Abstract
Background
Replication studies showed conflicting effects of ABCG2 and SLC2A9 polymorphisms on gout and serum urate. This meta-analysis therefore aimed to pool their effects across studies.
Methods
Studies were located from MEDLINE and Scopus from inception to 17th June 2018. Observational studies in adults with any polymorphism in ABCG2 or SLC2A9, and outcome including gout, hyperuricemia, and serum urate were included for pooling. Data extractions were performed by two independent reviewers. Genotype effects were pooled stratified by ethnicity using a mixed-effect logistic model and a multivariate meta-analysis for dichotomous and continuous outcomes.
Results
Fifty-two studies were included in the analysis. For ABCG2 polymorphisms, mainly studied in Asians, carrying 1–2 minor-allele-genotypes of rs2231142 and rs72552713 were respectively about 2.1–4.5 and 2.5–3.9 times higher odds of gout than non-minor-allele-genotypes. The two rs2231142-risk-genotypes also had higher serum urate about 11–18 μmol/l. Conversely, carrying 1–2 minor alleles of rs2231137 was about 36–57% significantly lower odds of gout. For SLC2A9 polymorphisms, mainly studied in Caucasians, carrying 1–2 minor alleles of rs1014290, rs6449213, rs6855911, and rs7442295 were about 25–43%, 31–62%, 33–64%, and 35–65% significantly lower odds of gout than non-minor-allele-genotypes. In addition, 1–2 minor-allele-genotypes of the latter three polymorphisms had significantly lower serum urate about 20–49, 21–51, and 18–54 μmol/l than non-minor-allele-genotypes.
Conclusions
Our findings should be useful in identifying patients at risk for gout and high serum urate and these polymorphisms may be useful in personalized risk scores.
Trial registration
PROSPERO registration number: CRD42018105275.
Similar content being viewed by others
Introduction
Hyperuricemia, defined as serum urate > 7 mg/dl (or 416.4 μmol/l) [1], can lead to gout [1] and increased risk of renal disease, diabetes, hypertension, and cardiovascular disease [2]. Genome-wide association studies (GWAS) have shown that many single nucleotide polymorphisms (SNPs) of ATP-binding cassette sub-family G member 2 gene (ABCG2) and the solute carrier family 2 member 9 (SLC2A9) are associated with serum urate and gout [3,4,5,6]. Both ABCG2 and SLC2A9 are located on chromosome 4 [7]. Many more individual studies have replicated the findings of GWASs and these were summarized in systematic reviews of the effects of ABCG2 [8,9,10,11,12,13] and SLC2A9 [12, 14,15,16] polymorphisms on gout. The most updated review of ABCG2 [12] included 12 individual studies, indicating that carrying the A allele of rs2231142 increased the risk of gout by about 1.8–2.3 times relative to the C allele in Asian and non-Asian populations. The most recent review of SLC2A9 [15] included 13 studies indicating that rs6449213 (C versus T), rs16890979 (T versus C) and rs1014290 (C versus T) significantly decreased the risk of gout whereas rs3733591 increased the risk of gout only in Asian and Maori populations and Solomon Islanders.
There have been 12 studies of ABCG2-rs2231142 on gout published after these reviews and most reviews did not pool genotype effects, which could lead to suggest mode of gene effects. In addition, the effects of ABCG2 and SLC2A9 polymorphisms on serum urate have never been systematically reviewed. Therefore, an updated systematic review and meta-analysis regarding the influence of SNPs in ABCG2 and SLC2A9 on gout, hyperuricemia and serum urate is needed.
Methods
Search terms and strategies
Studies were located from MEDLINE (via PubMed search engine) and Scopus databases from inception to 17th June 2018. The search terms and strategies were constructed based on study genes (i.e., ABCG2 and SLC2A9 genes) and outcomes (i.e., gout, hyperuricemia and serum urate); see more details in Additional file 1. This study was registered in PROSPERO number CRD42018105275.
Inclusion/exclusion criteria
Screening of titles and abstracts were performed by TL and randomly checked by SR. Any type of observational study was selected as follows: studies in adults, with any polymorphism in ABCG2 or SLC2A9, and any outcome including gout, hyperuricemia, and serum urate. Studies with insufficient data and unavailable full-texts were excluded.
Data extraction
Two of 3 authors (i.e., TL, SR, ST, and NS) independently extracted data. Extracted information included study characteristics (study setting, study design), participants (ethnicity, age, gender, body mass index (BMI), estimated glomerular filtration rate (eGFR), co-morbidity, medications, and alcohol consumption), genes (ABCG2/SLC2A9/both, allele/genotype, and frequency/summary data), and outcomes (gout and its diagnostic criteria, hyperuricemia and its definition, and serum urate). In addition, data for pooling (frequency data for allele/genotype and outcome, and mean and standard deviation (SD) of urate by allele/genotype) were also extracted. Missing data was requested from authors. Any disagreement was resolved by discussion and consensus.
Risk of bias assessment
Risk of bias was independently assessed by two reviewers (TL and SR) using a risk-of-bias tool [17] which consisted of 4 domains, i.e., information bias, confounding bias, selective reporting, and Hardy-Weinberg equilibrium (HWE). Each question was answered as yes, no, and unclear representing low/no, possible/high, and unclear risk of bias due to insufficient information, respectively. Any disagreement was resolved by discussion and consensus with the team.
Study genes and outcome of interest
Data for the following polymorphisms were collected: ABCG2 gene (rs2231142 C > A, rs72552713 C > T and rs2231137 C > A) and SLC2A9 gene (rs1014290 G > A, rs3733591 G > A, rs6449213 G > A, rs6855911 G > A, rs7442295 G > A, rs12510549 G > A, rs16890979 G > A, and rs734553 T > G). Outcomes of interest were gout, hyperuricemia, and serum urate defined according to original studies.
Statistical analysis
Statistical analysis was performed using methods previously described [18]. HWE was checked using an exact test and only studies which complied with HWE were considered in the analysis. Prevalence of the minor allele of each SNP was pooled, and stratified by ethnicity. Gene effects were assessed as follows:
Per-allele approach
For hyperuricemia and gout, allele effect (i.e., odds ratio (OR)) for minor allele a versus A along with 95% confidence interval (CI) was estimated. Mean difference (MD) of urate, in μmol/l, for a versus A was estimated. Heterogeneity was explored using Cochrane’s Q test and I2. A random-effect model was used for pooling ORs if heterogeneity was present (P-value < 0.10 or I2 ≥ 25%), otherwise a fixed-effect model was applied. Sources of heterogeneity (e.g. age, gender, co-morbidity, etc.) were explored using meta-regression if the data were available, and subgroup analyses were then performed accordingly.
