed by others
Data availability
RNAseq data for this study is deposited in the NCBI Gene Expression Omnibus (GEO) (Accession number: GSE261312).
References
Suhail Y, Cain MP, Vanaja K, Kurywchak PA, Levchenko A, Kalluri R, Kshitiz. Systems biology of cancer metastasis. Cell Syst. 2019. https://doi.org/10.1016/J.CELS.2019.07.003.
Kennel KB, Burmeister J, Schneider M, Taylor CT. The PHD1 oxygen sensor in health and disease. J Physiol. 2018. https://doi.org/10.1113/JP275327.
Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010. https://doi.org/10.1038/ONC.2009.441.
Liao C, Zhang Q. Understanding the oxygen-sensing pathway and its therapeutic implications in diseases. Am J Pathol. 2020. https://doi.org/10.1016/J.AJPATH.2020.04.003.
Wang GL, Semenza GL. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci USA. 1993. https://doi.org/10.1073/PNAS.90.9.4304.
Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006. https://doi.org/10.1016/J.CMET.2006.02.002.
Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC, Phillips GM, Cline GW, Phillips AJ, Medzhitov R. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014. https://doi.org/10.1038/NATURE13490.
Kirito K, Hu Y, Komatsu N. HIF-1 prevents the overproduction of mitochondrial ROS after cytokine stimulation through induction of PDK-1. Cell Cycle. 2009. https://doi.org/10.4161/CC.8.17.9544.
Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci USA. 2010. https://doi.org/10.1073/PNAS.0914433107.
Feng Y, Xiong Y, Qiao T, Li X, Jia L, Han Y. Lactate dehydrogenase a: a key player in carcinogenesis and potential target in cancer therapy. Cancer Med. 2018. https://doi.org/10.1002/CAM4.1820.
Manzoor AA, Schroeder T, Dewhirst MW. One-stop-shop tumor imaging: buy hypoxia, get lactate free. J Clin Invest. 2008. https://doi.org/10.1172/JCI35543.
Kshitiz AJ, Suhail Y, Chang H, Hubbi ME, Hamidzadeh A, Goyal R, Liu Y, Sun P, Nicoli S, Dang CV, Levchenko A. Lactate-dependent chaperone-mediated autophagy induces oscillatory HIF-1α activity promoting proliferation of hypoxic cells. Cell Syst. 2022. https://doi.org/10.1016/J.CELS.2022.11.003.
Ordway B, Gillies RJ, Damaghi M. Extracellular acidification induces lysosomal dysregulation. Cells. 2021. https://doi.org/10.3390/CELLS10051188.
Brisson L, Bański P, Sboarina M, Dethier C, Danhier P, Fontenille MJ, Van Hée VF, Vazeille T, Tardy M, Falces J, Bouzin C, Porporato PE, Frédérick R, Michiels C, Copetti T, Sonveaux P. Lactate dehydrogenase b controls lysosome activity and autophagy in cancer. Cancer Cell. 2016. https://doi.org/10.1016/J.CCELL.2016.08.005.
Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019. https://doi.org/10.1038/s41587-019-0201-4.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence Alignment/Map format and SAMtools. Bioinform. 2009. https://doi.org/10.1093/BIOINFORMATICS/BTP352.
Liao Y, Smyth GK, Shi W. Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinform. 2014. https://doi.org/10.1093/BIOINFORMATICS/BTT656.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014. https://doi.org/10.1186/S13059-014-0550-8/FIGURES/9.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc: Ser B (Methodol). 1995. https://doi.org/10.1111/J.2517-6161.1995.TB02031.X.
Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2021. https://doi.org/10.1101/060012.
Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000. https://doi.org/10.1093/NAR/28.1.27.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005. https://doi.org/10.1073/PNAS.0506580102/SUPPL_FILE/06580FIG7.JPG.
Place TL, Domann FE, Case AJ. Limitations of oxygen delivery to cells in culture: an underappreciated problem in basic and translational research. Free Radic Biol Med. 2017. https://doi.org/10.1016/J.FREERADBIOMED.2017.10.003.
Pias SC. How does oxygen diffuse from capillaries to tissue mitochondria? Barriers and pathways. J Physiol. 2021. https://doi.org/10.1113/JP278815.
Boonen RACM, Vreeswijk MPG, van Attikum H. CHEK2 variants: linking functional impact to cancer risk. Trends Cancer. 2022. https://doi.org/10.1016/J.TRECAN.2022.04.009.
