Abstract
Background
Breast density and single-nucleotide polymorphisms (SNPs) have both been associated with breast cancer risk. To determine the extent to which these two breast cancer risk factors are associated, we investigate the association between a panel of validated SNPs related to breast cancer and quantitative measures of mammographic density in a cohort of Caucasian and African-American women.
Methods
In this IRB-approved, HIPAA-compliant study, we analyzed a screening population of 639 women (250 African American and 389 Caucasian) who were tested with a validated panel assay of 12 SNPs previously associated to breast cancer risk. Each woman underwent digital mammography as part of routine screening and all were interpreted as negative. Both absolute and percent estimates of area and volumetric density were quantified on a per-woman basis using validated software. Associations between the number of risk alleles in each SNP and the density measures were assessed through a race-stratified linear regression analysis, adjusted for age, BMI, and Gail lifetime risk.
Results
The majority of SNPs were not found to be associated with any measure of breast density. SNP rs3817198 (in LSP1) was significantly associated with both absolute area (p = 0.004) and volumetric (p = 0.019) breast density in Caucasian women. In African-American women, SNPs rs3803662 (in TNRC9/TOX3) and rs4973768 (in NEK10) were significantly associated with absolute (p = 0.042) and percent (p = 0.028) volume density respectively.
Conclusions
The majority of SNPs investigated in our study were not found to be significantly associated with breast density, even when accounting for age, BMI, and Gail risk, suggesting that these two different risk factors contain potentially independent information regarding a woman’s risk to develop breast cancer. Additionally, the few statistically significant associations between breast density and SNPs were different for Caucasian versus African American women. Larger prospective studies are warranted to validate our findings and determine potential implications for breast cancer risk assessment.
Similar content being viewed by others
Background
Breast cancer is currently the most commonly diagnosed cancer and the second leading cause of cancer death in women in the US [1]. Recently, there has been focus on the personalization of breast cancer screening recommendations [2] based on measurable factors known to influence an individual woman’s risk for breast cancer [3]. Of these, breast density has emerged as one of the strongest risk factors for breast cancer [4-15], which can potentially allow for substantial improvements in breast cancer risk estimation.
Mammographic density, the most broadly used measure of breast density, represents the relative amount of radiographically-opaque fibroglandular tissue versus radiographically-translucent adipose tissue in the breast. Commonly measured via visual assessment either qualitatively using the American College of Radiology Breast Imaging-Reporting and Data System (BI-RADS) density categories [16,17], or quantitatively as percent density (PD%) using semi-automated tools [4,18], it has been shown to lead to improvements in breast cancer risk assessment [19-23]. More recently, fully-automated tools have also been developed [13,24,25] which hold the promise to provide more accurate quantitative estimates of density for breast cancer risk evaluation.
To date, the etiological pathways underlying the increase in breast cancer risk due to the presence of dense tissue are not yet clearly understood [26,27]. Breast density is thought to have a polygenic basis [28,29], and identifying which genes are involved in the formation of the dense tissue could elucidate potential pathways linking breast density and breast cancer formation. Genome-wide association studies have identified multiple low and moderate penetrance breast cancer susceptibility loci in women, commonly referred to as single nucleotide polymorphisms (SNPs), associated with both overall and sub-type specific risk [30] that may be useful in breast cancer risk assessment [31-35]. As such, it would be important to determine whether such genetic risk factors are associated with breast density or whether they are potentially independent predictors of a woman’s risk to develop breast cancer.
In this context, we investigate associations between a panel of validated SNPs related to breast cancer risk and quantitative measures of mammographic density in a race-stratified cohort. Given the increasing interest in identifying which measures of breast density are most related to breast cancer risk [36], we evaluate these associations using both area and volumetric density measures. Ultimately, understanding the relationship between breast density and genetic risk factors for breast cancer could provide further insight into the etiological pathways driving the association between breast density and cancer risk. Furthermore, by exploring these associations we can begin to understand how these risk factors relate to each other and how they could be leveraged jointly in breast cancer risk assessment, should they contain independent information.
Methods
Study population
In this University of Pennsylvania Institutional Review Board (IRB) approved, HIPAA compliant study, we retrospectively identified a cohort of women aged 40 years or older from our routine breast screening population who had also been prospectively recruited by a separate, IRB-approved, HIPAA compliant clinical study at our institution investigating the added value of genomic markers in breast cancer risk prediction [37]. For the purposes of our study, informed consent was waived, as this was a retrospective analysis and these women were already consented for research purposes in the original study [37] at the time of their recruitment. Each of these women was imaged as part of their routine screening with a full-field digital mammography (FFDM) system (Selenia Dimensions, Hologic Inc.) under a standard protocol. From a total of 810 women originally recruited, a total of 670 had raw (i.e., “FOR PROCESSING”) digital images available on record for quantitative analysis. All these women were interpreted as negative (BI-RADS 1 or 2 screening outcome), and confirmed with at least 1 year follow-up. Information regarding each woman’s current age, demographic and reproductive history, height, weight and race was collected via self-report. Gail lifetime risk, the probability that a woman will develop invasive or in situ breast cancer in a specified time period, was estimated using the National Cancer Institute’s on-line Breast Cancer Risk Assessment Tool [38]. Specifically, the Gail model uses a woman’s current age, age at menarche, age at first live birth, benign breast disease history and family history as predictor variables. In addition, height and weight information was further used to compute body mass index (BMI), categorized as normal weight (BMI < 25 kg/m2), overweight (25 kg/m2 ≤ BMI < 30 kg/m2) and obese (BMI ≥ 30 kg/m2). Race information was categorized as Caucasian, African-American or Other; however, given the relatively small number of women who identify as “Other” (N = 31), only women who identified as either Caucasian (N = 389) or African-American (N = 250) were included in this study.