Per-genotype approach
ORs (i.e., OR1 (aa versus AA) and OR2 (Aa versus AA)) and mean differences (MDs) (MD1 (aa versus AA) and MD2 (Aa versus AA)) were estimated for dichotomous and continuous outcomes, respectively. Heterogeneity was explored as above. Aggregated genotype and outcome data were expanded to individual patient data (IPD). A one-stage approach with a mixed-effect logistic regression was applied by fitting genotypes (i.e., aa versus AA and Aa versus AA) on outcome, and pooled ORs were estimated. For serum urate, a multivariate (mv) meta-analysis was applied to pool MD1 and MD2 across studies. Mode of gene effects (i.e., dominant, recessive, additive) was then determined by estimating lambda (λ) (i.e., logOR2/logOR1 or MD2/MD1) using model-free Bayesian approach [19].
Sensitivity analysis was performed by including studies with HWE disequilibrium. Publication bias was assessed using funnel plots and Egger’s tests. If any of these suggested asymmetry, a contour enhanced-funnel was constructed to determine whether asymmetry was caused by publication bias or heterogeneity.
STATA software version 15.1 and WinBUGS version 1.4.3 were used for analyses. The level of significance was < 0.05 except for the heterogeneity test, in which < 0.10 was used.
Results
Identifying studies
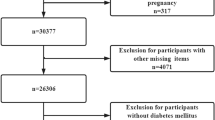
Among 2087 identified studies, 52 studies were eligible, which included one study manually added from the reference, see Fig. 1. Among them, 34, 5, and 22 studies had outcomes of gout, hyperuricemia, and serum urate, respectively.
The characteristics of 52 studies (i.e., 30 ABCG2 studies [20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49] and 32 SLC2A9 studies [24,25,26, 28, 31, 33, 39, 40, 43, 45, 50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71]) with a total of 63 sub-studies are described in Additional file 2. Most studies were cohorts (50%), and split roughly evenly between Asians (46%) and Caucasians (42%). The mean ages and BMIs ranged from 37 to 66 years and from 22.6 to 34.6 kg/m2, respectively. Of 34 studies on gout, most studies (68%) used the criteria of American College of Rheumatology [72] for diagnosis. All studies used the cut-off of 416.4 μmol/l (or 7 mg/dl) for defining hyperuricemia.
Risk of bias assessment
The risk of bias was low for population stratification and selective outcome report (see Additional file 3 A-B). Twenty sub-studies were at high risk of bias in ascertainment of genotyping because they did not clearly describe the methods they used. Furthermore, 17 sub-studies had high risk of bias in ascertainment of outcome mostly due to no information clearly mentioned.
Gout
Among 34 studies, 3 SNPs in ABCG2 (i.e., rs2231142, rs72552713, and rs2231137) and 7 SNPs in SLC2A9 (i.e., rs1014290, rs3733591, rs6449213, rs16890979, rs6855911, rs7442295, and rs12510549) had sufficient data for pooling as described in Additional file 4.1. Minor allele prevalences of these polymorphisms were pooled, see Additional file 4.2.
Genotype effects of ABCG2 polymorphisms (i.e., rs2231142 (N = 21), rs72552713.
(N = 7), and rs2231137 (N = 6)) were estimated stratifying by Asians and Caucasians, see Fig. 2, Table 1, and Additional file 4.1. In Asian populations, carrying the homozygous minor and heterozygous genotypes of rs2231142 and rs72552713 were higher risk of gout than the homozygous major genotypes with pooled OR1 and OR2 of 4.53 (4.10, 5.00) and 2.10 (1.95, 2.26) for rs2231142; and 3.86 (2.30, 9.76) and 2.46 (1.93, 3.18) for rs72552713. Sensitivity analysis by including a study that did not comply with HWE for rs2231142 [42] did not change the results (data not shown). Likewise, rs2231142 also carried higher risk of gout in Caucasians with pooled OR1 and OR2 of 3.24 (2.39, 4.41) and 1.64 (1.47, 1.82). Conversely, carrying homozygous minor and heterozygous genotypes of rs2231137 carried lower risk of gout in Asians with pooled OR1 and OR2 of 0.43 (0.34, 0.55) and 0.64 (0.56, 0.72), respectively.
Effects of 7 polymorphisms in SLC2A9 (i.e., rs1014290 (N = 7), rs3733591 (N = 7), rs6449213 (N = 5), rs6855911 (N = 5), rs16890979 (N = 4), rs7442295 (N = 3), and rs12510549 (N = 3)) on gout were assessed, see Fig. 2, Table 1, and Additional file 4.1. Pooled minor allele prevalences are reported in Additional file 4.2. Among Asian studies, effects of 3 polymorphisms (i.e., rs3733591, rs1014290, and rs6855911) on gout were pooled indicating that homozygous minor and heterozygous genotypes of rs3733591 carried higher risks of gout, i.e. 1.55 (1.17, 2.06) and 1.29 (1.07, 1.55) times higher than homozygous major genotype. Conversely, carrying homozygous minor and heterozygous genotypes of rs1014290 and rs6855911 carried lower risk, but only the former SNP was significant with pooled OR1 and OR2 of 0.42 (0.36, 0.49) and 0.65 (0.58, 0.72), respectively.
Among these SLC2A9 polymorphisms, only rs1014290 and rs6855911 were significantly associated with gout in Caucasians with pooled OR1 and OR2 for homozygous minor and heterozygous genotypes of 0.57 (0.37, 0.87) and 0.75 (0.63, 0.89) for rs1014290, and 0.36 (0.28, 0.46) and 0.67 (0.59, 0.77) for rs6855911; the effects of rs3733591 were not significant. Three additional polymorphisms, studied only in Caucasians, were also significantly associated with gout, i.e., rs6449213, rs7442295, and rs12510549. Homozygous minor and heterozygous genotypes of these polymorphisms carried lower risk than homozygous major genotype, with pooled OR1 and OR2 of 0.38 (0.29, 0.50) and 0.69 (0.62, 0.78) for rs6449213; 0.35 (0.28, 0.46) and 0.65 (0.57, 0.74) for rs7442295; and 0.44 (0.34, 0.58) and 0.66 (0.58, 0.76) for rs12510549, see Fig. 2, Table 1, and Additional file 4.1.
