Abstract
Background
In a previous study, using a molecular approach, we reported the presence of P. vivax in Namibia. Here, we have extended our investigation to the Duffy antigen genetic profile of individuals of the same cohort with and without Plasmodium infections.
Methods
Participants with P. vivax (n = 3), P. falciparum (n = 23) mono-infections and co-infections of P. vivax/P. falciparum (n = 4), and P. falciparum/P. ovale (n = 3) were selected from seven regions. Participants with similar age but without any Plasmodium infections (n = 47) were also selected from all the regions. Duffy allelic profile was examined using standard PCR followed by sequencing of amplified products. Sequenced samples were also examined for the presence or absence of G125A mutation in codon 42, exon 2.
Results
All individuals tested carried the − 67 T > C mutation. However, while all P. vivax infected participants carried the c.G125A mutation, 7/28 P. falciparum infected participants and 9/41 of uninfected participants did not have the c.G125A mutation. The exon 2 region surrounding codon 42, had a c.136G > A mutation that was present in all P. vivax infections. The odds ratio for lack of this mutation with P. vivax infections was (OR 0.015, 95% CI 0.001–0.176; p = 0.001).
Conclusion
We conclude that P. vivax infections previously reported in Namibia, occurred in Duffy negative participants, carrying the G125A mutation in codon 42. The role of the additional mutation c.136 G > A in exon 2 in P. vivax infections, will require further investigations.
Similar content being viewed by others
Background
The life cycle of Plasmodium species in human hosts is initiated when an infected Anopheles mosquito injects sporozoites into the skin [1,2,3]. A fraction of the sporozoites move from the skin to circulation and take residence in the liver [4]. Here, the sporozoites multiply extensively to generate thousands of merozoites. Next, liver merozoites enter the circulation and infect red blood cells (RBCs) and initiate the erythrocytic cycle [5]. In RBCs, erythrocytic schizogony goes through a ring stage (immature trophozoites), mature trophozoites and merozoites stages, which is repeated overtime. This stage is responsible for the clinical symptoms of infection, due in part to RBCs lysis and release of their contents into the blood stream leading to a pro-inflammatory immune response [6]. Plasmodium vivax (P. vivax) is unique in that it has preference for immature reticulocytes during its erythrocytic cycle [7].
Duffy antigen receptor for chemokines (DARC) also called cluster of differentiation 234 (CD234) or atypical chemokine receptor (ACKR) is a receptor for a family of proinflammatory chemokines [8,9,10]. It was discovered to be a receptor for P. knowlesi and P. vivax and on the surface of RBCs in the 1970’s [11]. It is also present on the surface of endothelial cells [12]. The DARC glycoprotein is encoded by the FY gene. Two codominant alleles FY*A and FY*B exist. These alleles produce the respective antigens Fya and Fyb, which differ by a point mutation at position c.125G > A [13]. The Duffy negative/null antigen Fy(a−b−) is associated with a cI-67 T > C mutation on the GATA-1 transcription factor binding motif of the Fy*A/B genes [14]. The mutation on Fyb is commonly seen in sub-Saharan Africans, while the Fya mutation is common in Papua New Guineans [15, 16]. The null mutation on the Fyb allele was thought to be responsible for resistance of invasion of reticulocytes by P. vivax in sub-Saharan Africans [17]. However, there have been cumulative evidence using microscopy and/or molecular tools for the presence of P. vivax parasites in Duffy negative individuals in Sub-Saharan Africans [18, 19]. Since most of these studies were performed with subjects who had mostly no travel history to known endemic areas, this raises the question as to whether P. vivax is now emerging, having adapted to invade reticulocytes independent of DARC. Alternatively, it is possible that P. vivax infection did always occur in Duffy negative individuals, but its diagnosis was overlooked.
As a first approach to assess the prevalence of P. vivax infection in Duffy negative infected people, active case detection needs to be done across sub-Saharan Africa. This will be the basis of the understanding of P. vivax bionomics allowing comprehensive studies of invasion of reticulocytes and survival in its human host.