Genotyping and SNP selection
For each woman, information regarding the genotype of 12 SNPs were obtained from a commercially available assay based on Illumina Infinium II whole-genome genotyping (deCODE BreastCancer, deCODE genetics, Inc.) [37]. The deCODE SNP assay includes 12 genetic loci, specifically 2q35 (rs13387042), MRPS30 (rs4415084), FGFR2 (rs1219648), TNRC9/TOX3 (rs3803662), 8q24 (rs13281615), LSP1 (rs3817198), MAP3K1 (rs889312), NEK10 (rs4973768), 1p11 (rs11249433), RAD51L1 (rs999737), COX11/STXBP4 (rs6504950), and CASP8 (rs1045485), which have been consistently associated with either overall or subtype specific cancer risk, the risk for metastatic disease or age at diagnosis [39-49]. Details of the 12 SNPs investigated in our study are provided in Table 1.
Breast density assessment
Breast density was measured using fully-automated methods. Area-based absolute and percent mammographic density was assessed on a per-image basis using a previously validated, fully-automated algorithm [24]. Briefly, the software automatically delineates the breast region in a digital mammogram from background air and the pectoral muscle. The breast is then subdivided into regions of similar x-ray attenuation via an unsupervised clustering technique, which are then classified into dense and non-dense regions using a support vector machine classifier. The absolute aggregate area of the regions classified as dense, D A , is divided by the total breast area, B A , to obtain a woman’s breast percent density (PD%) using equation 1:
These area density estimates acquired per image were averaged across each individual woman’s left and right mediolateral-oblique (MLO) and craniocaudal (CC) screening images in order to obtain a per-woman estimate of both absolute area of dense tissue and PD% for further analysis.
Absolute fibroglandular breast tissue volume and volumetric percent density were also automatically assessed on a per-image basis using fully-automated, FDA-cleared software (Quantra™ v.2.0, Hologic, Inc.) which is based on the widely validated Highnam and Brady method adapted for digital mammography [50,51]. Briefly, this method quantifies the total amount of breast and fibroglandular tissue present within each image pixel via a model of the image acquisition physics and known anatomical properties of the breast and dense tissue. The sum of the breast tissue volume, B V , and fibroglandular dense tissue volume, D V , are then used to calculate the relative volumetric percent density (VD%) seen mammographically via equation 2:
As with the area density measures, the individual volumetric density estimates acquired per-image were averaged to obtain corresponding per-woman estimates of absolute fibroglandular tissue volume and VD%.
Statistical analysis
Differences in age, BMI, Gail lifetime risk, and breast density distributions between the Caucasian and African-American cohorts were assessed using two-sided t-tests with unequal variances for continuous variables and the Chi-squared test for categorical variables at an α = 0.05 significance level. Pearson’s correlation coefficient was used to assess associations between absolute dense area, absolute dense volume, PD% and VD%. Associations between the four breast density measures and each SNP were then assessed with linear regression, in which we adjusted for age, BMI, and Gail lifetime risk by including them as additional covariates in the regression model to determine the significance of the change in density due to the differences in SNP genotype between women in the presence of these additional explanatory variables.
For all analyses, breast density measures were first log-transformed to approximate a normal distribution as has been done in prior works investigating the genetic basis of breast density [29] as well as the association between breast density and risk [13]. The risk allele frequency of each SNP was coded as an ordinal variable (i.e., 0, 1 or 2). In this way, category 0 represents those women homozygous for the common allele of that particular SNP, category 1 represents heterozygous women and category 2 represents women homozygous for the high risk allele. The age and Gail lifetime risk covariates were treated as continuous variables, while BMI category was treated as an ordinal variable. Missing BMI data was handled via race-stratified, standard multiple imputation [52], which replaces missing values with values based on the posterior probability derived from known values [53] within each racial group. For this study, a total of 25 imputations were used, which is greater than the suggested minimum number of 20 [54]. The regression coefficient, confidence interval, and p-value of each SNP was recorded, using the standard α = 0.05 level threshold for significance. Bonferroni correction [55] was also applied to account for multiple comparisons, yielding a second, more stringent significance level cutoff of p = 0.004 (i.e., α = 0.05 divided by 12, the total number of SNPs). In order to assess potential joint associations to breast density, multivariable regression analysis was also performed considering all SNPs and adjusting for age, BMI, and Gail lifetime risk as additional covariates in the regression model. Lastly, to assess the amount of variation in breast density explained by the combination of SNP, age, BMI and Gail lifetime risk, we also computed and report the coefficient of determination, R2, for each regression model with a significant association to a breast density measure, using a recently proposed method for datasets with multiple imputation [56]. Lastly, given the strong relationship between BMI and breast density, we performed a complete-data analysis to assess whether the associations found in the imputation analysis are maintained when only analyzing those women with known BMI at a lower statistical power. All statistical analyses were performed with STATA 13.1 (StataCorp, College Station, Texas, USA).