The mode of gene effects (λ) were estimated suggesting that effects of ABCG2 polymorphisms were mostly additive effects, except for rs72552713 which might be between an additive or dominant effect, see Table 1. Likewise, the mode of SLC2A9 effects on gout might be mostly additive effects, see Table 1.
Effects of ABCG2 and SLC2A9 polymorphisms on gout were homogenous to highly heterogeneous with I2 values of 0 to 75.2% and 39.2 to 62.9% in Asians and Caucasians for ABCG2; and 0 to 68.1% and 0 to 49.4% in Asians and Caucasians for SLC2A9 polymorphisms, see Table 1. Sources of heterogeneity were explored for ABCG2 polymorphisms (i.e., rs2231142 and rs2231137), see Additional file 4.3. Sub-group analysis by percent male ≥90% versus < 90% indicated stronger effects particularly for OR1 in percent male ≥90% with the OR1 of 5.32 (4.75, 5.97) and 6.14 (0.72, 52.13) for rs2231142 in Asians and Caucasians.
Likewise, sources of heterogeneity were explored for SLC2A9 polymorphisms; again, percent male was a potential source of variation for rs1014290 and rs3733591 in Asians, and rs7442295 in Caucasians, see Additional file 4.3. Their effects were even higher in studies with a high percent of males, with pooled OR1 of 0.39 (0.33, 0.46) for rs1014290 and 2.52 (1.71, 3.73) for rs3733591 in Asians, and 0.19 (0.09, 0.41) for rs7442295 in Caucasians. Publication bias was assessed by Egger’s tests and funnel plots, see Additional file 4.4–4.5, and none of these polymorphisms had evidence of publication bias.
Hyperuricemia
Effects of rs2231142 (in ABCG2) on hyperuricemia were assessed (N = 5, see Additional file 5.1). Carrying homozygous minor and heterozygous genotypes carried a higher risk of hyperuricemia than carrying homozygous major genotype with pooled OR1 and OR2 of 2.25 (1.80, 2.81) and 1.56 (1.35, 1.80), respectively, see Fig. 3, Table 1, and Additional file 5.1. The estimated λ was 0.527, suggesting an additive gene effect, see Table 1. Sensitivity analysis by including 1 Asian study that did not comply with HWE [42] did not materially change the results (data not shown). These gene effects were moderately heterogeneous (I2 = 44.6–67.2%), with BMI and percent male as potential sources, see Additional file 5.2. There was no evidence of publication bias from Egger’s tests and funnel plots, see Additional file 5.3–5.4.
Serum urate
Among 22 studies, 1 SNP in ABCG2 (i.e., rs2231142) and 5 SNPs in SLC2A9 (i.e., rs1014290, rs6449213, rs6855911, rs7449253, and rs734553) had sufficient data for pooling, see Additional file 6.1. An additional polymorphism not studied for gout was rs734553 and the pooled minor allele prevalence was 0.30 (0.28, 0.32). For rs2231142 (in ABCG2), homozygous minor and heterozygous genotypes had higher urate levels than homozygous major genotype, with pooled MD1 and MD2 of 18.41 (8.68, 28.14) and 11.49 (4.10, 18.89) respectively in Asians; and 50.43 (32.66, 68.20) and 19.46 (14.19, 24.73) respectively in Caucasians, see Fig. 4, Table 2, and Additional file 6.1. Sensitivity analysis by including 1 Asian study that did not comply with HWE [39] did not materially change the results (data not shown). In contrast, homozygous minor and heterozygous genotypes of rs1014290 (in SLC2A9) decreased urate more than homozygous major genotype, with pooled MD1 and MD2 of − 21.47 (− 30.97, − 11.96) and − 8.16 (− 16.63, 0.31), respectively in Asians, and − 32.33 (− 52.61,-12.05) and − 9.87 (− 25.27,5.54), respectively in Caucasians, see Fig. 4, Table 2, and Additional file 6.1.
Four additional SLC2A9 polymorphisms were studied in Caucasians only. Homozygous minor and heterozygous genotypes of these SNPs reduced urate levels compared to homozygous major genotypes, with pooled MD1 and MD2 of − 49.29 (− 59.85, − 38.74) and − 20.43 (− 28.91, − 11.94) for rs6449213; − 51.23 (− 57.22, − 45.25) and − 20.65 (− 23.83, − 17.47) for rs6855911; − 54.30 (− 58.78, − 49.82) and − 54.30 (− 58.78, − 49.82) for rs7442295; − 29.29 (− 63.90, 5.33) and − 20.68 (− 47.08, 5.71) for rs734553, see Fig. 4, Table 2, and Additional file 6.1. The mode of effects of these SLC2A9-SNPs were most likely to be an additive effect, see Table 2.
Effects of ABCG2 and SLC2A9 SNPs on serum urate were homogeneous to highly heterogeneous, i.e., I2 ranged from 59.1 to 63.1% and 0 to 68.7% in Asians and Caucasians for ABCG2-rs2231142; 65.7 to 67.9% and 0 to 88.5% in Asians and Caucasians for SLC2A9 SNPs, see Table 2. In Caucasians, type of population accounted for some of the heterogeneity, see Additional file 6.2. After excluding one Caucasian study in type 2 diabetes (T2D) patients, sub-group analysis yielded stronger effects of these SNPs in a general population with pooled MD1 and MD2 of − 40.16 (− 47.74, − 32.57) and − 15.80 (− 23.36, − 8.24) for rs1014290; and − 55.67 (− 82.77, − 28.57)) and − 29.00 (− 41.82, − 16.19) for rs734553. The effects of rs734553 became significant and strongest after excluding the study in the T2D population. Publication bias was assessed by Egger’s tests and funnel plots, see Additional file 6.3–6.4. Publication bias may be present for the effects of rs6449213 in Caucasians, as suggested by asymmetry of the funnel and the contour enhanced-funnel plot (data not shown). The pooled effect sizes of ABCG2 and SLC2A9 SNPs on all outcomes are summarized in Table 3.