We recently reported the identification of P. vivax in Namibia [20]. Here, we have followed up to assess the Duffy status of the subjects who were infected. Our results add to the growing evidence that P. vivax infected individuals can be Duffy negative.
Methods
Study sites and population selection
The details of the sample selection are as previously published [20]. Samples from the following regions respectively grouped into infected and uninfected, were included: Kunene (0:8), Omusati (3:8), Oshana (2:10), Ohangwena (7:5), Kavango West (2:2), Kavango East (17:7) and Zambezi (Caprivi) (2:7). Plasmodium infected participants previously published were selected and uninfected with similar ages were also selected. The Plasmodium infected participants totaled 33 while 47 were uninfected. The Plasmodium infected participants consisted of the following categories: P. vivax mono-infection (n = 3), P. falciparum mono-infection (n = 23), P. vivax/P. falciparum mixed infection (n = 4) and P. falciparum/P. ovale mixed infection (n = 3).
Ethics statement
The study was approved by the Ministry of Health and Social Services (MOHSS) Ethical Committee, Namibia. All parents/guardians provided informed consent on behalf of all participants. Where necessary, assent was also obtained from the child before sample collection.
Blood sample collection
The details of the procedure are as published previously [20]. In brief, an aliquot of 1.5–2.5 ml venous blood was collected into EDTA tubes and centrifuged at 3000 rpm for 5 min to separate the buffy coat, plasma and red blood cells into separate tubes. These were then stored at − 20 °C and later transferred to − 80 °C until analyzed.
Laboratory analyses
DNA extraction
Genomic DNA was extracted from pelleted red blood cells using the automated Hamilton Star Microlab Workstation (Hamilton Bonaduz AG, Bonaduz, Switzerland) with the Machery and Nagel 96 blood DNA extraction kit. The starting blood sample was 200 μl of the packed and thawed red blood cells (RBCs), and final DNA elution volume of 120 μl sterile PCR-grade water.
Molecular detection of DARC
All detection assays were single-plexed and run in a high throughput 96 well plate Applied BioSystem GeneAmp 9700 PCR system, Singapore. Primers were ordered from Eurogentec, Liege, Belgium. All PCR amplification reactions were carried out in a total volume of 50 μl. For the standard PCR amplification reaction, 2.5 μl of DNA extracted from pelleted red blood cells was used, together with the KOD hotstart enzyme and the reaction mix was obtained from Sigma Chemical (Merck, Darmstadt, Germany). The amplified products were analyzed on 1.5% agarose gels by electrophoresis, followed by visualization on a UVP Geldoc-it Imager TS 310 (Cambridge, UK) after ethidium bromide staining. The amplified products were subsequently sent to Inqaba Biotech™, Pretoria, South Africa, for sequencing on both strands. Sequencing was done 2 × to confirm the observed mutation. The PCR cycling parameters for the primary amplifications were as follows: Initial denaturation at 95 °C for 2 min, then 30 cycles each of denaturation at 95 °C for 20 s, annealing at 62 °C for 10 s, extension at 70 °C for 11 s and a final hold at 4 °C. The primers had a final concentration of 0.2 µM with sequences of: Forward: CTCATTAGTCCTTGGCTCTTAC and Reverse: AGCTGCTTCCAGGTTGGCAC, and AGCTGCTTCCAGGTTGGCAT. The amplified products were 711 bp.
Statistical analysis
Data were entered in an Excel data sheet and IBM Corp SPSS version 26 (IBM Corp. Released 2019. IBM Armonk, NY: USA) was used for analysis. Descriptive statistics and appropriate measures of central tendency were provided for relevant demographic covariates. Multinomial logistic regression was used to assess association of a mutation with a risk of infection with a Plasmodium parasite. A p value < 0.05 was considered significant.
