Abstract
Background
In intensive care unit (ICU), infection and colonization by resistant Gram-negative bacteria increase costs, length of stay and mortality. Extended-spectrum beta-lactamase − producing Enterobacteriaceae (ESBL-E) is a group of pathogens increasingly encountered in ICU setting. Conditions that promote ESBL-E acquisition are not completely understood. The increasing incidence of infections related to ESBL-E and the unsolved issues related to ESBL-E cross-transmission, prompted us to assess the rates of referred and acquired cases of ESBL-E in ICU and to assess patient-to-patient cross-transmission of ESBL-E using a multimodal microbiological analysis.
Methods
During a 5-month period, all patients admitted to a medical ICU were tested for ESBL-E carriage. A rectal swab was performed at admission and then twice a week until discharge or death. ESBL-E strains were analyzed according to antibiotic susceptibility pattern, rep-PCR (repetitive-element Polymerase chain reaction) chromosomal analysis, and plasmid PCR (Polymerase chain reaction) analysis of ESBL genes. Patient-to-patient transmission was deemed likely when 2 identical strains were found in 2 patients hospitalized simultaneously in the ICU.
Results
Among the 309 patients assessed for ESBL-E carriage on admission, 25 were found to carry ESBL-E (importation rate: 8 %). During follow-up, acquisition was observed among 19 of them (acquisition rate: 6.5 %). Using the multimodal microbiological approach, we found only one case of likely patient-to-patient ESBL-E transmission.
Conclusions
In unselected ICU patients, we found rather low rates of ESBL-E referred and acquired cases. Only 5 % of acquisitions appeared to be related to patient-to-patient transmission. These data highlight the importance of jointly analyzing phenotypic profile and molecular data to discriminate strains of ESBL-E.
Similar content being viewed by others
Background
In intensive care unit (ICU), Gram-negative bacterial resistance to antibiotic therapy increases costs, length of stay and mortality [1, 2]. One major mechanism of resistance is related to extended-spectrum beta-lactamase (ESBL) production. Since the first report in mid-1980s, the incidence of ESBL-producing Enterobacteriaceae (ESBL-E) has been increasing worldwide [3]. “Old” ESBL (derived from SHV and TEM families) were, until the year 2000, essentially related to Klebsiella pneumoniae and Escherichia coli, which were responsible for nosocomial infections, mostly in the ICU setting [4, 5]. Escherichia coli thereafter became dominant among ESBL-E. Interestingly, the change of dominant species occurred concomitantly with the emergence of enzymes that belong to the CTX-M family. These “new” ESBL have superseded the TEM- and SHV-related enzymes, and their incidence is currently increasing in the community setting [6, 7].
ESBL-E community carriage and/or hospital acquisition rates vary worldwide. In Madagascar, more than 10 % of healthy volunteers carry an ESBL-E strain [8]. In Spain, ESBL-E carriage increases between 1991 and 2003 of 1–5 % among ambulatory patients and 1–12 % among hospitalized patients [9]. In France, carriage of ESBL-E is about 1 % in healthy volunteers [10] and up to 6 % in patients admitted to a medical ward [11].
Acquisition can be due to transmission from one patient to another via health care worker’s hands. This pattern is largely accepted for glycopeptide-resistant Enterococcus (GRE), and prevention programs designed to minimize cross-transmission, have reduced this mode of acquisition [12–15]. Programs designed to prevent the spread of “old” ESBLs are less convincing and even discordant with “new” ESBLs epidemiology [16, 17]. Other patterns of acquisition include antibiotic pressure [18], and the use of antibiotics in food animal breeding [19]. Regarding the environment, some authors report possible GRE and Methicilin Resistant Staphylococcus aureus (MRSA) contamination from bedroom furniture and medical devices [20, 21], which can be decreased by reinforced environmental cleaning [22].
The relative contributions of all these factors to ESBL-E acquisition are incompletely understood [23]. Contact isolation measures are usually applied to ESBL-E carriers [14] but are potentially harmful for patients and their effectiveness is even debated [24].
The increasing incidence of infections related to community-acquired or nosocomial ESBL-E and the issues raised by data on patient-to-patient transmission, prompted us to assess colonization and acquisition rates of ESBL-E and to characterize ESBL-E cross-transmission using microbiological multimodal analysis.