Discussion
We conducted a systematic review and meta-analysis to assess associations between SNPs in ABCG2 and SLC2A9 and gout, hyperuricemia and serum urate, stratified by ethnicity. ABCG2-SNPs were common in both Asians and Caucasians with a minor allele frequency of 11 to 31%, except for rs72552713 which was very rare in Asians. SLC2A9-SNPs were common in both Asians (6–41%) and Caucasians (18–40%), except for rs6855911 and rs16890979 which were rare in Asians (1–6%). For ABCG2, rs2231142 significantly increased risk of gout by about 2 to 4 times and 1.6 to 3 times in Asians and Caucasians respectively for carrying heterozygous and homozygous minor genotypes. In addition, carrying homozygous/heterozygous minor genotypes of rs72552713 also increased risk of gout by about 2.5–3.9 times in Asians. By contrast, carrying homozygous minor or heterozygous genotypes of rs2231137 reduced risk of gout by about 36–57% in Asians. Likewise, most SLC2A9-SNPs (i.e., rs1014290, rs6449213, rs6855911, rs7442295, and rs12510549) showed significantly lower risk of gout by about 25–65% in Asians and/or Caucasians, except for rs6855911 which was not significant in Asians. Carrying homozygous minor or heterozygous genotypes of rs2231142-ABCG2 also increased the risk of hyperuricemia in Asians and serum urate in both Asians and Caucasians. In addition, all SLC2A9-SNPs, except rs1014290 and rs734553, also significantly increased serum urate level.
Regarding ABCG2, risk effects of rs2231142 were consistently found on gout, hyperuricemia and increased serum urate in Asians. The risk effects were strong on gout compared to weaker effects on hyperuricemia and modest effects on urate level. There might be other mechanisms of rs2231142 that lead to gout occurrence without raising serum urate level. In Caucasians, there were weaker risk effects of rs2231142 on gout, but stronger risk effects on serum urate.
For SLC2A9, rs6855911, rs6449213 and rs7442295 could significantly lower risk of gout (OR1 of 0.35–0.38 and OR2 of 0.65–0.69) and serum urate (MD1 of − 54.30 to − 49.29 and MD2 of − 20 .65 to − 18.34) in Caucasians. The causative SNP might be any of these 3 SNPs because there was high linkage disequilibrium between rs6449213 and rs7442295 (r2 = 0.88) [31]. rs1014290 also lowered the risk of gout in Caucasians and Asians, and its effects on serum urate were significant in Asians and for MD1 in Caucasians; the lack of significance in MD2 in Caucasians is likely due to the low numbers of included studies in the latter given similar risk effects.
The percentage of males might be a potential source of heterogeneity in the effects of ABCG2 and SLC2A9 on gout, since the risk or protective effects were consistently stronger in men; this may indicate sex-specific differences in pathological mechanisms or in the handling of urate. T2D might be another source of heterogeneity (for the outcome of serum urate); excluding the study with T2D Caucasians [26], made the effects of rs1014290 and rs734553 stronger in the remaining/general population.
Considering the effect of ABCG2 SNPs on gout, effects of rs2231142 were consistent with the recent updated meta-analyses that pooled allele [12] and genotype [11] effects. Our pooling had 12 additional studies and stratified by Asian and Caucasian ethnicity because of different allele frequencies across these populations. Our results for SLC2A9 SNPs and gout were also generally consistent with the recent updated meta-analysis, but we were able to pool additional effects of rs1014290, rs6449213 and rs7449213 in Caucasians and also rs6855911 in Asians. Some previous reviews reported only pooled allele effects [12, 15] and some reported many different types of pooling effects [14, 16]. However, we are the first to report both pooled homozygous and heterozygous effects of these SLC2A9 SNPs on gout. Furthermore, we also explored the effects of rs72552713 on gout, rs2231142 on hyperuricemia, and SNPs of both genes on serum urate level. Risk of bias in ascertainment of genotyping and outcome were observed. However, publication bias was not evidenced in most analyses.
The ABCG2 protein is a urate efflux transporter responsible for urate excretion [42, 46]. In vitro studies showed that rs2231142 reduced ABCG2 protein expression [42, 73,74,75], ATPase activity [76] and the urate transport [21, 42, 46] resulting in higher accumulation of urate whereas rs2231137 did not change the level of ABCG2 protein expression [42, 73, 75] and the urate transport activity [21, 42]. The proposed mechanism by which rs2231137 lowers the risk of gout is still unknown. The effect of rs72552713 on increasing the risk of gout is supported by the fact that it is a nonsense variant causing a stop codon that prematurely ends protein translation of ABCG2; Matsuo et al. [42] also reported the absence of ABCG2 protein expression and the almost entirely removed urate transport activity due to rs72552713 in the functional analysis. It was also found that the combination of rs2231142 and rs72552713 attributed to a higher risk of hyperuricemia than other typical risk factors including obesity, alcohol consumption and increasing age [77]. ABCG2 variants were associated with gout/hyperuricemia in both Asians and Caucasians [21, 78]. Particularly, individuals of European descent who carried ABCG2 variants had a significantly earlier onset of gout/hyperuricemia and the family history of gout was significantly more frequent among them [78]. These findings are supported by a recent study showing that pediatric-onset gout/hyperuricemia patients had higher minor allele frequency of rs2231142 than those of adult-onset patients and normouricemic controls [79].
The glucose transporter 9 (GLUT9) protein, regulated by the SLC2A9 gene, is a urate reuptake transporter [3]. It was discovered that SLC2A9 polymorphisms cause renal hypouricemia type 2 by reducing the urate transport activity leading to lowering renal urate reabsorption and lower serum urate [80,81,82]. Therefore, SLC2A9 could be another promising target for lowering serum urate and the risk of gout. However, one study found that SLC2A9 variants including rs16890979 and rs3733591 did not significantly change the urate transport activity in Xenopus oocyte model [83]. Therefore, the findings regarding the functional mechanisms underlying the GWAS-identified association of SLC2A9 variants on urate transport are still not consistent. Our pooled results showed a trend of decreasing the risk of gout and serum urate in most SLC2A9 variants, except for the effect of rs3733591 on gout in Asians. More association and functional studies are required to confirm/explain this. In addition, a recent GWAS found that ABCG2, SLC2A9, ALDH2, and novel loci induce asymptomatic hyperuricemia into gout development [84]. This emphasizes the strong influence of ABCG2 and SLC2A9 variants on gout development and points out the importance of identifying high risk patients having these genetic variants.