Results
Caucasian women were slightly older (p = 0.03), had a lower overall BMI (p < 0.001), and a higher Gail lifetime risk (p < 0.001) than African-American women. When comparing breast density between the two groups, Caucasian women were denser in terms of their percent density both by the area (p < 0.001) and volumetric (p = 0.003) metrics, while African-American women had a greater absolute volume of fibroglandular tissue (p < 0.001). No significant difference was seen between the two groups in terms of absolute area density (p = 0.90). A summary of the demographic and imaging characteristics for the women in our study cohort is shown in Table 2. Statistically significant (p ≤ 0.009) correlations were observed between all the quantitative breast density estimates (Additional file 1: Table S1). Absolute and percent area density had the strongest correlation (r = 0.70, p < 0.001), while absolute and percent volume density had the weakest correlation (r = 0.10, p = 0.009). Figure 1 provides illustrative examples of the dense tissue segmentations in digital mammograms of four representative Caucasian women in our study.
Area-density segmentations on right, mediolateral-oblique view mammograms for various SNP genotypes. Four Caucasian women with negative screening exams and different genotypes of rs3817198 in LSP1 (a,b) and rs1045485 in CASP8 (c,d). Mammograms in the left column (a,c) represent women who are homozygous for the common allele for each SNP, while mammograms in the right column (b,d) are from women who are homozygous for the risk allele for each SNP. Overall, SNPs, age, Gail life-time risk and BMI were found to only explain a small fraction of the variability in breast density between women. For reference, each woman’s age and overall PD% score are provided as annotations on each image.
When assessing associations between area-based density measures and SNPs (Table 3), only one SNP, rs3817198, was found to be significantly associated to absolute area density in Caucasian women at the Bonferroni level (p = 0.004, R2 = 0.07, Figure 2a). This SNP was not found to have a similar association in African-American women (p = 0.175). When assessing associations between volumetric density measures and SNPs, no SNP was found to be significant at the Bonferroni corrected level (Table 4). However rs3817198 was found to be significantly associated with the absolute volume of dense tissue at the standard significance level in Caucasian women (p = 0.019, R2 = 0.14, Figure 2b), while it was not significant at either level in African-American women (p = 0.792). In contrast, a different SNP, rs3803662, was found to be significantly associated at the standard significance level to absolute volume of dense tissue in African-American women (p = 0.043, R2 = 0.16, Figure 2c). In addition, SNP rs4973768 was found to be significantly associated with volumetric percent density at the standard significance level in African-American women (p = 0.028, R2 = 0.12), but not in Caucasian women (p = 0.680, Figure 2d). Finally, the difference in density score by risk-allele count for those density measures significantly associated with SNPs were confirmed to vary monotonically (Table 5).
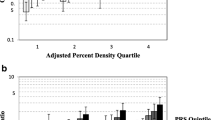
Box plots illustrating the distribution of breast density measures to significantly associated SNP genotypes. a) Absolute dense tissue area versus SNP rs3817198 in Caucasian women; b) Absolute volume of dense tissue versus SNP rs3817198 in Caucasian women; c) Absolute volume of dense tissue versus SNP rs3803662 in African-American women; and d) Percent volumetric density (VD%) versus SNP rs4973768 in African-American women. All box plots provide the median (red-line), interquartile range (blue box), 95% confidence interval (black whiskers) and outliers beyond the 95% confidence interval (red plus-signs).
When investigating joint associations between the entire panel of SNPs and each breast density measure through multivariable analysis (Additional files 2, 3, 4, 5: Tables S2-S5), rs3817198 remained significantly associated to absolute dense area (p = 0.003) and absolute dense volume (p = 0.026) in Caucasian women, and also became significantly associated with area percent density (p = 0.044). SNP rs3803662 also retained its significance in terms of its association with absolute volume density in African-American women (p = 0.048); while rs4973768 ceased to be significantly associated with volumetric percent density (p = 0.059). Lastly, complete-data analysis (Additional files 6, 7: Tables S6-S7) showed similar overall trends as the multiple imputation analysis with rs3817198 remaining significantly associated (p ≤ 0.05) with absolute measures of breast density in Caucasian women, although SNPs rs3803662 and rs4973768 only approached significance (p ≤ 0.1) with absolute volume density and volume percent density, respectively, in African-American women, likely due to the decreased sample size in the complete-data analysis leading to a loss of statistical power.
Discussion
We evaluated potential associations between a panel of validated breast cancer-related SNPs and quantitative measures of volumetric and area-based breast density in a cohort of Caucasian and African-American women. We found that the majority of the SNPs evaluated are not associated with breast density, and that those SNPs that are associated with breast density explain only a small fraction of the total variability in density after accounting for age, BMI and Gail lifetime risk (R2: 7%-16%). Specifically, SNP rs3817198 (in LSP1) was associated with absolute measures of area and volume density in Caucasian women, while in African-American women different SNPs, namely rs3803662 (in TNRC9/TOX3) and rs4973768 (in NEK10) were associated with absolute volume of dense tissue and percent volume density, respectively.