Methods
Study design and patient population
This study was approved by the Comité de Protection des Personnes de l’Hôpital Saint-Antoine.
We assessed in a multimodal analysis, microbiological samples collected during routine screening for multidrug-resistant bacteria in the medical ICU of a 660-bed tertiary teaching hospital, during a period of 5 consecutive months (March 15th to August 15th, 2011). The medical ICU has 3 units containing each 6 single beds. Two physicians are in charge of a Unit. A nurse cares for 3 patients. All patients admitted to the medical ICU were given information on the study and their (or next of kin) oral consent was obtained. Every patient underwent rectal swab screening for ESBL-E carriage at admission and then twice a week until ICU discharge. Enhanced hygiene measures (protective gowns, gloves, ESBL-E announcing stickers) were applied in the case of patients colonized and/or infected by ESBL-E and preventively in patients considered at risk for ESBL-E carriage.
Microbiological methods
Screening for ESBL-E was performed by inoculating rectal swabs on selective medium supplemented with ceftazidime (bioMérieux, Marcy l’Etoile, France). After 24 h at 37 °C, the species were identified by MALDI-TOF (Matrix-Assisted Laser Desorption/Ionisation, time-of-flight mass spectrometry) analysis. Antibiotic susceptibility was tested using the standard agar diffusion method on Mueller-Hinton agar (Bio-Rad, Marnes-la-Coquette, France) according to CA-SFM 2012 guidelines. The following antibiotics (bioRad) were tested: amoxicillin, amoxicillin-clavulanate, ticarcillin, piperacillin, piperacillin-tazobactam, cefalotine, cefoxitine, moxalactam, cefotaxime, ceftazidime, aztreonam, cefepime, imipenem, ertapenem, meropenem, gentamicin, tobramycin, netilmicin amikacin, nalidixic acid, ofloxacin, ciprofloxacin and sulfamethoxazole. The double-disk synergy method was used to confirm ESBL production [25]. All ESBL-E − producing isolates were stored at −80 °C.
Molecular methods
Clonal relationships of ESBL-E isolates were investigated using the DiversiLab® fingerprinting system (bioMérieux, Marcy l'Etoile, France), a commercially available repetitive-element (rep)-PCR tool [26] successfully used for the typing of ESLB-E. This technique is faster than and as discriminating as pulsed-field gel electrophoresis [27, 28]. After thawing, strains were isolated and incubated at 37 ° C for 24 h. DNA was extracted using an UltraClean DNA isolation kit® (MO-BIO, USA). The DNA solutions were then normalized at a concentration of 35 ng/mL. For the study, we used specific rep-PCR kits respectively for Escherichia coli, Enterobacter ssp., and Klebsiella spp. species. PCR cycling parameters for all the kits were similar: an initial denaturation at 94 °C for 2 min, 35 cycles of denaturation at 94 °C for 30 s, hybridization (at 55 °C for Enterobacter spp. and Klebsiella spp. and at 50 °C for Escherichia coli) for 30 s, extension at 70 °C for 90 s and a final extension at 70 °C for 3 min. DNA amplicons were separated by a bioanalyzer (Agilent Technologies, Les Ulis, France).
DiversiLab® software was used for analysis of the results. This used the Pearson correlation coefficient to determine distance matrices and the UPGMA method (Unweighted Pair Group Method with Arithmetic Mean) to create dendrograms. Reports were automatically generated including dendrogram, electropherogram, virtual gel images and scatter plots to aid in data interpretation. A cluster was defined by a set of strains having both a similarity coefficient equal to or greater than 95 % and difference less than 2 peaks on the electropherogram.
In order to complement the results, all ESBL genes were characterized. DNA preparation and multiplex PCRs, previously developed to detect the most frequent widespread beta-lactamase genes encoding the ESBL (SHV-, TEM- and CTX-M-types), were performed as previously described [29]. PCR products were purified using the ExoSap purification kit (Illustra EXOSTAR-1 Step, Duscher, Brumath, France). All PCR products were subjected to bidirectional DNA sequencing using the BigDye terminator 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, USA). Each sequence was aligned using Applied Biosystems SeqScape® software. The nucleotide sequences and deduced protein sequences were analyzed with the BLAST and FASTA programs of the National Centre for Biotechnology Information.