Results
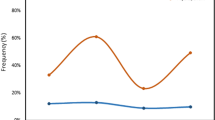
The median ages of the Plasmodium infected and uninfected individuals were respectively, 5 years (25–75 percentile, 2–8 years) and 6 years (25–75 percentile, 4–7 years). Data was obtained for all samples at the − 67 T > C locus. Similarly, data was obtained for Plasmodium infected participants at the c.125G > A and c136G > A locus. However, in uninfected participants, 41/47 sequence reads were obtained at the c.125G > A and c.136G > A locus (Table 1). P. vivax infected participants (5/7) were Duffy negative (− 67 T > C mutation) as were all other Plasmodium infected individuals tested (Fig. 1A). The remaining two P. vivax samples could not be examined because of insufficient DNA. All P. vivax infected participants had the c.125G > A mutation (Fig. 1B). However, 9/41 of the Plasmodium uninfected participants (6 samples reads were not good within the region) and 7/28 of the P. falciparum infected participants (Table 1), had the FY*A genotype (G at c.125 in exon 2) (Fig. 1B). There was a c.136 G > A mutation in exon 2 that was present in all P. vivax infections (5/7). The odds ratio for lack of the mutation with P. vivax infection was (OR 0.015, 95% CI 0.001–0.176; p = 0.001).
Discussion
The present study compliments our previous investigation reporting the presence of P. vivax in Namibia [20]. The data clearly shows that the P. vivax infections previously observed in children, occurred in Duffy negative individuals. The observed mutation at c.136G > A occurring together with P. vivax Duffy negative infections in the FY*B allele needs to be examined further, as to whether it has any significant role in the infection dynamics of P. vivax. This mutation has not been previously published, to the best of our knowledge. Other rare DARC polymorphisms that have been reported are the c.265C > T mutation in the FY*B allele leading to the FY*X allele, which has a reduced expression of the gene by 90% [21], and the c.298G > A mutation resulting in a codon change from Ala100Thr [22], that reduces the expression of the Duffy antigen in erythrocytes. It is also interesting that none of the P. vivax infected participants carried a FY*A allele. P. vivax malaria is less benign than previously thought and considering that its life cycle is complicated by hypnozoites and early gametocyte release [23], the reports of the emergence of P. vivax in sub-Saharan Africa are a cause for concern. In Southern America it has been observed that recurrent infections of P. vivax frequently occur, even in subjects who had received full radical cure with primaquine [24, 25]. This shows that elimination of P. vivax is a difficult task requiring the National Malaria Control Programs (NMCPs) in African countries to plan accordingly.
The recent detections of P. vivax infections in Duffy-negative individuals suggest that the resistance associated with Duffy antigen negativity by P. vivax to reticulocyte infection is incomplete. [18]. The precise function of DARC on erythrocytes has not been deciphered. In general, however, it is known to be associated with the modulation of chemokine levels locally and systemically, to dampen inflammatory response [26, 27]. As to how this can impact parasite invasion and survival in the reticulocyte or erythrocyte is unknown. It has been reported that plasma and serum chemokine levels differ between individuals with FY*A and FY*B alleles [28]. The FY*B allele was observed to have a lower expression of the Duffy antigen than FY*A allele [29], although there was no effect on the binding affinities for chemokines. More data is required, examining new mutations in individuals infected or not infected with Plasmodium species to further understand the links with chemokine expression and inflammatory responses and how that relates to P. vivax entry into reticulocytes.
Conclusion
The emergence of P. vivax infections in Duffy negative individuals in sub-Saharan Africa is a cause for concern. Namibia joins the list of these countries, which requires serious attention on the malaria elimination and eradication agenda of sub-Saharan Africa and partners.
Availability of data and materials
The datasets generated and/or analyzed during the current study are not publicly available due restrictions imposed by the Ethical Committee of the Ministry of Health and Social Services, but are available from the corresponding author on reasonable request.