Our study has some strengths. We pooled genotype effects stratified by ethnicity for different allele frequencies. We considered all relevant outcomes, i.e., gout, hyperuricemia, and serum urate and mode of gene effects were estimated. Although some gene effects were heterogeneous, we could identify that male gender and diabetes might be potential sources of heterogeneity. However, we could not avoid some limitations. Genotype frequencies in some studies [23, 24, 26, 27, 32, 34, 38, 48, 53, 56, 61, 67, 68, 70] were missing and thus were estimated assuming HWE. Some of these SNPs were in high linkage disequilibrium, so haplotype effects should be further assessed to determine which polymorphism had the largest causal effect. Other important confounders (e.g. diuretic use, dietary intake and alcohol consumption) of gout/urate should be considered in assessing any association, but these data were not available. Only a few number of studies conducted in African Americans and other indigenous people, so further studies in these populations should be conducted.
In conclusion, our study suggested that the ABCG2-rs2231142 SNP increased the risk of gout and serum urate in both Asians and Caucasians. In addition, ABCG2-rs72552713 also increased the risk of gout whereas ABCG2-rs2231137 reduced the risk in Asians. Furthermore, SLC2A9-SNPs (i.e., rs1014290, rs6449213, rs6855911, rs7442295, and rs12510549) significantly lowered the risk of gout in Asians and/or Caucasians. Further studies in other populations especially African Americans are still needed. Our findings may be helpful in creating more accurate risk stratification models.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Abbreviations
- ABCG2 :
-
ATP-binding cassette sub-family G member 2
- BMI:
-
Body mass index
- CI:
-
Confidence interval
- eGFR:
-
Estimated glomerular filtration rate
- GWAS:
-
Genome-wide association study
- HWE:
-
Hardy-Weinberg equilibrium
- MD:
-
Mean difference
- OR:
-
Odds ratio
- SD:
-
Standard deviation
- SLC2A9 :
-
Solute carrier family 2 member 9
- SNP:
-
Single nucleotide polymorphism
- T2D:
-
Type 2 diabetes
References
Pittman JR, Bross MH. Diagnosis and management of gout. Am Fam Physician. 1999;59(7):1799–806.
Johnson RJ, Kang DH, Feig D, Kivlighn S, Kanellis J, Watanabe S, et al. Is there a pathogenetic role for uric acid in hypertension and cardiovascular and renal disease? Hypertension. 2003;41(6):1183–90.
Dehghan A, Kottgen A, Yang Q, Hwang SJ, Kao WL, Rivadeneira F, et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372(9654):1953–61.
Okada Y, Sim X, Go MJ, Wu JY, Gu D, Takeuchi F, et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nat Genet. 2012;44(8):904–9.
Kottgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. 2013;45(2):145–54.
Yang B, Mo Z, Wu C, Yang H, Yang X, He Y, et al. A genome-wide association study identifies common variants influencing serum uric acid concentrations in a Chinese population. BMC Med Genet. 2014;7:10.
Kolz M, Johnson T, Sanna S, Teumer A, Vitart V, Perola M, et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 2009;5(6):e1000504.
Qiu Y, Liu H, Qing Y, Yang M, Tan X, Zhao M, et al. The ABCG2 gene Q141K polymorphism contributes to an increased risk of gout: a meta-analysis of 2185 cases. Mod Rheumatol. 2014;24(5):829–34.
Lv X, Zhang Y, Zeng F, Yin A, Ye N, Ouyang H, et al. The association between the polymorphism rs2231142 in the ABCG2 gene and gout risk: a meta-analysis. Clin Rheumatol. 2014;33(12):1801–5.
Li R, Miao L, Qin L, Xiang Y, Zhang X, Peng H, et al. A meta-analysis of the associations between the Q141K and Q126X ABCG2 gene variants and gout risk. Int J Clin Exp Pathol. 2015;8(9):9812–23.
Dong Z, Guo S, Yang Y, Wu J, Guan M, Zou H, et al. Association between ABCG2 Q141K polymorphism and gout risk affected by ethnicity and gender: a systematic review and meta-analysis. Int J Rheum Dis. 2015;18(4):382–91.
Chen Y, Ying Y, Shi S, Li L, Huang Y, Ye H, et al. A systematic review and meta-analysis of 4 candidate polymorphisms with the risk of gout. Int J Clin Exp Med. 2016;9(8):16025–33.
Wu L, He Y, Zhang D. Meta-analysis on relationship between single nucleotide polymorphism of rs2231142 in ABCG2 gene and gout in east Asian population. Zhonghua Liu Xing Bing Xue Za Zhi. 2015;36(11):1291–6.
Meng Q, Yue J, Shang M, Shan Q, Qi J, Mao Z, et al. Correlation of GLUT9 polymorphisms with gout risk. Medicine (Baltimore). 2015;94(44):e1742.
Zhang X, Yang X, Wang M, Li X, Xia Q, Xu S, et al. Association between SLC2A9 (GLUT9) gene polymorphisms and gout susceptibility: an updated meta-analysis. Rheumatol Int. 2016;36(8):1157–65.
Lee YH, Seo YH, Kim JH, Choi SJ, Ji JD, Song GG. Associations between SLC2A9 polymorphisms and gout susceptibility : a meta-analysis. Z Rheumatol. 2017;76(1):64–70.
Thakkinstian A, McKay GJ, McEvoy M, Chakravarthy U, Chakrabarti S, Silvestri G, et al. Systematic review and meta-analysis of the association between complement component 3 and age-related macular degeneration: a HuGE review and meta-analysis. Am J Epidemiol. 2011;173(12):1365–79.
Thakkinstian A, McElduff P, D'Este C, Duffy D, Attia J. A method for meta-analysis of molecular association studies. Stat Med. 2005;24(9):1291–306.