Definitions
According to the results of culture and clinical data (date of admission and discharge, location and transfer within the unit, and previous admissions), each ESBL-producing strain was classified as referred or acquired in the unit. Referred cases were those that met any of the following criteria: patient with ESBL-producing strain found at admission; patient with ESBL-producing strain found from a clinical sample within 48 h following ICU admission; and/or history of colonization and/or infection with an ESBL-producing strain (defined by positive rectal swab and/or positive microbiologic exam for ESBL-E at a previous stay). The rate of referred cases was computed for patients who underwent sampling at admission. ESBL-E acquired cases were defined by the identification 72 h after admission of one ESBL-E strain among patients with negative admission screening. Identification of several novel ESBL-E strains with different beta-lactamases at the same sampling was considered as several acquisitions. Acquisition rate was computed for patients in whom admission sampling had been performed. ESBL-E acquired cases occurring among patients carrying referred strains was defined as identification 72 h after admission and during follow-up of an ESBL-E strain different from the one identified on admission sampling. Cross-transmission (patient-to-patient transmission) was recorded when strains isolated among 2 patients were similar in both beta-lactamase gene characterization and rep-PCR analysis. Moreover, the 2 patients ICU stays had to overlap.
Statistics
Quantitative values were expressed as the median with the interquartile range [25–75 %]. Rates for referred cases and acquired cases were calculated considering only patients whose screening samples at admission were available. For identification of cross-transmission, all ESBL-E strains isolated during the study were considered and acquisitions were studied, one by one, on the basis of antibiotic susceptibility pattern, ESBL-type rep PCR profile, and the patient’s period of ICU stay. The Mann–Whitney test was used to compare quantitative values. Proportions were compared using the Chi-2 test and/or the Fisher exact test, as appropriate.
Results
Demographic data
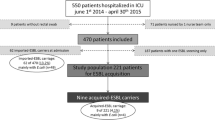
During the 5-month study period, 432 patients were admitted to the medical ICU (age 64 years [51–76], male/female ratio 56 %, IGSII 38 [27–52]) (Table 1). They were mainly admitted for medical reasons (90 %): respiratory failure (54 %), septic shock (21 %), coma (14 %), postoperative major surgery (5 %), and others pathologies (6 %). Patients came from a medical ward (54 %), from surgery (5 %), and from home (41 %). Length of stay was 5 days [3–7], and mortality was 18 %.
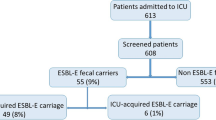
ESBL-E referred cases/acquired cases (Fig. 1)
Patients
On admission, rectal swabs were performed in 309/432 patients (72 %). Twenty-five were positive for ESBL-E (referred cases rate 8 %) with 25 strains isolated from 25 patients. During follow-up of these 309 patients, 20 acquisitions were observed with 20 strains isolated from 19 patients (acquired cases: 6.5 %). Acquisition was observed in 3/19 of patients known to carry an ESBL-E upon admission. Acquisition occurred after 7 days [4–15] and ICU length of stay was significantly higher in patients who acquired ESBL-E (12 vs. 4 days, p < 0.005). Twelve patients had a history of colonization related in their records. For six of them, an ESBL-E was identified on a rectal swab during ICU stay. Among the 123 patients for whom admission samples were not available (and who therefore were not taken into account in computing rates of imported cases at admission and acquired cases), subsequent follow-up revealed carriage of ESBL-E strains in 10 of them.
Strains
Fifty-two ESBL-producing strains of Enterobacteriaceae including E. coli (n = 27), E. cloacae (n = 11), K. pneumoniae (n = 10), and K. oxytoca (n = 4) were isolated during the study.
Phenotypic analysis
Phenotypic classification of the strains was based on Antibiotic susceptibility profile defined as: I: gentamicin- and amikacin-sensitive; II: gentamicin-resistant and amikacin-sensitive; III: gentamicin-sensitive and amikacin-resistant; a: ciprofloxacin-sensitive; b: ciprofloxacin-resistant. This allowed discriminating 5 different antibiotic resistance patterns for E. coli, 4 for E. cloacae, 5 for K. pneumoniae, and 2 for K. oxytoca (Additional file 1).
