Abstract
Purpose
Readmission of patients colonized with antimicrobial-resistant bacteria (AMRB) is important in the nosocomial dynamics of AMRB. We assessed the duration of colonization after discharge from the intensive care unit (ICU) with highly resistant Enterobacteriaceae (HRE), methicillin-resistant Staphylococcus aureus (MRSA), and vancomycin-resistant enterococci (VRE).
Methods
Data were obtained from a cluster-randomized trial in 13 ICUs in 8 European countries (MOSAR-ICU trial, 2008–2011). All patients were screened on admission and twice weekly for AMRB. All patients colonized with HRE, MRSA, or VRE and readmitted to the same ICU during the study period were included in the current analysis. Time between discharge and readmission was calculated, and the colonization status at readmission was assessed. Because of interval-censored data, a maximum likelihood analysis was used to calculate the survival function, taking censoring into account. A nonparametric two-sample test was used to test for differences in the survival curves.
Results
The MOSAR-ICU trial included 14,390 patients, and a total of 64,997 cultures were taken from 8,974 patients admitted for at least 3 days. One hundred twenty-five unique patients had 141 episodes with AMRB colonization and at least 1 readmission. Thirty-two patients were colonized with two or more AMRBs. Median times until clearance were 4.8 months for all AMRB together, 1.4 months for HRE, <1 month for MRSA, and 1.5 months for VRE. There were no significant differences between the survival curves.
Conclusion
Fifty percent of the patients had lost colonization when readmitted 2 or more months after previous ICU discharge.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The increasing prevalence of antimicrobial-resistant bacteria (AMRB) in health care facilities places a heavy load on infection control policies. Readmission of patients colonized with AMRB is an important factor in the nosocomial dynamics of AMRB. It creates a ‘feedback loop’ where pathogens are reintroduced into the ward and can colonize or infect new patients [1]. Yet, AMRB carriage disappears after some time in most patients after discharge. In many hospitals AMRB-positive patients are ‘flagged’ in patient systems in order to allow quick identification at readmission [2–4]. Naturally, the feasibility of this infection control measure would be enhanced if patients could be safely ‘deflagged.’ However, little is known about duration of colonization with AMRB after hospital discharge.
Several studies have assessed the duration of carriage of methicillin-resistant Staphylococcus aureus (MRSA) in varying settings [2, 4–8], yielding large differences in colonization duration. Results vary from 50 % clearance in about 1 month [2] to a half-life of MRSA persistence of 40 months [4].
For highly resistant Enterobacteriaceae (HRE) and vancomycin-resistant enterococci (VRE) estimates of colonization duration are even scarcer, but also non-consistent [9–18]. In Thailand [10], a median duration of outpatient colonization with ESBL-producing bacteria of 98 days was observed, while a mean duration of carriage of 9 months was reported among adopted children from Mali [13]. For VRE, the reported median colonization time was 7 weeks [15], and in another study VRE was still recovered from 60 % of carriers of a particular outbreak strain, but only from 20 % of carriers of non-epidemic strains after 6 months [14].
Knowledge about the time until clearance of AMRB is of great importance for understanding nosocomial dynamics and for predicting effects of interventions. Colonization is a better indicator of bacterial dynamics than infection, since colonization only leads to infection in a small group but contributes significantly to the epidemiology of these bacteria [19]. Therefore, we assessed the duration of colonization after discharge from the intensive care unit (ICU) with HRE, MRSA, and VRE.
This work was presented in part at the 22nd ECCMID 2012, London, UK, 2 April 2012, abstract O126 [20].
Methods
Study design and study population
Data were obtained from a cluster-randomized trial in 13 ICUs in 8 European countries (MOSAR-ICU trial): France (three ICUs), Greece (two ICUs), Italy, Latvia, Luxembourg, Portugal (two ICUs), Slovenia (two ICUs), and Spain. Data were prospectively collected from May 2008 until April 2011. The trial consisted of a 6-month baseline period (phase 1), followed by implementation of a hand hygiene improvement program and unit-wide chlorhexidine body washing in months 7–26 (phase 2), and surveillance screening for AMRB carriage at ICU admission (followed by contact precautions for AMRB carriers) with ICUs randomized to PCR-based or chromogenic agar-based screening methods in months 13–26 (phase 3). During the whole study period, all patients expected to stay for more than 2 days were screened on admission and twice weekly for HRE, MRSA, and VRE. More details can be found in the paper of Derde et al. [21].
Written approval of the study protocol was obtained from each institution’s review board or national ethics committee. A waiver for informed consent was granted for all participants since the study was considered to involve no more than minimal risk of harm to patients (ClinicalTrials.gov Identifier: NCT00976638).