Hubbi ME, Kshitiz GDM, Rey S, Wong CC, Luo W, Kim D-H, Dang CV, Levchenko A, Semenza GL. A nontranscriptional role for HIF-1α as a direct inhibitor of DNA replication. Sci Signal. 2013. https://doi.org/10.1126/scisignal.2003417.
Benjamin D, Robay D, Hindupur SK, Pohlmann J, Colombi M, El-Shemerly MY, Maira SM, Moroni C, Lane HA, Hall MN. Dual inhibition of the lactate transporters MCT1 and MCT4 is synthetic lethal with metformin due to NAD+ depletion in cancer cells. Cell Rep. 2018. https://doi.org/10.1016/J.CELREP.2018.11.043.
Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, Esparza LA, Reya T, Zhan L, YanxiangGuo J, White E, Rabinowitz JD. Glucose feeds the TCA cycle via circulating lactate. Nat. 2017. https://doi.org/10.1038/NATURE24057.
Wang N, Liu W, Zheng Y, Wang S, Yang B, Li M, Song J, Zhang F, Zhang X, Wang Q, Wang Z. CXCL1 derived from tumor-associated macrophages promotes breast cancer metastasis via activating NF-κB/SOX4 signaling. Cell Death Dis. 2018. https://doi.org/10.1038/s41419-018-0876-3.
Shafi AA, McNair CM, McCann JJ, Alshalalfa M, Shostak A, Severson TM, Zhu Y, Bergman A, Gordon N, Mandigo AC, Chand SN, Gallagher P, Dylgjeri E, Laufer TS, Vasilevskaya IA, Schiewer MJ, Brunner M, Feng FY, Zwart W, Knudsen KE. The circadian cryptochrome, CRY1, is a pro-tumorigenic factor that rhythmically modulates DNA repair. Nat Commun. 2021. https://doi.org/10.1038/s41467-020-20513-5.
Guo L, Li W, Zhu X, Ling Y, Qiu T, Dong L, Fang Y, Yang H, Ying J. PD-L1 expression and CD274 gene alteration in triple-negative breast cancer: implication for prognostic biomarker. Springerplus. 2016. https://doi.org/10.1186/S40064-016-2513-X.
Bajikar SS, Wang CC, Borten MA, Pereira EJ, Atkins KA, Janes KA. Tumor suppressor inactivation of GDF11 occurs by precursor sequestration in triple-negative breast cancer. Dev Cell. 2017. https://doi.org/10.1016/J.DEVCEL.2017.10.027.
Yin P, Wang W, Gao J, Bai Y, Wang Z, Na L, Sun Y, Zhao C. Fzd2 contributes to breast cancer cell mesenchymal-like stemness and drug resistance. Oncol Res. 2020. https://doi.org/10.3727/096504020X15783052025051.
Siletz A, Kniazeva E, Jeruss JS, Shea LD. Transcription factor networks in invasion-promoting breast carcinoma-associated fibroblasts. Cancer Microenviron. 2013. https://doi.org/10.1007/s12307-012-0121-z.
Furue M (2011) Epithelial tumor, invasion and stroma. Ann Dermatol
De Wever O, Mareel M. Role of tissue stroma in cancer cell invasion. J Pathol. 2003. https://doi.org/10.1002/path.1398.
Liu S, Suhail Y, Novin A, Perpetua L, Kshitiz. Metastatic transition of pancreatic ductal cell adenocarcinoma is accompanied by the emergence of pro-invasive cancer-associated fibroblasts. Cancers (Basel). 2022. https://doi.org/10.3390/CANCERS14092197.
Afzal J, Du W, Novin A, Liu Y, Wali K, Murthy A, Garen A, Wagner G, Kshitiz. Paracrine HB-EGF signaling reduce enhanced contractile and energetic state of activated decidual fibroblasts by rebalancing SRF-MRTF-TCF transcriptional axis. Front Cell Dev Biol. 2022. https://doi.org/10.3389/FCELL.2022.927631.
Novin A, Suhail Y, Ajeti V, Goyal R, Wali K, Seck A, Jackson A, Kshitiz. Diversity in cancer invasion phenotypes indicates specific stroma regulated programs. Hum Cell. 2021. https://doi.org/10.1007/S13577-020-00427-6.
Kshitiz AJ, Suhail Y, Ahn EH, Goyal R, Hubbi ME, Hussaini Q, Ellison DD, Goyal J, Nacev B, Kim DH, Lee JH, Frankel S, Gray K, Bankoti R, Chien AJ, Levchenko A. Control of the interface between heterotypic cell populations reveals the mechanism of intercellular transfer of signaling proteins. Integr Biol (Camb). 2015. https://doi.org/10.1039/C4IB00209A.