Molecular analysis
Molecular typing derived from rep-PCR analysis discriminated several clusters respectively among E coli, E cloacae, K pneumoniae, K oxytoca isolates.
-
For the 27 E. coli isolates, (Fig. 2), 17 clusters were individualized: 13 of them with a single isolate, whereas clusters 6, 7, 9 and 14 contained respectively 3, 2, 2, and 7 isolates.
-
For E. cloacae, (Fig. 3); molecular typing discriminated 7 clusters: 5 clusters each containing only one isolate, one cluster with 2 isolates, and one cluster observed with 4 isolates.
-
Three of the 10 K. pneumoniae isolates were part of the same cluster whereas each remaining seven were from a different cluster (Fig. 4).
-
The 4 K. oxytoca isolates each showed a distinct cluster (Fig. 5).
Our results showed CTX-M enzymes to be the most frequent ESBL-types (Additional file 1). ESBL for E. coli included 21 CTX-M (8 CTX-M-15, 7 CTX-M-1, 1 CTX-M-3, and 5 CTX-M-14), 2 TEM-type enzymes and 2 SHV-12. For Klebsiella species, 8/10 K. pneumoniae produced CTX-M-15 whereas the 4 K. oxytoca produced SHV-12. Finally, 10/11 E. cloacae were CTX-M-15 producers.
Integrating all molecular results has allowed showing that in the same cluster, analysis of the genes encoding ESBL resistance is required to differentiate strains there between. For example, on E. coli, cluster 14 contains 1 strain with resistance encoded by CTX-M-27, 5 strains with resistance encoded by CTX-M-15, and 1 strain with resistance encoded by CTX-M-14). Moreover, we observed strains of the same species with similar resistance profiles that did not derive from the same cluster (Additional file 1).
Cross-transmission
Eleven ESBL-E acquisitions out of 19 (19 patients) corresponded to an ESBL-E that was found neither on hygiene sampling nor on clinical sampling performed during the study period, which makes patient-to-patient transmission very unlikely.
One patient acquired an E. coli strain, which belonged to a cluster, found on admission sampling in another patient admitted to our ICU 10 days after. Whereas they belonged to the same cluster, these 2 strains exhibited both a different pattern of antibiotic susceptibility to aminoglycosides and a different ESBL type.
An E. cloacae acquisition was observed in one patient, 13 days before the admission of another patient to the same unit from whom admission screening samples indicated the same strain (Cluster 7), making patient-to-patient cross-transmission very unlikely.
Four ESBL-Es were acquired during the study period (similar cluster and ESBL analysis) in the same unit (one E. coli (Cluster 14), 2 E. cloacae (Cluster 6), one K. pneumoniae (Cluster 1). However, there was no overlap in the ICU stays of these patients, suggesting that patient-to-patient transmission was very unlikely.
E. coli (E18) was found in follow-up sampling in one patient (acquisition). This cluster (14) was found in samples from another (E22) patient. However, molecular analysis individualized 2 different ESBL-types (CTX-M-27 and CTX-M-15).
Finally, according to molecular (rep PCR and ESBL PCR) and phenotypic typing and bearing in mind geographic and temporal compatibility, only one case of highly probable patient-to-patient transmission occurred during the study (K. pneumoniae, Cluster 1).
Discussion
In this study, we assessed the rates of referred ESBL-E at admission and ESBL-E acquisition during ICU stay. Our study complements the few ones that have investigated Enterobacteriaceae acquisition and cross-transmission using a multimodal approach [16, 17, 30].
In our study, ESBL-E carriage at admission and acquisition during ICU stay were rare, occurring in 8 % and 6 % of patients, respectively. Using phenotypic and molecular typing, and considering geographic and temporal compatibility, we observed only one possible case of patient-to-patient transmission among the 19 patients who acquired an ESBL-E during their ICU stay.