All patients colonized with HRE, MRSA, or VRE in at least one of the two last cultures during the first admission and at least one readmission to the same ICU were included in our study. HRE included Enterobacteriaceae suspected to harbor extended-spectrum beta-lactamase (ESBL): Escherichia coli (E. coli), Klebsiella, Enterobacter, Serratia, and Citrobacter (KESC), and Proteus, Providencia, and Morganella (PPM) spp. The date of discharge was taken as the start of the ‘at risk’ period for clearance. This ‘at risk’ period for clearance was calculated as the time between discharge and readmission. At readmission, we analyzed the first two available cultures for every patient and labeled a patient as colonized if at least one of these cultures was positive or decolonized if both cultures were negative. We assumed admission was non-informative with regard to colonization status. We performed analyses for all bacteria together (looking at colonization as the event of interest, regardless of the type of bacteria the patients were colonized with) and for all different bacterial types separately. When a patient was colonized with more than one bacterial species in the same admission, these were counted as separate colonization episodes. If a patient was admitted multiple times while colonized, then each readmission was treated as a new episode.
Microbiological tests
Swabs were obtained from the anterior nares (for detection of MRSA), the perineal area (for detection of HRE and VRE), and wounds (if present, for detection of HRE, MRSA, and VRE). Samples were frozen at −70/−80 °C for a maximum of 2 months before processing in phase 1 and 2, and samples were analyzed instantly in phase 3. The media used for detection of AMRB were Brilliance ESBL 2 for detection of HRE, BBL CHROMagar MRSA II for detection of MRSA, and BBL Enterococcosel Agar with 8 μg/ml vancomycin for detection of VRE.
Statistical analyses
The time between discharge and readmission varied between patients. Naturally, it was impossible to determine exact times until clearance for patients non-colonized upon readmission, resulting in interval-censored data [22]. In Mathematica 7.0 (Wolfram Research, Inc., Mathematica, Version 7.0, Champaign, IL), we used a maximum likelihood analysis to calculate the survival function, taking censoring into account. A more detailed description of the method can be found in the paper of Goggins and Finkelstein [22]. If a patient lost colonization, then the day of readmission would not be taken as the day of clearance (the last day of the interval, as would happen using the more conventional Kaplan-Meier method), but rather a day in the interval, calculated to be most likely the day of clearance. Furthermore, with the maximum likelihood analysis, sensitivity of the test could be assessed. Graphs were constructed to visualize the survival functions for the different bacteria. A nonparametric two-sample test was used to test for significant differences in the survival curves. This method was developed by Andersen and Rønn [23], and a more detailed description can be found in their paper.
In sensitivity analyses, we determined the effects of detection bias by assuming that colonization remained until discharge, and thus included all cases with at least one positive culture any time during admission, and by analyzing the first readmission culture instead of the first two. Also, we analyzed the data excluding wound cultures since they were taken infrequently and were dependent on the presence of wounds. Furthermore, we investigated different study phases separately (baseline versus intervention period) to determine whether decolonization therapy (chlorhexidine body washings), improved hand hygiene, and surveillance screening with contact precautions of identified carriers influenced colonization duration. Finally, we checked whether there was a difference between patients who were readmitted from their home and those coming from health care facilities (including hospital wards and long-term care facilities).
Results
The MOSAR-ICU trial included 14,390 patients, and a total of 64,997 cultures were taken from 8,974 patients admitted for at least 3 days. There were 926 patients with at least one readmission who had—in all—2,111 admissions during the study period. One hundred twenty-five of these 926 patients were colonized with AMRB in at least one of the last two cultures during their first admission and had a subsequent readmission during the study period and were included in the analysis (Fig. 1). Details on patient characteristics can be found in Table 1.
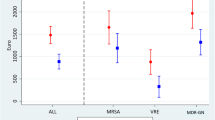
When analyzing all AMRB together, 141 episodes of colonization were recorded, including 32 episodes of colonization with two or more organisms, 101 episodes of HRE colonization (further divided into 34 for E. coli, 73 for the KESC, and 8 for the PPM group), 48 episodes of colonization with MRSA, and 19 episodes of colonization with VRE. The median times to clearance were around 1–2 months for all bacterial types (Table 2; Fig. 2). The nonparametric two-sample test revealed no significant differences between the survival curves of the different bacteria.
For AMRB, the median time to clearance was 4.8 months, which was longer than for individual species. This can be explained by the fact that, in this case, we looked at colonization as the event of interest, regardless of the type of bacteria with which the patients were colonized. When a patient was readmitted and colonized with another type of bacterium than in his/her previous admission, this was still counted as ‘colonized.’ In our study, in 88 out of 98 cases in which a patient was still colonized at readmission, this was with (at least) one of the bacteria with which he/she was colonized in the previous admission. Ten cases were colonized with another type of bacterium. The same applies to HRE: we regarded a patient, colonized with HRE at discharge, as ‘still colonized’ at readmission if he/she was colonized with any type of HRE. In 62 out of 65 cases in which a patient was still colonized with an HRE at readmission, this was with (at least) one of the bacteria with which he/she was colonized in the previous admission. For comparison, in the Supplementary information data are analyzed using the Kaplan-Meier method. Here, interval censoring is not taken into account. For some bacteria, the results are comparable (e.g., all AMRB together and all HRE together), but for example for MRSA and VRE the time to clearance is higher than the numbers derived using the maximum likelihood method.