Previous studies investigating associations between SNPs and breast density have primarily focused on investigating associations with area-based measures of mammographic density [27,57-62]. These studies have shown a consistent association between breast PD% and SNP rs3817198 in LSP1 in Caucasian women [27,57-62], as also observed in our study; individual studies have also shown associations between PD% and TNRC9/TOX3-rs12443621 [57,58] and ZNF365-rs10995190 [61]. Few studies have also investigated the association between measures of absolute dense area and validated breast cancer risk loci [27,60]. Of these, Vachon et al. [27] observed that a second SNP rs3803662 in TNRC9/TOX3, a gene also identified in our study, is associated with the absolute amount of dense area. Finally, a recent meta-analysis by Varghese et al. suggested that density has a polygenic basis that likely overlaps at least partially with the genetic basis of breast cancer [28]; although not specifically focusing on which genes and SNPs drive this association, two of the strongest associations were observed with rs10995190 and rs10509168, two SNPs in gene ZNF365 which have also been associated with breast cancer risk [63]. However, these SNPs were not included in the panel assessed in our study, therefore limiting our ability to directly compare with our findings.
While informative, previous studies investigating associations between density and breast cancer susceptibility SNPs have been limited in different aspects: First, most have relied on semi-automated, reader-based [27,57-61] or visual [62] estimates of density, which are known to be sensitive to inter-reader variability [64,65] and may have introduced bias affecting the observed associations. In addition, they have primarily focused on area-based measures of the dense tissue. Given that these measures are a projection estimate of the true volume of fibroglandular tissue, volumetric assessment may provide a more accurate representation of the fibroglandular tissue [66].
In addition, few studies have investigated such associations in African-American women, a population with lower breast cancer incidence but higher mortality rate than Caucasian women [67]. As a result, only some of the SNPs in the panel used in this study have also been validated independently as breast cancer risk factors in African-American women, often with mixed results [48,68-74]. For example, the T allele of rs3803662 (16q12, TOX3), which we have found to be significantly associated with breast density, has also been shown to be significantly associated with a decreased breast cancer risk in African American women but an increased risk in Caucasian women [72], although the finding in African-American women has not been consistently replicated [68-70,75]. In contrast, rs4973768 in NEK10, which we found to be associated with volumetric percent density in the African-American cohort, has not been found to be associated with breast cancer risk in African-American women [70,72,75]. Regarding the panel as a whole, recent work by our group has found evidence that the 12 SNPs are jointly associated with breast cancer risk in African American women referred for biopsy [76]. Overall, this may suggest that not only may the genetic basis of breast cancer risk vary by race, but the genetic basis of breast density may vary by race as well, potentially allowing for complementary information about breast cancer risk to be ascertained when both genetic and radiographic risk factor information is considered in tandem. Larger studies would be needed to validate this hypothesis.
Although limited by a small sample size, one potentially interesting observation in our study is that the association between SNPs and breast density appears to differ by race, with different SNPs being significant in the two groups even when accounting for age, BMI and Gail lifetime risk. One potential explanation for this observation may be that although there is a large intra-racial variation in density relative to the mean inter-racial differences (Table 2), the genetic basis of density itself may perhaps partially differ in some respects between women of different races, similar to how tumor biology also tends to differ by race [77]. Another possible explanation may be that the total amount of glandular tissue, captured by volumetric density measures, and the spatial distribution of the dense tissue within the breast, captured by area-based measures, could reflect different aspects of the parenchymal pattern originally described by Wolfe [6,14], and thus may represent different aspects of risk related to breast density. Given these open questions, the exploration of potential racial differences in the biology of the different measures of breast density may be worth exploring in future, larger studies.
Although association studies such as ours cannot directly inform on or assess underlying biological processes, they do have value in identifying potential pathways of interest that could be interrogated in subsequent studies through hypothesis generation. For example, LSP1 is thought to play a role in mediating neutrophil activation and chemotaxis, and is expressed in both lymphocytes and endothelium [78], suggesting density may perhaps be, in part, a radiophenotype of genetic risk factors for breast cancer involving tissue vascularization. NEK protein kinases such as NEK10 are thought to play a role in cell cycle regulation [79] and may be related to breast density via factors related to cellular proliferation. Lastly, the protein encoded by TOX3/TNCR9 contains high-mobility-group motif used in altering chromatin structure [80], and thus may be potentially associated with density via some relationship with DNA transcription. Ultimately, a better understanding of the biological pathways could lead to a better understanding of breast oncogenesis as well as the development of better risk assessment tools.
Our study has certain limitations. First, we performed retrospective analysis using data from a single institution. The sample size was also relatively small, which may have limited our power to detect more subtle associations between individual SNPs and density, especially in the context of the race-stratified analysis and the adjustment performed to account for established covariates (i.e., age, BMI, Gail risk). In addition, we only investigated a panel of 12 low-penetrance SNPs associated with breast cancer, while many more risk loci have been recently identified [81]. To fully explore these associations, additional candidate genes related to breast density will also need to be investigated. Furthermore, ancestry informative markers were unfortunately not available for our study to account for population stratification within our Caucasian and African-American sub-cohorts beyond what was already accounted for by a race-stratified analysis. Although not likely to be a major confounder in our study given that genome-wide association studies for breast density have been performed in several populations in which there was little evidence of population stratification [28], they may potentially be of use to account for potential ethnic differences in relatively less-studied African-American or Asian populations and may help explain some of the large intra-racial variation in density relative to the mean inter-racial differences seen in Table 2. Lastly, although breast density is the most common descriptor of the breast parenchyma, genetic variants may also drive other differences in mammographic parenchymal patterns such as texture beyond what can be described by density alone, as previously suggested for the high-penetrance BRCA 1/2 genes [82] and additional SNPs such as rs451632 in the UGT2B gene cluster [83]. Given that parenchymal texture has been shown to be a potentially strong risk factor for breast cancer independent of density [8,84], such texture features may offer another surrogate marker by which the risk conferred by SNPs could manifest radiographically and thus should be considered by future research studies. Overall, larger prospective studies that include parenchymal texture measures as potential radiographic phenotypes of the risk for breast cancer conferred by a more comprehensive panel of known genetic risk factors are warranted to independently validate our findings.