ESBL-E imported cases accounted for less than 10 % in the ICU. This rate is in keeping with data published during the last decade. In 2007, Buke et al. reported at admission ESBL-E carriage in less than 8 % of patients who stayed for more than a month in a French hospital [31]. In Spain, carriage of ESBL-E among hospitalized patients was 8.2 % in 2010 [32]. The acquisition rate was 6 % in the present study. Despite recently observed emergence of community-acquired ESBL-E, in-hospital ESBL-E acquisition remained stable over years. This could suggest that either hygiene measures are effective or that the role attributed to patient-to-patient transmission of ESBL-E in the acquisition pattern is weak [3]. In the present study, using a multimodal microbiological approach, the observed acquisition rate was very low. Indeed, among 309 patients screened on admission to the ICU, cross-transmission was considered likely in only one patient. This result is certainly due to many issues. First, the present study was conducted during a period of very low endemicity. In consequence, we cannot exclude the possibility that different results may be observed during a period of high endemicity or during an outbreak. In addition, isolation measures applied in a preventive manner could have modified the cross-transmission rate [33].
A recently published study using a similar methodology reported possible patient-to-patient transmission for only 3 patients in 69 cases of ESBL-E acquisition [34]. In an older French study, cross-transmission rate was higher with over 85 % of strains acquired, but this study was only based on antibiotic susceptibility [35].
We compared both phenotypic analysis, based on antibiotic susceptibility, and molecular analysis. We confirmed that the capacity of antibiotic susceptibility analysis to discriminate similar strains is weak. Moreover, we confirm that strain with similar antibiotic susceptibility patterns and which belong to the same cluster (relying on rep-PCR) can differ only by their ESBL production. This is the case for 2 pairs of E. coli isolates (18 and 22) and (12 and 13) that carried (CTX-M 27 and CTX-M 15) and (SHV 12 and CTX-M 14), respectively. These observations are consistent with the fact that ESBL genetic support is mainly extrachromosomal (plasmid-related). As reported by several other authors, we observed this phenomenon only for E. coli strains [16, 17]. Our data highlight the importance-combined analysis of phenotypic profile and molecular data to discriminate ESBL-E strains.
The present study, however, has several limitations. First, despite its worldwide use, the rectal swab is of limited sensitivity in detecting ESBL-E carriage. This issue is well documented for vancomycin-resistant enterococci (VRE), for which probability of detection is inversely proportional to the number of colony-forming units in rectal swabs [36]. The risk of false-negative results with rectal swabs is probably lower for ESBL-E due to the virulence and larger inocula observed with these bacteria [23]. However, Harris et al. reported a non-detection rate of 69 % for E. coli and Klebsiella spp. [37]. In the present study, when rectal swabs were considered by laboratory technicians as containing insufficient amount of stool, they were discarded and rectal sampling was immediately re performed.
Antimicrobial exposure could have contributed to change duration of carriage and limited the detection during the study. Among patients with at least one positive rectal swab, 48 % had successively several negative rectal swabs. For 66 % of them, active antibiotics on isolated ESBL-E were administered when negative screening samples were observed. Duration of colonization by the ESBL-E strain is difficult to assess. One study showed that the median duration of carriage of ESBL-producing Klebsiella pneumoniae was 160 days after hospital discharge [38]. This study showed a significant variability between patients. Haverkate et al. showed that the median duration of carriage of highly resistant ESBL Enterobacteriaceae was 1.4 months in patients colonised with a MDRO during a previous stay in the ICU [39]. Currently, there is no formal data to recommend routine screening to assess the persistence of carriage.
Second, potential increased risk of acquisition, transmission and carriage which antimicrobial may contribute exposure was not looked for.
Third, sampling at admission was analysable in only 70 % of patients. The pertinence of the carriage rate we observed would have been probably greater with a higher percentage of patients sampled on admission. However, we included 86 % of all patients in the cross-transmission analysis, a percentage similar to that in most studies designed to characterize multiresistant strains cross-transmission.
Median length of stay was short (4 days), which is usual length of stay of patients admitted to our unit. However, we observed that patients who acquired ESBL-E had a significantly longer ICU stay (12 days). A similar study in patients who spend longer in the ICU may, therefore, indicate a different acquisition rate. Furthermore, criteria as those used to define cross-transmission can be appropriate for an ICU with short median lengths of stay but are not necessarily applicable in other ICU settings.