Abbreviations
- RBCs:
-
Red blood cells
- DARC:
-
Duffy antigen receptor for chemokines
- CD234:
-
Cluster of differentiation 234
- ACKR:
-
Atypical chemokine receptor
- GATA:
-
DNA binding sequence by transcription factors
- FY:
-
Duffy glycoprotein gene
- MOHSS:
-
Ministry of Health and Social Services
- EDTA:
-
Ethylene-diamine tetra acetic acid
- PCR:
-
Polymerase chain reaction
- KOD:
-
A DNA polymerase
- UVP:
-
Ultra violet protection
- NMCP:
-
National Malaria Control Program
References
Beier JC. Malaria parasite development in mosquitoes. Annu Rev Entomol. 1998;43:519–43.
Frevert U. Sneaking in through the back entrance: the biology of malaria liver stages. Trends Parasitol. 2004;20(9):417–24.
Kappe SH, Kaiser K, Matuschewski K. The Plasmodium sporozoite journey: a rite of passage. Trends Parasitol. 2003;19(3):135–43.
Cowman AF, Kappe SH. Microbiology. Malaria’s stealth shuttle. Science. 2006;313(5791):1245–6.
Cowman AF, Berry D, Baum J. The cellular and molecular basis for malaria parasite invasion of the human red blood cell. J Cell Biol. 2012;198(6):961–71.
Bannister L, Mitchell G. The ins, outs and roundabouts of malaria. Trends Parasitol. 2003;19(5):209–13.
Malleret B, Li A, Zhang R, Tan KS, Suwanarusk R, Claser C, et al. Plasmodium vivax: restricted tropism and rapid remodeling of CD71-positive reticulocytes. Blood. 2015;125(8):1314–24.
Demogines A, Truong KA, Sawyer SL. Species-specific features of DARC, the primate receptor for Plasmodium vivax and Plasmodium knowlesi. Mol Biol Evol. 2012;29(2):445–9.
Horuk R. The Duffy antigen receptor for chemokines DARC/ACKR1. Front Immunol. 2015;5(6):279.
Bachelerie F, Ben-Baruch A, Burkhardt AM, Combadiere C, Farber JM, Graham GJ, et al. International Union of Basic and Clinical Pharmacology. [corrected]. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol Rev. 2013;66(1):1–79.
Miller LH, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science. 1975;189(4202):561–3.
Chaudhuri A, Polyakova J, Zbrzezna V, Williams K, Gulati S, Pogo AO. Cloning of glycoprotein D cDNA, which encodes the major subunit of the Duffy blood group system and the receptor for the Plasmodium vivax malaria parasite. Proc Natl Acad Sci U S A. 1993;90(22):10793–7.
Iwamoto S, Omi T, Kajii E, Ikemoto S. Genomic organization of the glycoprotein D gene: Duffy blood group Fya/Fyb alloantigen system is associated with a polymorphism at the 44-amino acid residue. Blood. 1995;85(3):622–6.
Tournamille C, Colin Y, Cartron JP, Le Van KC. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10(2):224–8.
Parasol N, Reid M, Rios M, Castilho L, Harari I, Kosower NS. A novel mutation in the coding sequence of the FY*B allele of the Duffy chemokine receptor gene is associated with an altered erythrocyte phenotype. Blood. 1998;92(7):2237–43.
Hoher G, Fiegenbaum M, Almeida S. Molecular basis of the Duffy blood group system. Blood Transfus. 2018;16(1):93–100.
Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype FyFy. N Engl J Med. 1976;295(6):302–4.
Gunalan K, Niangaly A, Thera MA, Doumbo OK, Miller LH. Plasmodium vivax infections of Duffy-negative erythrocytes: historically undetected or a recent adaptation? Trends Parasitol. 2018;34(5):420–9.
Menard D, Barnadas C, Bouchier C, Henry-Halldin C, Gray LR, Ratsimbasoa A, et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci U S A. 2010;107(13):5967–71.