Minelli C, Thompson JR, Abrams KR, Thakkinstian A, Attia J. The choice of a genetic model in the meta-analysis of molecular association studies. Int J Epidemiol. 2005;34(6):1319–28.
Chen CJ, Tseng CC, Yen JH, Chang JG, Chou WC, Chu HW, et al. ABCG2 contributes to the development of gout and hyperuricemia in a genome-wide association study. Sci Rep. 2018;8(1):3137.
Higashino T, Takada T, Nakaoka H, Toyoda Y, Stiburkova B, Miyata H, et al. Multiple common and rare variants of ABCG2 cause gout. RMD Open. 2017;3(2):e000464.
Stiburkova B, Pavelcova K, Zavada J, Petru L, Simek P, Cepek P, et al. Functional non-synonymous variants of ABCG2 and gout risk. Rheumatology (Oxford). 2017;56(11):1982–92.
Yu KH, Chang PY, Chang SC, Wu-Chou YH, Wu LA, Chen DP, et al. A comprehensive analysis of the association of common variants of ABCG2 with gout. Sci Rep. 2017;7(1):9988.
Li Z, Zhou Z, Hou X, Lu D, Yuan X, Lu J, et al. Replication of gout/urate concentrations GWAS susceptibility loci associated with gout in a Han Chinese population. Sci Rep. 2017;7(1):4094.
Zheng C, Yang H, Wang Q, Rao H, Diao Y. Association analysis of five SNP variants with gout in the Minnan population in China. Turk J Med Sci. 2016;46(2):361–7.
Bartakova V, Kuricova K, Pacal L, Nova Z, Dvorakova V, Svrckova M, et al. Hyperuricemia contributes to the faster progression of diabetic kidney disease in type 2 diabetes mellitus. J Diabetes Complicat. 2016;30(7):1300–7.
Oetjens MT, Bush WS, Denny JC, Birdwell K, Kodaman N, Verma A, et al. Evidence for extensive pleiotropy among pharmacogenes. Pharmacogenomics. 2016;17(8):853–66.
Tu HP, Chung CM, Min-Shan Ko A, Lee SS, Lai HM, Lee CH, et al. Additive composite ABCG2, SLC2A9 and SLC22A12 scores of high-risk alleles with alcohol use modulate gout risk. J Hum Genet. 2016;61(9):803–10.
Kannangara DR, Phipps-Green AJ, Dalbeth N, Stamp LK, Williams KM, Graham GG, et al. Hyperuricaemia: contributions of urate transporter ABCG2 and the fractional renal clearance of urate. Ann Rheum Dis. 2016;75(7):1363–6.
Cheng ST, Wu S, Su CW, Teng MS, Hsu LA, Ko YL. Association of ABCG2 rs2231142-A allele and serum uric acid levels in male and obese individuals in a Han Taiwanese population. J Formos Med Assoc. 2017;116(1):18–23.
Kim YS, Kim Y, Park G, Kim SK, Choe JY, Park BL, et al. Genetic analysis of ABCG2 and SLC2A9 gene polymorphisms in gouty arthritis in a Korean population. Korean J Intern Med. 2015;30(6):913–20.
Jiri M, Zhang L, Lan B, He N, Feng T, Liu K, et al. Genetic variation in the ABCG2 gene is associated with gout risk in the Chinese Han population. Clin Rheumatol. 2016;35(1):159–63.
Wan W, Xu X, Zhao DB, Pang YF, Wang YX. Polymorphisms of uric transporter proteins in the pathogenesis of gout in a Chinese Han population. Genet Mol Res. 2015;14(1):2546–50.
Matsuo H, Yamamoto K, Nakaoka H, Nakayama A, Sakiyama M, Chiba T, et al. Genome-wide association study of clinically defined gout identifies multiple risk loci and its association with clinical subtypes. Ann Rheum Dis. 2016;75(4):652–9.
Zhou D, Liu Y, Zhang X, Gu X, Wang H, Luo X, et al. Functional polymorphisms of the ABCG2 gene are associated with gout disease in the Chinese Han male population. Int J Mol Sci. 2014;15(5):9149–59.
Stiburkova B, Pavlikova M, Sokolova J, Kozich V. Metabolic syndrome, alcohol consumption and genetic factors are associated with serum uric acid concentration. PLoS One. 2014;9(5):e97646.
Tu HP, Ko AM, Chiang SL, Lee SS, Lai HM, Chung CM, et al. Joint effects of alcohol consumption and ABCG2 Q141K on chronic tophaceous gout risk. J Rheumatol. 2014;41(4):749–58.
Takeuchi F, Yamamoto K, Isono M, Katsuya T, Akiyama K, Ohnaka K, et al. Genetic impact on uric acid concentration and hyperuricemia in the Japanese population. J Atheroscler Thromb. 2013;20(4):351–67.
Hu M, Tomlinson B. Gender-dependent associations of uric acid levels with a polymorphism in SLC2A9 in Han Chinese patients. Scand J Rheumatol. 2012;41(2):161–3.
Tabara Y, Kohara K, Kawamoto R, Hiura Y, Nishimura K, Morisaki T, et al. Association of four genetic loci with uric acid levels and reduced renal function: the J-SHIPP Suita study. Am J Nephrol. 2010;32(3):279–86.
Yamagishi K, Tanigawa T, Kitamura A, Kottgen A, Folsom AR, Iso H. The rs2231142 variant of the ABCG2 gene is associated with uric acid levels and gout among Japanese people. Rheumatology (Oxford). 2010;49(8):1461–5.
Matsuo H, Takada T, Ichida K, Nakamura T, Nakayama A, Ikebuchi Y, et al. Common defects of ABCG2, a high-capacity urate exporter, cause gout: a function-based genetic analysis in a Japanese population. Sci Transl Med. 2009;1(5):5ra11.
Brandstatter A, Lamina C, Kiechl S, Hunt SC, Coassin S, Paulweber B, et al. Sex and age interaction with genetic association of atherogenic uric acid concentrations. Atherosclerosis. 2010;210(2):474–8.
Wang B, Miao Z, Liu S, Wang J, Zhou S, Han L, et al. Genetic analysis of ABCG2 gene C421A polymorphism with gout disease in Chinese Han male population. Hum Genet. 2010;127(2):245–6.