Finally, it should be highlighted that patient-to-patient transmission was low in our study. This can be related to several issues. Protective isolation measures were applied as soon as ESBL-E detection and immediately at admission in patients at high risk of carrying ESBL-E. In several cases of acquisition, patient-to-patient transmission initially deeming likely (because the strains were identical) was finally ruled out because there was no overlap of patient ICU stays. This type of acquisition could account for a possible acquisition from inanimate surfaces since ESBL-E can survive in such conditions for a long time [21].
Conclusions
In this observational study conducted in patients with a relatively short length of stay in the ICU, low rates of both ESBL-E carriage at admission and ESBL-E acquisition during ICU stay were observed. Using a multimodal microbiological approach, we identified only one possible case of patient-to-patient transmission of ESBL-E among the 19 acquisitions observed during ICU stay. These data highlight the importance of jointly analyzing phenotypic and molecular profiles, to discriminate strains of ESBL-E before assuming that they are identical.
Abbreviations
- CA-SFM:
-
comite de l’antibiogramme – societe française de microbiologie
- DNA:
-
deoxyribonucleic acid
- ESBL-E:
-
extended-spectrum beta-lactamase − producing Enterobacteriaceae
- ESBL:
-
extended-spectrum beta-lactamase
- ICU:
-
intensive care unit
- IGSII:
-
index gravity score
- IRB:
-
institutional review board
- GRE:
-
glycopeptide-resistant Enterococcus
- MALDI-TOF:
-
matrix-assisted laser desorption/ionisation, time-of-flight mass spectrometry
- MRSA:
-
methicillin resistant Staphylococcus aureus
- PCR:
-
polymerase chain reaction
- Rep-PCR:
-
repetitive-element PCR
- UPGMA:
-
unweighted pair group method with arithmetic mean
References
Tansarli GS, Karageorgopoulos DE, Kapaskelis A, Falagas ME. Impact of antimicrobial multidrug resistance on inpatient care cost: an evaluation of the evidence. Expert Review of Anti-Infective Therapy. 2013;11(3):321–31.
Shorr AF. Review of studies of the impact on Gram-negative bacterial resistance on outcomes in the intensive care unit. Crit Care Med. 2009;37(4):1463–9.
Conterno LO, Shymanski J, Ramotar K, Toye B, Zvonar R, Roth V. Impact and cost of infection control measures to reduce nosocomial transmission of extended-spectrum beta-lactamase-producing organisms in a non-outbreak setting. J Hosp Infect. 2007;65(4):354–60.
Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14(4):933–51.
Brun-Buisson C, Legrand P, Philippon A, Montravers F, Ansquer M, Duval J. Transferable enzymatic resistance to third-generation cephalosporins during nosocomial outbreak of multiresistant Klebsiella pneumoniae. Lancet. 1987;2(8554):302–6.
Coque TM, Baquero F, Canton R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill 2008 13(47):1-11.
Woerther PL, Angebault C, Lescat M, Ruppe E, Skurnik D, Mniai AE, Clermont O, Jacquier H, Costa AD, Renard M et al. Emergence and dissemination of extended-spectrum beta-lactamase-producing Escherichia coli in the community: lessons from the study of a remote and controlled population. J Infect Dis. 2010;202(4):515–23.
Herindrainy P, Randrianirina F, Ratovoson R, Ratsima Hariniana E, Buisson Y, Genel N, Decre D, Arlet G, Talarmin A, Richard V. Rectal carriage of extended-spectrum beta-lactamase-producing gram-negative bacilli in community settings in Madagascar. PLoS One 2011 6(7): e1122738.
Valverde A, Coque TM, Sanchez-Moreno MP, Rollan A, Baquero F, Canton R. Dramatic increase in prevalence of fecal carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae during nonoutbreak situations in Spain. J Clin Microbiol. 2004;42(10):4769–75.
Leflon-Guibout V, Blanco J, Amaqdouf K, Mora A, Guize L, Nicolas-Chanoine MH. Absence of CTX-M enzymes but high prevalence of clones, including clone ST131, among fecal Escherichia coli isolates from healthy subjects living in the area of Paris, France. J Clin Microbiol. 2008;46(12):3900–5.