Conclusion
In conclusion, the majority of the SNPs evaluated in our study were not found to be significantly associated with breast density. Although this may be due to the relatively small sample size of our study, and therefore limited power to detect more subtle associations, our observations suggest that these two risk factors may be capturing potentially independent information regarding a woman’s risk for breast cancer. As such our findings may have implications in the development of future breast cancer risk models by providing evidence that both SNPs and breast density could be considered simultaneously as risk predictors to potentially improve discriminatory capacity. Additionally, our study suggests that the associations between SNPs and breast density appear to differ between Caucasian and African American women. Larger prospective studies are warranted to further validate our findings and determine potential implications for breast cancer risk assessment. Ultimately, understanding the independent pathways that these different risk factors relate to breast cancer could lead to the development of improved risk assessment tools and prevention strategies.
Abbreviations
- BI-RADS:
-
Breast imaging-reporting and data system
- BMI:
-
Body mass index
- FFDM:
-
Full field digital mammography
- HIPAA:
-
Health insurance portability and accountability act
- IRB:
-
Institutional review board
- PD%:
-
Area percent density
- SNP:
-
Single nucleotide polymorphism
- VD%:
-
Volume percent density
References
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30.
Gail MH. Personalized estimates of breast cancer risk in clinical practice and public health. Stat Med. 2011;30(10):1090–104.
Hall P, Easton D. Breast cancer screening: time to target women at risk. Br J Cancer. 2013;108(11):2202–4.
Boyd NF, Guo H, Martin LJ, Sun L, Stone J, Fishell E, et al. Mammographic density and the risk and detection of breast cancer. N Engl J Med. 2007;356(3):227–36.
Vacek PM, Geller BM. A prospective study of breast cancer risk using routine mammographic breast density measurements. Cancer Epidemiol Biomarkers Prev. 2004;13(5):715–22.
Harvey JA, Bovbjerg VE. Quantitative assessment of mammographic breast density: relationship with breast cancer risk. Radiology. 2004;230(1):29–41.
Vachon CM, Brandt KR, Ghosh K, Scott CG, Maloney SD, Carston MJ, et al. Mammographic breast density as a general marker of breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16(1):43–9.
McCormack VA, dos Santos Silva I. Breast density and parenchymal patterns as markers of breast cancer risk: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2006;15(6):1159–69.
Keller B, Conant E, Oh H, Kontos D. Breast cancer risk prediction via area and volumetric estimates of breast density. In: Maidment ADA, Bakic PR, Gavenonis S, editors. 11th International Workshop on Breast Imaging (IWDM), Lecture Notes in Computer Science (LNCS), vol. 7361. Berlin Heidelberg: Springer-Verlag; 2012. p. 236–43.
Ding J, Warren R, Warsi I, Day N, Thompson D, Brady M, et al. Evaluating the effectiveness of using standard mammogram form to predict breast cancer risk: case–control study. Cancer Epidemiol Biomarkers Prev. 2008;17(5):1074–81.
Boyd N, Martin L, Gunasekara A, Melnichouk O, Maudsley G, Peressotti C, et al. Mammographic density and breast cancer risk: evaluation of a novel method of measuring breast tissue volumes. Cancer Epidemiol Biomarkers Prev. 2009;18(6):1754–62.
Aitken Z, McCormack VA, Highnam RP, Martin L, Gunasekara A, Melnichouk O, et al. Screen-film mammographic density and breast cancer risk: a comparison of the volumetric standard mammogram form and the interactive threshold measurement methods. Cancer Epidemiol Biomarkers Prev. 2010;19(2):418–28.
Shepherd JA, Kerlikowske K, Ma L, Duewer F, Fan B, Wang J, et al. Volume of mammographic density and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2011;20(7):1473–82.
Wolfe JN. Breast patterns as an index of risk for developing breast cancer. AJR Am J Roentgenol. 1976;126(6):1130–7.
Byrne C, Schairer C, Wolfe J, Parekh N, Salane M, Brinton LA, et al. Mammographic features and breast cancer risk: effects with time, age, and menopause status. J Natl Cancer Inst. 1995;87(21):1622–9.
D'Orsi CJ, Bassett LW, Berg WA, Feig SA, Jackson VP, Kopans DB: Breast imaging reporting and data system: ACR BI-RADS, Mammography 4th ed., Reston, Am. Col. of Rad. 2003.
Price ER, Hargreaves J, Lipson JA, Sickles EA, Brenner RJ, Lindfors KK, et al. The California breast density information group: a collaborative response to the issues of breast density, breast cancer risk, and breast density notification legislation. Radiology. 2013;269(3):887–92.