Stark K, Reinhard W, Grassl M, Erdmann J, Schunkert H, Illig T, et al. Common polymorphisms influencing serum uric acid levels contribute to susceptibility to gout, but not to coronary artery disease. PLoS One. 2009;4(11):e7729.
Woodward OM, Kottgen A, Coresh J, Boerwinkle E, Guggino WB, Kottgen M. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc Natl Acad Sci U S A. 2009;106(25):10338–42.
Phipps-Green AJ, Merriman ME, Topless R, Altaf S, Montgomery GW, Franklin C, et al. Twenty-eight loci that influence serum urate levels: analysis of association with gout. Ann Rheum Dis. 2016;75(1):124–30.
Urano W, Taniguchi A, Inoue E, Sekita C, Ichikawa N, Koseki Y, et al. Effect of genetic polymorphisms on development of gout. J Rheumatol. 2013;40(8):1374–8.
Wang Qiong WC, Xi-bo W, Zhi-min M, Ying C, Chang-gui L. Association between gout and polymorphisms of rs2231142 in ABCG2 in female Han Chinese. Prog Mod Biomed. 2014;14(13):4.
Tu HP, Min-Shan Ko A, Lee SS, Lee CP, Kuo TM, Huang CM, et al. Variants of ALPK1 with ABCG2, SLC2A9, and SLC22A12 increased the positive predictive value for gout. J Hum Genet. 2018;63(1):63–70.
Kobylecki CJ, Afzal S, Nordestgaard BG. Plasma urate, cancer incidence, and all-cause mortality: a mendelian randomization study. Clin Chem. 2017;63(6):1151–60.
Zhang XY, Geng TT, Liu LJ, Yuan DY, Feng T, Kang LL, et al. SLC2A9 and ZNF518B polymorphisms correlate with gout-related metabolic indices in Chinese Tibetan populations. Genet Mol Res. 2015;14(3):9915–21.
Laston SL, Voruganti VS, Haack K, Shah VO, Bobelu A, Bobelu J, et al. Genetics of kidney disease and related cardiometabolic phenotypes in Zuni Indians: the Zuni kidney project. Front Genet. 2015;6:6.
Mallamaci F, Testa A, Leonardis D, Tripepi R, Pisano A, Spoto B, et al. A genetic marker of uric acid level, carotid atherosclerosis, and arterial stiffness: a family-based study. Am J Kidney Dis. 2015;65(2):294–302.
Testa A, Mallamaci F, Spoto B, Pisano A, Sanguedolce MC, Tripepi G, et al. Association of a polymorphism in a gene encoding a urate transporter with CKD progression. Clin J Am Soc Nephrol. 2014;9(6):1059–65.
Voruganti VS, Kent JW Jr, Debnath S, Cole SA, Haack K, Goring HH, et al. Genome-wide association analysis confirms and extends the association of SLC2A9 with serum uric acid levels to Mexican Americans. Front Genet. 2013;4:279.
Lyngdoh T, Vuistiner P, Marques-Vidal P, Rousson V, Waeber G, Vollenweider P, et al. Serum uric acid and adiposity: deciphering causality using a bidirectional Mendelian randomization approach. PLoS One. 2012;7(6):e39321.
Hollis-Moffatt JE, Gow PJ, Harrison AA, Highton J, Jones PB, Stamp LK, et al. The SLC2A9 nonsynonymous Arg265His variant and gout: evidence for a population-specific effect on severity. Arthritis Res Ther. 2011;13(3):R85.
Liu WC, Hung CC, Chen SC, Lin MY, Chen LI, Hwang DY, et al. The rs1014290 polymorphism of the SLC2A9 gene is associated with type 2 diabetes mellitus in Han Chinese. Exp Diabetes Res. 2011;2011:527520.
Guan M, Zhou D, Ma W, Chen Y, Zhang J, Zou H. Association of an intronic SNP of SLC2A9 gene with serum uric acid levels in the Chinese male Han population by high-resolution melting method. Clin Rheumatol. 2011;30(1):29–35.
Cummings N, Dyer TD, Kotea N, Kowlessur S, Chitson P, Zimmet P, et al. Genome-wide scan identifies a quantitative trait locus at 4p15.3 for serum urate. Eur J Hum Genet. 2010;18(11):1243–7.
Urano W, Taniguchi A, Anzai N, Inoue E, Sekita C, Endou H, et al. Association between GLUT9 and gout in Japanese men. Ann Rheum Dis. 2010;69(5):932–3.
Hollis-Moffatt JE, Xu X, Dalbeth N, Merriman ME, Topless R, Waddell C, et al. Role of the urate transporter SLC2A9 gene in susceptibility to gout in New Zealand Maori, Pacific Island, and Caucasian case-control sample sets. Arthritis Rheum. 2009;60(11):3485–92.
Tu HP, Chen CJ, Tovosia S, Ko AM, Lee CH, Ou TT, et al. Associations of a non-synonymous variant in SLC2A9 with gouty arthritis and uric acid levels in Han Chinese subjects and Solomon islanders. Ann Rheum Dis. 2010;69(5):887–90.
Brandstatter A, Kiechl S, Kollerits B, Hunt SC, Heid IM, Coassin S, et al. Sex-specific association of the putative fructose transporter SLC2A9 variants with uric acid levels is modified by BMI. Diabetes Care. 2008;31(8):1662–7.
Stark K, Reinhard W, Neureuther K, Wiedmann S, Sedlacek K, Baessler A, et al. Association of common polymorphisms in GLUT9 gene with gout but not with coronary artery disease in a large case-control study. PLoS One. 2008;3(4):e1948.
Vitart V, Rudan I, Hayward C, Gray NK, Floyd J, Palmer CN, et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat Genet. 2008;40(4):437–42.
Doring A, Gieger C, Mehta D, Gohlke H, Prokisch H, Coassin S, et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat Genet. 2008;40(4):430–6.
Li S, Sanna S, Maschio A, Busonero F, Usala G, Mulas A, et al. The GLUT9 gene is associated with serum uric acid levels in Sardinia and chianti cohorts. PLoS Genet. 2007;3(11):e194.
Nakayama A, Nakaoka H, Yamamoto K, Sakiyama M, Shaukat A, Toyoda Y, et al. GWAS of clinically defined gout and subtypes identifies multiple susceptibility loci that include urate transporter genes. Ann Rheum Dis. 2017;76(5):869–77.