Ruppe E, Pitsch A, Tubach F, de Lastours V, Chau F, Pasquet B, et al. Clinical predictive values of extended-spectrum beta-lactamase carriage in patients admitted to medical wards. Eur J Clin Microbiol Infect Dis. 2012;31(3):319-25.
Derde LP, Cooper BS, Goossens H, Malhotra-Kumar S, Willems RJ, Gniadkowski M, Hryniewicz W, Empel J, Dautzenberg MJ, Annane D et al. Interventions to reduce colonisation and transmission of antimicrobial-resistant bacteria in intensive care units: an interrupted time series study and cluster randomised trial. Lancet Infect Dis. 2014;14(1):31–9.
Jarlier V, Trystram D, Brun-Buisson C, Fournier S, Carbonne A, Marty L, Andremont A, Arlet G, Buu-Hoi A, Carlet J et al. Curbing methicillin-resistant Staphylococcus aureus in 38 French hospitals through a 15-year institutional control program. Arch Intern Med. 2010;170(6):552–9.
Cooper BS, Stone SP, Kibbler CC, Cookson BD, Roberts JA, Medley GF, Duckworth G, Lai R, Ebrahim S. Isolation measures in the hospital management of methicillin resistant Staphylococcus aureus (MRSA): systematic review of the literature. BMJ. 2004;329(7465):533.
Lucet JC, Paoletti X, Lolom I, Paugam-Burtz C, Trouillet JL, Timsit JF, Deblangy C, Andremont A, Regnier B. Successful long-term program for controlling methicillin-resistant Staphylococcus aureus in intensive care units. Intensive Care Med. 2005;31(8):1051–7.
Harris AD, Kotetishvili M, Shurland S, Johnson JA, Morris JG, Nemoy LL, Johnson JK. How important is patient-to-patient transmission in extended-spectrum beta-lactamase Escherichia coli acquisition. Am J Infect Control. 2007;35(2):97–101.
Harris AD, Perencevich EN, Johnson JK, Paterson DL, Morris JG, Strauss SM, Johnson JA. Patient-to-patient transmission is important in extended-spectrum beta-lactamase-producing Klebsiella pneumoniae acquisition. Clin Infect Dis. 2007;45(10):1347–50.
Lee SO, Kim YS, Kim BN, Kim MN, Woo JH, Ryu J. Impact of previous use of antibiotics on development of resistance to extended-spectrum cephalosporins in patients with enterobacter bacteremia. Eur J Clin Microbiol Infect Dis. 2002;21(8):577–81.
Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawkey P, Heck M, Savelkoul P, Vandenbroucke-Grauls C, van der Zwaluw K et al. Extended-spectrum beta-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerging Infectious Diseases. 2011;17(7):1216–22.
Huang SS, Datta R, Platt R. Risk of acquiring antibiotic-resistant bacteria from prior room occupants. Arch Intern Med. 2006;166(18):1945–51.
Kramer A, Schwebke I, Kampf G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC infectious diseases. 2006;6:130.
Datta R, Platt R, Yokoe DS, Huang SS. Environmental cleaning intervention and risk of acquiring multidrug-resistant organisms from prior room occupants. Arch Intern Med. 2011;171(6):491–4.
Harris AD, McGregor JC, Furuno JP. What infection control interventions should be undertaken to control multidrug-resistant gram-negative bacteria? Clin Infect Dis. 2006;43 Suppl 2:S57–61.
Abad C, Fearday A, Safdar N. Adverse effects of isolation in hospitalised patients: a systematic review. J Hosp Infect. 2010;76(2):97–102.
Jarlier V, Nicolas MH, Fournier G, Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10(4):867–78.
Healy M, Huong J, Bittner T, Lising M, Frye S, Raza S, Schrock R, Manry J, Renwick A, Nieto R et al. Microbial DNA typing by automated repetitive-sequence-based PCR. J Clin Microbiol. 2005;43(1):199–207.
Pitout JD, Campbell L, Church DL, Wang PW, Guttman DS, Gregson DB. Using a commercial DiversiLab semiautomated repetitive sequence-based PCR typing technique for identification of Escherichia coli clone ST131 producing CTX-M-15. J Clin Microbiol. 2009;47(4):1212–5.