Byng JW, Yaffe MJ, Jong RA, Shumak RS, Lockwood GA, Tritchler DL, et al. Analysis of mammographic density and breast cancer risk from digitized mammograms. Radiographics. 1998;18(6):1587–98.
Chen J, Pee D, Ayyagari R, Graubard B, Schairer C, Byrne C, et al. Projecting absolute invasive breast cancer risk in white women with a model that includes mammographic density. J Natl Cancer Inst. 2006;98(17):1215–26.
Tice JA, Cummings SR, Ziv E, Kerlikowske K. Mammographic breast density and the Gail model for breast cancer risk prediction in a screening population. Breast Cancer Res Treat. 2005;94(2):115–22.
Tice JA, Cummings SR, Smith-Bindman R, Ichikawa L, Barlow WE, Kerlikowske K. Using clinical factors and mammographic breast density to estimate breast cancer risk: development and validation of a new predictive model. Ann Intern Med. 2008;148(5):337–47.
Barlow WE, White E, Ballard-Barbash R, Vacek PM, Titus-Ernstoff L, Carney PA, et al. Prospective breast cancer risk prediction model for women undergoing screening mammography. J Natl Cancer Inst. 2006;98(17):1204–14.
Nickson C, Arzhaeva Y, Aitken Z, Elgindy T, Buckley M, Li M, et al. Auto density: an automated method to measure mammographic breast density that predicts breast cancer risk and screening outcomes. Breast Cancer Res. 2013;15(5):R80.
Keller BM, Nathan DL, Wang Y, Zheng Y, Gee JC, Conant EF, et al. Estimation of breast percent density in raw and processed full field digital mammography images via adaptive fuzzy c-means clustering and support vector machine segmentation. Med Phys. 2012;39(8):4903–17.
Heine JJ, Cao K, Rollison DE. Calibrated measures for breast density estimation. Acad Radiol. 2011;18(5):547–55.
Kelemen LE, Sellers TA, Vachon CM. Can genes for mammographic density inform cancer aetiology? Nat Rev Cancer. 2008;8(10):812–23.
Vachon CM, Scott CG, Fasching PA, Hall P, Tamimi RM, Li J, et al. Common breast cancer susceptibility variants in LSP1 and RAD51L1 are associated with mammographic density measures that predict breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2012;21(7):1156–66.
Varghese JS, Thompson DJ, Michailidou K, Lindstrom S, Turnbull C, Brown J, et al. Mammographic breast density and breast cancer: evidence of a shared genetic basis. Cancer Res. 2012;72(6):1478–84.
Stone J, Gurrin LC, Byrnes GB, Schroen CJ, Treloar SA, Padilla EJ, et al. Mammographic density and candidate gene variants: a twins and sisters study. Cancer Epidemiol Biomarkers Prev. 2007;16(7):1479–84.
Burton H, Chowdhury S, Dent T, Hall A, Pashayan N, Pharoah P. Public health implications from COGS and potential for risk stratification and screening. Nat Genet. 2013;45(4):349–51.
Hajiloo M, Damavandi B, Hooshsadat M, Sangi F, Mackey JR, Cass CE, et al. Breast cancer prediction using genome wide single nucleotide polymorphism data. BMC Bioinformatics. 2013;14 Suppl 13:S3.
Dite GS, Mahmoodi M, Bickerstaffe A, Hammet F, Macinnis RJ, Tsimiklis H, et al. Using SNP genotypes to improve the discrimination of a simple breast cancer risk prediction model. Breast Cancer Res Treat. 2013;139(3):887–96.
Gail MH. Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J Natl Cancer Inst. 2008;100(14):1037–41.
Mealiffe ME, Stokowski RP, Rhees BK, Prentice RL, Pettinger M, Hinds DA. Assessment of clinical validity of a breast cancer risk model combining genetic and clinical information. J Natl Cancer Inst. 2010;102(21):1618–27.
Wacholder S, Hartge P, Prentice R, Garcia-Closas M, Feigelson HS, Diver WR, et al. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 2010;362(11):986–93.
Eng A, Gallant Z, Shepherd J, McCormack V, Li J, Dowsett M, et al. Digital mammographic density and breast cancer risk: a case inverted question markcontrol study of six alternative density assessment methods. Breast Cancer Res. 2014;16(5):439.
McCarthy AM, Armstrong K, Handorf E, Boghossian L, Jones M, Chen J, et al. Incremental impact of breast cancer SNP panel on risk classification in a screening population of white and African American women. Breast Cancer Res Treat. 2013;138(3):889–98.
Breast Cancer Risk Assessment Tool [http://www.cancer.gov/bcrisktool/]
Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39(3):352–8.
Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39(7):865–9.
Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–93.
Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39(7):870–4.
Stacey SN, Manolescu A, Sulem P, Thorlacius S, Gudjonsson SA, Jonsson GF, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40(6):703–6.
Garcia-Closas M, Hall P, Nevanlinna H, Pooley K, Morrison J, Richesson DA, et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4(4):e1000054.
Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat Genet. 2009;41(5):579–84.
Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41(5):585–90.