Mahfudzah A, Nazihah MY, Wan Syamimee WG, Huay Lin T, Wan Rohani WT. Suggestive evidence of Slc2a9 polymorphisms association in gouty malay males. Int Med J Malaysia. 2015;14(2):35–9.
Wallace SL, Robinson H, Masi AT, Decker JL, McCarty DJ, Yu TF. Preliminary criteria for the classification of the acute arthritis of primary gout. Arthritis Rheum. 1977;20(3):895–900.
Imai Y, Nakane M, Kage K, Tsukahara S, Ishikawa E, Tsuruo T, et al. C421A polymorphism in the human breast cancer resistance protein gene is associated with low expression of Q141K protein and low-level drug resistance. Mol Cancer Ther. 2002;1(8):611–6.
Furukawa T, Wakabayashi K, Tamura A, Nakagawa H, Morishima Y, Osawa Y, et al. Major SNP (Q141K) variant of human ABC transporter ABCG2 undergoes lysosomal and proteasomal degradations. Pharm Res. 2009;26(2):469–79.
Tamura A, Wakabayashi K, Onishi Y, Takeda M, Ikegami Y, Sawada S, et al. Re-evaluation and functional classification of non-synonymous single nucleotide polymorphisms of the human ATP-binding cassette transporter ABCG2. Cancer Sci. 2007;98(2):231–9.
Mizuarai S, Aozasa N, Kotani H. Single nucleotide polymorphisms result in impaired membrane localization and reduced atpase activity in multidrug transporter ABCG2. Int J Cancer. 2004;109(2):238–46.
Nakayama A, Matsuo H, Nakaoka H, Nakamura T, Nakashima H, Takada Y, et al. Common dysfunctional variants of ABCG2 have stronger impact on hyperuricemia progression than typical environmental risk factors. Sci Rep. 2014;4:5227.
Toyoda Y, Mančíková A, Krylov V, Morimoto K, Pavelcová K, Bohatá J, et al. Functional characterization of clinically-relevant rare variants in ABCG2 identified in a gout and hyperuricemia cohort. Cells. 2019;8(4):363.
Stiburkova B, Pavelcova K, Pavlikova M, Ješina P, Pavelka K. The impact of dysfunctional variants of ABCG2 on hyperuricemia and gout in pediatric-onset patients. Arthritis Res Ther. 2019;21(1):77.
Matsuo H, Chiba T, Nagamori S, Nakayama A, Domoto H, Phetdee K, et al. Mutations in glucose transporter 9 gene SLC2A9 cause renal hypouricemia. Am J Hum Genet. 2008;83(6):744–51.
Dinour D, Gray NK, Campbell S, Shu X, Sawyer L, Richardson W, et al. Homozygous SLC2A9 mutations cause severe renal hypouricemia. J Am Soc Nephrol. 2010;21(1):64–72.
Mancikova A, Krylov V, Hurba O, Sebesta I, Nakamura M, Ichida K, et al. Functional analysis of novel allelic variants in URAT1 and GLUT9 causing renal hypouricemia type 1 and 2. Clin Exp Nephrol. 2016;20(4):578–84.
Hurba O, Mancikova A, Krylov V, Pavlikova M, Pavelka K, Stibůrková B. Complex analysis of urate transporters SLC2A9, SLC22A12 and functional characterization of non-synonymous allelic variants of GLUT9 in the Czech population: no evidence of effect on hyperuricemia and gout. PLoS One. 2014;9(9):e107902.
Kawamura Y, Nakaoka H, Nakayama A, Okada Y, Yamamoto K, Higashino T, et al. Genome-wide association study revealed novel loci which aggravate asymptomatic hyperuricaemia into gout. Ann Rheum Dis. 2019;78(10):1430–7.
Acknowledgments
The authors would like to thank Mr. Stephen Pinder for proof-reading this article.
Funding
This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
TL designed the study, performed acquisition of data, analysis and interpretation of data, and drafting the article. SR performed acquisition, analysis and interpretation of data. ST and NS performed acquisition of data. AI and JA performed interpretation of data. AT designed the study and performed interpretation of data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethics approval and consent to participate are not required for this study.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
Search strategies. (DOCX 26 kb)
Additional file 2.
Characteristics of included studies investigating associations between ABCG2 and SLC2A9 polymorphisms and urate. (DOCX 146 kb)
Additional file 3.
Risk of bias assessment. (DOCX 26 kb)
Additional file 4.
Gout. 4.1. Data used for pooling effects of ABCG2 and SLC2A9 polymorphisms on gout. 4.2. Pooled prevalence of minor allele of ABCG2 and SLC2A9 polymorphisms. 4.3. Exploring source of heterogeneity for ABCG2 and SLC2A9 polymorphisms on gout.4.4. Egger’s tests for ABCG2 and SLC2A9 polymorphisms on gout. 4.5. Funnel plots for ABCG2 and SLC2A9 polymorphisms on gout. (DOCX 662 kb)
Additional file 5.
Hyperuricemia. 5.1. Data used for pooling effects of ABCG2-rs2231142 on hyperuricemia. 5.2. Exploring source of heterogeneity for ABCG2-rs2231142 on hyperuricemia. 5.3. Egger’s tests for ABCG2-rs2231142 on hyperuricemia. 5.4. Funnel plots of ABCG2-rs2231142 on hyperuricemia in Asians. A) OR1 in Asians B) OR2 in Asians. (DOCX 77 kb)
Additional file 6.
Serum urate. 6.1. Data used for pooling mean difference of ABCG2 and SLC2A9 polymorphisms on serum urate. 6.2. Exploring source of heterogeneity for ABCG2 and SLC2A9 polymorphisms on serum urate. 6.3. Egger’s tests for ABCG2 and SLC2A9 polymorphisms on serum urate. 6.4. Funnel plots for ABCG2 and SLC2A9 polymorphisms on serum urate. (DOCX 412 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lukkunaprasit, T., Rattanasiri, S., Turongkaravee, S. et al. The association between genetic polymorphisms in ABCG2 and SLC2A9 and urate: an updated systematic review and meta-analysis. BMC Med Genet 21, 210 (2020). https://doi.org/10.1186/s12881-020-01147-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-020-01147-2