Nemoy LL, Kotetishvili M, Tigno J, Keefer-Norris A, Harris AD, Perencevich EN, Johnson JA, Torpey D, Sulakvelidze A, Morris JG, Jr. et al. Multilocus sequence typing versus pulsed-field gel electrophoresis for characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolates. J Clin Microbiol. 2005;43(4):1776–81.
Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. The Journal of Antimicrobial Chemotherapy. 2010;65(3):490–5.
Decre D, Gachot B, Lucet JC, Arlet G, Bergogne-Berezin E, Regnier B. Clinical and bacteriologic epidemiology of extended-spectrum beta-lactamase-producing strains of Klebsiella pneumoniae in a medical intensive care unit. Clin Infect Dis. 1998;27(4):834–44.
Buke C, Armand-Lefevre L, Lolom I, Guerinot W, Deblangy C, Ruimy R, Andremont A, Lucet JC. Epidemiology of multidrug-resistant bacteria in patients with long hospital stays. Infect Control Hosp Epidemiol. 2007;28(11):1255–60.
Paniagua R, Valverde A, Coque TM, Baquero F, Canton R. Assessment of prevalence and changing epidemiology of extended-spectrum beta-lactamase-producing Enterobacteriaceae fecal carriers using a chromogenic medium. Diagn Microbiol Infect Dis. 2010;67(4):376–9.
Laurent C, Rodriguez-Villalobos H, Rost F, Strale H, Vincent JL, Deplano A, Struelens MJ, Byl B. Intensive care unit outbreak of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae controlled by cohorting patients and reinforcing infection control measures. Infect Control Hosp Epidemiol. 2008;29(6):517–24.
Gardam MA, Burrows LL, Kus JV, Brunton J, Low DE, Conly JM, Humar A. Is surveillance for multidrug-resistant enterobacteriaceae an effective infection control strategy in the absence of an outbreak? J Infect Dis. 2002;186(12):1754–60.
Lucet JC, Regnier B. [Enterobacteria producing extended spectrum beta-lactamases]. Pathol Biol (Paris). 1998;46(4):235–43.
D’Agata EM, Gautam S, Green WK, Tang YW. High rate of false-negative results of the rectal swab culture method in detection of gastrointestinal colonization with vancomycin-resistant enterococci. Clin Infect Dis. 2002;34(2):167–72.
Harris AD, Nemoy L, Johnson JA, Martin-Carnahan A, Smith DL, Standiford H, Perencevich EN. Co-carriage rates of vancomycin-resistant Enterococcus and extended-spectrum beta-lactamase-producing bacteria among a cohort of intensive care unit patients: implications for an active surveillance program. Infect Control Hosp Epidemiol. 2004;25(2):105–8.
Bird J, Browning R, Hobson RP, MacKenzie FM, Brand J, Gould IM. Multiply-resistant Klebsiella pneumoniae: failure of spread in community-based elderly care facilities. J Hosp Infect. 1998;40(3):243–7.
Haverkate MR, Derde LP, Brun-Buisson C, Bonten MJ, Bootsma MC. Duration of colonization with antimicrobial-resistant bacteria after ICU discharge. Intensive Care Med. 2014;40(4):564–71.
Acknowledgements
There was no funding source.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MA collected patients’ data and wrote the manuscript. DM, NB, CP, HAO, JLB collected patients’ data. AL and FB performed bacteriological analysis. DD performed bacteriological analysis and wrote the manuscript. GO and BG coordinated the care of patients, provided intellectual position in the in design of the study and the drafting of the manuscript and made relevant corrections to the final version of the manuscript. EM collected patients’ data and wrote the manuscript. All authors read and approved the final manuscript.
Additional file
Additional file 1:
Characterization of ESBL-E. (DOCX 92 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Alves, M., Lemire, A., Decré, D. et al. Extended-spectrum beta-lactamase − producing enterobacteriaceae in the intensive care unit: acquisition does not mean cross-transmission. BMC Infect Dis 16, 147 (2016). https://doi.org/10.1186/s12879-016-1489-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-016-1489-z