Fu Y-P, Edvardsen H, Kaushiva A, Arhancet JP, Howe TM, Kohaar I, Porter-Gill P, Shah A, Landmark-Høyvik H, Fosså SD: Research NOTCH2 in breast cancer: association of SNP rs11249433 with gene expression in ER-positive breast tumors without TP53 mutations. 2010.
Rebbeck TR, DeMichele A, Tran TV, Panossian S, Bunin GR, Troxel AB, et al. Hormone-dependent effects of FGFR2 and MAP3K1 in breast cancer susceptibility in a population-based sample of post-menopausal African-American and European-American women. Carcinogenesis. 2009;30(2):269–74.
Huijts P, Vreeswijk M, Kroeze-Jansema K, Jacobi CE, Seynaeve C, Krol-Warmerdam E, et al. Clinical correlates of low-risk variants in FGFR2, TNRC9, MAP3K1, LSP1 and 8q24 in a Dutch cohort of incident breast cancer cases. Breast Cancer Res. 2007;9(6):R78.
Highnam R, Jeffreys M, McCormack V, Warren R, Davey Smith G, Brady M. Comparing measurements of breast density. Phys Med Biol. 2007;52(19):5881–95.
Hartman K, Highnam R, Warren R, Jackson V. Volumetric Assessment of Breast Tissue Composition from FFDM Images. In: Krupinski EA, editor. 9th International Workshop on Digital Mammography (IWDM), Lecture Notes in Computer Science (LNCS), 5116. Springer-Verlag: Berlin Heidelberg; 2008. p. 33–9.
Rubin DB. Inference and missing data. Biometrika. 1976;63(3):581–92.
Rubin DB. Multiple imputation after 18+ years. J Am Stat Assoc. 1996;91(434):473–89.
Sterne JA, White IR, Carlin JB, Spratt M, Royston P, Kenward MG, et al. Multiple imputation for missing data in epidemiological and clinical research: potential and pitfalls. BMJ. 2009;338:b2393.
Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43(1):223–5.
Harel O. The estimation of R 2 and adjusted R 2 in incomplete data sets using multiple imputation. J Appl Stat. 2009;36(10):1109–18.
Tamimi RM, Cox D, Kraft P, Colditz GA, Hankinson SE, Hunter DJ. Breast cancer susceptibility loci and mammographic density. Breast Cancer Res. 2008;10(4):R66.
Woolcott CG, Maskarinec G, Haiman CA, Verheus M, Pagano IS, Le Marchand L, et al. Association between breast cancer susceptibility loci and mammographic density: the multiethnic cohort. Breast Cancer Res. 2009;11(1):R10.
Lee E, Haiman CA, Ma H, Van Den Berg D, Bernstein L, Ursin G. The role of established breast cancer susceptibility loci in mammographic density in young women. Cancer Epidemiol Biomarkers Prev. 2008;17(1):258–60.
Odefrey F, Stone J, Gurrin LC, Byrnes GB, Apicella C, Dite GS, et al. Common genetic variants associated with breast cancer and mammographic density measures that predict disease. Cancer Res. 2010;70(4):1449–58.
Lindstrom S, Vachon CM, Li J, Varghese J, Thompson D, Warren R, et al. Common variants in ZNF365 are associated with both mammographic density and breast cancer risk. Nat Genet. 2011;43(3):185–7.
Fernandez-Navarro P, Pita G, Santamarina C, Moreno MP, Vidal C, Miranda-Garcia J, et al. Association analysis between breast cancer genetic variants and mammographic density in a large population-based study (Determinants of Density in Mammographies in Spain) identifies susceptibility loci in TOX3 gene. Eur J Cancer. 2013;49(2):474–81.
Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42(6):504–7.
Ooms EA, Zonderland HM, Eijkemans MJ, Kriege M, Mahdavian Delavary B, Burger CW, et al. Mammography: interobserver variability in breast density assessment. Breast. 2007;16(6):568–76.
Amir E, Freedman OC, Seruga B, Evans DG. Assessing women at high risk of breast cancer: a review of risk assessment models. J Natl Cancer Inst. 2010;102(10):680–91.
Kopans DB. Basic physics and doubts about relationship between mammographically determined tissue density and breast cancer risk. Radiology. 2008;246(2):348–53.
DeSantis C, Naishadham D, Jemal A. Cancer statistics for African Americans, 2013. CA Cancer J Clin. 2013;63(3):151–66.
Barnholtz-Sloan JS, Shetty PB, Guan X, Nyante SJ, Luo J, Brennan DJ, et al. FGFR2 and other loci identified in genome-wide association studies are associated with breast cancer in African-American and younger women. Carcinogenesis. 2010;31(8):1417–23.
Ruiz-Narvaez EA, Rosenberg L, Cozier YC, Cupples LA, Adams-Campbell LL, Palmer JR. Polymorphisms in the TOX3/LOC643714 locus and risk of breast cancer in African-American women. Cancer Epidemiol Biomarkers Prev Publ Am Assoc Cancer Res Am Soc Prev Oncol. 2010;19(5):1320–7.
Huo D, Zheng Y, Ogundiran TO, Adebamowo C, Nathanson KL, Domchek SM, et al. Evaluation of 19 susceptibility loci of breast cancer in women of African ancestry. Carcinogenesis. 2012;33(4):835–40.