Haiyambo DH, Uusiku P, Mumbengegwi D, Pernica JM, Bock R, Malleret B, et al. Molecular detection of P. vivax and P. ovale foci of infection in asymptomatic and symptomatic children in Northern Namibia. PLoS Negl Trop Dis. 2019;13(5): e0007290.
Olsson ML, Hansson C, Avent ND, Akesson IE, Green CA, Daniels GL. A clinically applicable method for determining the three major alleles at the Duffy (FY) blood group locus using polymerase chain reaction with allele-specific primers. Transfusion. 1998;38(2):168–73.
Tournamille C, Le Van KC, Gane P, Cartron JP, Colin Y. Molecular basis and PCR-DNA typing of the Fya/fyb blood group polymorphism. Hum Genet. 1995;95(4):407–10.
Price RN, Tjitra E, Guerra CA, Yeung S, White NJ, Anstey NM. Vivax malaria: neglected and not benign. Am J Trop Med Hyg. 2007;77(6 Suppl):79–87.
Orjuela-Sanchez P, da Silva NS, da Silva-Nunes M, Ferreira MU. Recurrent parasitemias and population dynamics of Plasmodium vivax polymorphisms in rural Amazonia. Am J Trop Med Hyg. 2009;81(6):961–8.
Baird JK. Severe and fatal vivax malaria challenges “benign tertian malaria” dogma. Ann Trop Paediatr. 2009;29(4):251–2.
Jilma-Stohlawetz P, Homoncik M, Drucker C, Marsik C, Rot A, Mayr WR, et al. Fy phenotype and gender determine plasma levels of monocyte chemotactic protein. Transfusion. 2001;41(3):378–81.
Fukuma N, Akimitsu N, Hamamoto H, Kusuhara H, Sugiyama Y, Sekimizu K. A role of the Duffy antigen for the maintenance of plasma chemokine concentrations. Biochem Biophys Res Commun. 2003;303(1):137–9.
Schnabel RB, Baumert J, Barbalic M, Dupuis J, Ellinor PT, Durda P, et al. Duffy antigen receptor for chemokines (Darc) polymorphism regulates circulating concentrations of monocyte chemoattractant protein-1 and other inflammatory mediators. Blood. 2010;115(26):5289–99.
Xiong Z, Cavaretta J, Qu L, Stolz DB, Triulzi D, Lee JS. Red blood cell microparticles show altered inflammatory chemokine binding and release ligand upon interaction with platelets. Transfusion. 2011;51(3):610–21.
Acknowledgements
The authors acknowledge the support of the National Malaria Control Program of Namibia (NMCP), the work of nurses Mrs Foibe Kalipi and Lucia Nghishongwa in sample collection. We acknowledge all subjects who agreed to participate in the work. The authors thank the Singapore Immunology Network (SIgN) flow cytometry core headed by Dr Anis Larbi, particularly Ivy Low, Seri Mustafah, and Nurhidaya Shadan. WWARN is acknowledged for forging the collaboration with SIgN.
Funding
Merck Global Health Institute.
Author information
Authors and Affiliations
Contributions
IKQ: Conceptualization. IKQ, DHH, PU, DM, LA, RB, BM, LR: Protocol/project development. IKQ: Project administration. IKQ, PU, DM, LA, BM, LR, RB: Data collection and management. IKQ, LA, DHH, BM, LR: Data analysis. IKQ, DHH, LA, LR, DHH, BM, PU, DM, RB, DHH: Manuscript writing/editing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the MOHSS Ethical Committee, Namibia. All methods were performed in accordance with the prevalent guidelines and regulations. All parents/guardians provided informed consent on behalf of all participants. Where necessary, assent was also obtained from the child before sample collection.
Consent for publication
Not applicable.
Competing interests
None.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Haiyambo, D.H., Aleksenko, L., Mumbengegwi, D. et al. Children with Plasmodium vivax infection previously observed in Namibia, were Duffy negative and carried a c.136G > A mutation. BMC Infect Dis 21, 856 (2021). https://doi.org/10.1186/s12879-021-06573-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-021-06573-y