De Neve J, Gerds TA. On the interpretation of the hazard ratio in Cox regression. Biom J. 2020. https://doi.org/10.1002/BIMJ.201800255.
Ilaslan E, Sajek MP, Jaruzelska J, Kusz-Zamelczyk K. Emerging roles of NANOS RNA-binding proteins in cancer. Int J Mol Sci. 2022. https://doi.org/10.3390/IJMS23169408.
Strumane K, Bonnomet A, Stove C, Vandenbroucke R, Nawrocki-Raby B, Bruyneel E, Mareel M, Birembaut P, Berx G, Van Roy F. E-cadherin regulates human Nanos1, which interacts with p120ctn and induces tumor cell migration and invasion. Cancer Res. 2006. https://doi.org/10.1158/0008-5472.CAN-05-3096.
Lee CH, Pao JB, Lu TL, Lee HZ, Lee YC, Liu CC, Huang CY, Lin VC, Yu CC, Yin HL, Huang SP, Bao BY. Prognostic value of prostaglandin-endoperoxide synthase 2 polymorphisms in prostate cancer recurrence after radical prostatectomy. Int J Med Sci. 2016. https://doi.org/10.7150/IJMS.16259.
Singh-Ranger G, Salhab M, Mokbel K. The role of cyclooxygenase-2 in breast cancer: review. Breast Cancer Res Treat. 2008. https://doi.org/10.1007/s10549-007-9641-5.
Patani N, Douglas-Jones A, Mansel R, Jiang W, Mokbel K. Tumour suppressor function of MDA-7/IL-24 in human breast cancer. Cancer Cell Int. 2010. https://doi.org/10.1186/1475-2867-10-29/FIGURES/4.
Qi SM, Cheng G, Cheng XD, Xu Z, Xu B, Zhang WD, Qin JJ. Targeting USP7-Mediated Deubiquitination of MDM2/MDMX-p53 pathway for cancer therapy: are we there yet? Front Cell Dev Biol. 2020. https://doi.org/10.3389/FCELL.2020.00233/BIBTEX.
Lin P, Li J, Ye F, Fu W, Hu X, Shao Z, Song C. KCNN4 induces multiple chemoresistance in breast cancer by regulating BCL2A1. Am J Cancer Res. 2020;10:3302.
Chadet S, Allard J, Brisson L, Lopez-Charcas O, Lemoine R, Heraud A, Lerondel S, Guibon R, Fromont G, Le Pape A, Angoulvant D, Jiang LH, Murrell-Lagnado R, Roger S. P2x4 receptor promotes mammary cancer progression by sustaining autophagy and associated mesenchymal transition. Oncogene. 2022. https://doi.org/10.1038/s41388-022-02297-8.
Wicks EE, Semenza GL. Hypoxia-inducible factors: cancer progression and clinical translation. J Clin Invest. 2022. https://doi.org/10.1172/JCI159839.
Terry S, Engelsen AST, Buart S, Elsayed WS, Venkatesh GH, Chouaib S. Hypoxia-driven intratumor heterogeneity and immune evasion. Cancer Lett. 2020. https://doi.org/10.1016/J.CANLET.2020.07.004.
Qiu GZ, Jin MZ, Dai JX, Sun W, Feng JH, Jin WL. Reprogramming of the tumor in the hypoxic niche: the emerging concept and associated therapeutic strategies. Trends Pharmacol Sci. 2017. https://doi.org/10.1016/j.tips.2017.05.002.
Omer S, Karunagaran D, Suraishkumar GK. The role of circadian and redox rhythms in cancer hypoxia. Adv in Redox Res. 2021. https://doi.org/10.1016/J.ARRES.2021.100018.
Skala MC, Fontanella A, Lan L, Izatt JA, Dewhirst MW. Longitudinal optical imaging of tumor metabolism and hemodynamics. J Biomed Opt. 2010. https://doi.org/10.1117/1.3285584).
Bader SB, Dewhirst MW, Hammond EM. Cyclic hypoxia: an update on its characteristics, methods to measure it and biological implications in cancer. Cancers (Basel). 2021. https://doi.org/10.3390/CANCERS13010023.
Jamali S, Klier M, Ames S, Felipe Barros L, McKenna R, Deitmer JW, Becker HM. Hypoxia-induced carbonic anhydrase IX facilitates lactate flux in human breast cancer cells by non-catalytic function. Sci Rep. 2015. https://doi.org/10.1038/srep13605.