Barnholtz-Sloan JS, Raska P, Rebbeck TR, Millikan RC. Replication of GWAS "Hits" by race for breast and prostate cancers in European Americans and African Americans. Front Genet. 2011;2:37.
Hutter CM, Young AM, Ochs-Balcom HM, Carty CL, Wang T, Chen CT, et al. Replication of breast cancer GWAS susceptibility loci in the Women's health initiative African American SHARe study. Cancer Epidemiol Biomarkers Prev Publ Am Assoc Cancer Res Am Soc Prev Oncol. 2011;20(9):1950–9.
Zheng W, Cai Q, Signorello LB, Long J, Hargreaves MK, Deming SL, et al. Evaluation of 11 breast cancer susceptibility loci in African-American women. Cancer Epidemiol Biomarkers Prev Publ Am Assoc Cancer Res Am Soc Prev Oncol. 2009;18(10):2761–4.
Ruiz-Narvaez EA, Rosenberg L, Rotimi CN, Cupples LA, Boggs DA, Adeyemo A, et al. Genetic variants on chromosome 5p12 are associated with risk of breast cancer in African American women: the black women's health study. Breast Cancer Res Treat. 2010;123(2):525–30.
Chen F, Chen GK, Millikan RC, John EM, Ambrosone CB, Bernstein L, et al. Fine-mapping of breast cancer susceptibility loci characterizes genetic risk in African Americans. Hum Mol Genet. 2011;20(22):4491–503.
McCarthy A, Keller B, Kontos D, Boghossian L, McGuire E, Bristol M, et al. The use of the Gail model, body mass index and SNPs to predict breast cancer among women with abnormal (BI-RADS 4) mammograms. Breast Cancer Res. 2015;17(1):1.
Amend K, Hicks D, Ambrosone CB. Breast cancer in African-American women: differences in tumor biology from European-American women. Cancer Res. 2006;66(17):8327–30.
Chen MB, Li C, Shen WX, Guo YJ, Shen W, Lu PH. Association of a LSP1 gene rs3817198T>C polymorphism with breast cancer risk: evidence from 33,920 cases and 35,671 controls. Molecular biology reports. 2011;38(7):4687–95.
Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–34.
Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40(1):17–22.
Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45(4):353–61. 361e351-352.
Huo Z, Giger ML, Olopade OI, Wolverton DE, Weber BL, Metz CE, et al. Computerized analysis of digitized mammograms of BRCA1 and BRCA2 gene mutation carriers. Radiology. 2002;225(2):519–26.
Li H, Giger ML, Sun C, Ponsukcharoen U, Huo D, Lan L, et al. Pilot study demonstrating potential association between breast cancer image-based risk phenotypes and genomic biomarkers. Med Phys. 2014;41(3):031917.
Manduca A, Carston MJ, Heine JJ, Scott CG, Pankratz VS, Brandt KR, et al. Texture features from mammographic images and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2009;18(3):837–45.
Acknowledgements
This work was supported in part by the National Cancer Institute (1U54CA163313-01 and UC2CA148310), the American Cancer Society (RSGHP-CPHPS-119586), and the Susan G. Komen for the Cure Foundation (SAC100003 and PDF14299284).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
BMK, AMM, JC, KA, SMD and DK declare that they have no competing interests related to this work. EFC serves on the scientific advisory board of Hologic, Inc. (Bedford, MA) and is clinical reader for Image Matrix (Philadelphia, PA).
Authors’ contributions
BMK carried out the image analytics and breast density estimation, performed the statistical analysis and drafted the manuscript. AMM and JC were involved in the statistical analysis and interpretation of the data. KA and SMD were involved in the acquisition of the genetic data and in critically revising the manuscript for important intellectual content. SMD and EFC were involved in the study conception, design, and interpretation of findings. DK was involved in the study conception, design, and interpretation of findings and in drafting the manuscript. All authors read, revised and approved the final manuscript.
Additional files
Additional file 1: Table S1.
Pair-wise Pearson correlations between the quantitative breast density measures considered in this work.
Additional file 2: Table S2.
Race-stratified, multivariable regression model between absolute area density and the SNP panel, age, BMI and Gail lifetime risk.
Additional file 3: Table S3.
Race-stratified, multivariable regression model between area percent density and the SNP panel, age, BMI and Gail lifetime risk.
Additional file 4: Table S4.
Race-stratified, multivariable regression model between absolute dense tissue volume and the SNP panel, age, BMI and Gail lifetime risk.
Additional file 5: Table S5.
Race-stratified, multivariable regression model between volume percent density and the SNP panel, age, BMI and Gail lifetime risk.
Additional file 6: Table S6.
Regression analysis between number of SNP risk alleles and absolute and relative percent area density measures in Caucasian and African-American women after adjusting for age, BMI and Gail lifetime risk for those women with known BMI (i.e., complete data analysis).
Additional file 7: Table S7.
Regression analysis between number of SNP risk alleles and absolute and relative percent volume density measures in Caucasian and African-American women after adjusting for age, BMI and Gail lifetime risk for those women with known BMI (i.e., complete data analysis).
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Keller, B.M., McCarthy, A.M., Chen, J. et al. Associations between breast density and a panel of single nucleotide polymorphisms linked to breast cancer risk: a cohort study with digital mammography. BMC Cancer 15, 143 (2015). https://doi.org/10.1186/s12885-015-1159-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-015-1159-3