Wang ZH, Peng WB, Zhang P, Yang XP, Zhou Q. Lactate in the tumour microenvironment: from immune modulation to therapy. EBioMedicine. 2021. https://doi.org/10.1016/J.EBIOM.2021.103627.
Eggleton GP, Eggleton| P, Hill A V The Coefficient of Diffusion of Lactic Acid through Muscle
Ju LK. Water-in-oil cultivation technology for viscous xanthan gum fermentation. Bioprocess Value-Added Prod Renew Resour Tech App. 2006. https://doi.org/10.1016/B978-044452114-9/50016-5.
Hubbi ME, Luo W, Baek JH, Semenza GL. MCM proteins are negative regulators of hypoxia-inducible factor 1. Mol Cell. 2011. https://doi.org/10.1016/J.MOLCEL.2011.03.029.
Fata JE, Werb Z, Bissell MJ. Regulation of mammary gland branching morphogenesis by the extracellular matrix and its remodeling enzymes. Breast Cancer Res. 2004. https://doi.org/10.1186/BCR634/FIGURES/3.
Gray RS, Cheung KJ, Ewald AJ. Cellular mechanisms regulating epithelial morphogenesis and cancer invasion. Curr Opin Cell Biol. 2010. https://doi.org/10.1016/J.CEB.2010.08.019.
Karthik S, Djukic T, Kim JD, Zuber B, Makanya A, Odriozola A, Hlushchuk R, Filipovic N, Jin SW, Djonov V. Synergistic interaction of sprouting and intussusceptive angiogenesis during zebrafish caudal vein plexus development. Sci Rep. 2018. https://doi.org/10.1038/s41598-018-27791-6.
Kshitiz AJ, Maziarz JD, Hamidzadeh A, Liang C, Erkenbrack EM, Nam H, Haeger JD, Pfarrer C, Hoang T, Ott T, Spencer T, Pavličev M, Antczak DF, Levchenko A, Wagner GP. Evolution of placental invasion and cancer metastasis are causally linked. Nat Ecol Evol. 2019. https://doi.org/10.1038/S41559-019-1046-4.
Suhail Y, Afzal J, Kshitiz. Evolved resistance to placental invasion secondarily confers increased survival in melanoma patients. J Clin Med. 2021. https://doi.org/10.3390/JCM10040595.
Pantel AR, Ackerman D, Lee SC, Mankoff DA, Gade TP. Imaging cancer metabolism: underlying biology and emerging strategies. J Nucl Med. 2018. https://doi.org/10.2967/JNUMED.117.199869.
Cheung SM, Husain E, Masannat Y, Miller ID, Wahle K, Heys SD, He J. Lactate concentration in breast cancer using advanced magnetic resonance spectroscopy. Br J Cancer. 2020. https://doi.org/10.1038/s41416-020-0886-7.
Acknowledgements
We thank National Cancer Institute R37CA248161 for funding the research presented in the manuscript.
Funding
National Cancer Institute, 1R37CA248161, Kshitiz
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
13577_2024_1046_MOESM1_ESM.tif
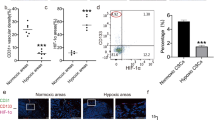
Supplementary file1 Mcm5 nuclear levels in hypoxia are reversed by lactate. Representative immunofluorescence images of MDA-MB-231 cells showing Mcm5 levels (green), and DAPI (blue) marking nuclei in normoxia, DMOG, hypoxia, as well as hypoxia with lactate, and NH4H2PO4, a weak acid control for lactate. Quantification in Figure 1G. immunofluorescence images of MDA-MB-231 cells showing Mcm5 levels (green), and DAPI (blue) marking nuclei in normoxia, DMOG, hypoxia, as well as hypoxia with lactate, and NH4H2PO4, a weak acid control for lactate. Quantification in Figure 1G. (TIF 5469 KB)
13577_2024_1046_MOESM2_ESM.tif
Supplementary file2 Lactate can reduce hypoxia-induced HIF-1a levels. Representative immunofluorescence images of MDA-MB-231 cells showing HIF-1α levels (green), and DAPI (blue) marking nuclei in normoxia, DMOG, hypoxia, as well as hypoxia with lactate, and NH4H2PO4, a weak acid control for lactate. Quantification in Figure 2C; images also shown in 2B for limited conditions. (TIF 3494 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, Y., Suhail, Y., Novin, A. et al. Lactate in breast cancer cells is associated with evasion of hypoxia-induced cell cycle arrest and adverse patient outcome. Human Cell 37, 768–781 (2024). https://doi.org/10.1007/s13577-024-01046-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-024-01046-1