Sensitivity analyses
Including all cases with at least one positive culture any time during admission instead of only looking at the last two samples of the first admission period did not significantly influence the results (data not shown). Restricting the analyses to the first culture of the readmission (instead of the first two cultures) also did not change results (data not shown). Furthermore, separate analyses of the different study phases or excluding wound cultures did not significantly change results (data not shown). Finally, there were no significant differences between patients being readmitted from home or from a health care facility.
Discussion
For all antimicrobial-resistant bacterial species, 50 % of the patients had lost colonization when readmitted 2 or more months after the previous ICU admission. Although this study was performed on a selection of hospital patients, i.e., patients admitted to ICUs, the results are of critical importance since these patients are especially prone to colonization and (subsequent) infection [19, 24]. From the figures, it seems that the percentage of patients still colonized is stable for a period of time (especially for all AMRB and HRE). One could hypothesize that there are two distinct populations: the majority, which clears colonization with AMRB quickly, and a minority (around 40 %) with longer persistence. More research is needed to ascertain this.
The analysis of all AMRB can be seen as a special case. Here, we did not assess the type(s) of bacterium a patient was colonized with, but only colonization status with any AMRB. Naturally, longer times to clearance were found, since the type of bacterium at readmission is not necessarily the same as the one at discharge from the previous admission. This analysis shows that there might be a risk group consisting of patients who are prone to become colonized with any AMRB. What these specific risk factors are was not addressed in this study, since it was only applicable to ten patients.
Our study revealed shorter clearance times than previously reported, especially for MRSA [4–8], which may have resulted from several methodological differences. For example, for the calculation of the colonization duration of MRSA, Scanvic et al. [5] only included patients readmitted more than 3 months after discharge, while we included all readmitted patients, regardless of time since discharge. Although Larsson et al. [6] found a median time to clearance of MRSA of 179 days (5.9 months), they also demonstrated that 43 % of the cases were colonized less than 2 months, which is more similar to our results. In one of the first studies on colonization duration of MRSA, Sanford et al. [4] estimated the half-life of MRSA persistence in readmitted carriers to be 41 months. However, plasmid analysis and information on phage types indicated that only a part of the cases of persistent MRSA carriage had continued carriage of the same strain and that the remainder represented acquisition of a new strain. Mattner et al. [7] indicate that they tend to overestimate the duration of colonization by regarding a patient as ‘still colonized’ if the readmission swabs were missed. This could partly explain the differences between their median time to clearance (549 days) and the time found in this study. However, our results are consistent with the study of Robicsek et al. [2]. They also found that 50 % of the people lost colonization quickly (within 1 month). However, the colonization rate decreased slowly thereafter.
Another reason for the differences with these studies is that four of them used the Kaplan-Meier method to estimate the median time to clearance [5–8]. As we pointed out in the methods section, the exact time of clearance is usually unknown, especially when readmission cultures are used or when sampling is infrequent. As this will result in interval-censored data, we used a maximum likelihood analysis to calculate the survival function, with censoring taken into account. This will yield more rapid clearance times than a Kaplan-Meier estimate. Also, we used a very specific patient population. Moreover, as our results are based on the results of growth of cultures on selective media and no genetic confirmation was used, patients colonized upon readmission with a new strain might be misclassified as still being colonized.
Our estimates on colonization duration with HRE were lower than findings in another study [18]. This can be explained by differences in study populations (ICU versus whole hospital), the fact that only patients readmitted after 3 months or more were included in the other study, and the use of the Kaplan-Meier method. Our estimates of duration of colonization are probably more accurate for the first months, but since the other study covered a period of 14 years, colonization and readmission after more than 1 or 2 years might have been captured better.
At readmission, we focussed on the first two cultures. We required one or both of the readmission cultures to be positive in order to label a patient (still) colonized. This strategy was chosen to prevent false-negative results of the first readmission culture, which may occur because of sampling errors depending on the swabbing technique used and the site swabbed [4, 25–28]. Perirectal swabs were used in this study for detection of HRE and VRE, but several publications suggest that the sensitivity of rectal and perirectal swabs is comparable [25, 27]. Different practices in ICUs of all countries included could induce different results. However, an extensive protocol was provided to all study sites to standardize all aspects of the main study [21]. Samples were not placed in enrichment broth before plating, which might have influenced the sensitivity of the test. Furthermore, the samples were frozen and stored during baseline (although for a maximum of 2 months), which might have influenced the results. Survival of gram-positive and -negative bacteria during freezing and storage seems quite good [29, 30], although the amount of bacteria recovered from frozen suspensions may be reduced [31]. This could also have an impact on the colonization times. If the sensitivity of the tests were indeed reduced, shorter colonization times would be found.
In the intervention period of the MOSAR trial, decolonization therapy (body washings with chlorhexidine) was implemented. No mupirocin was used. Body washing with chlorhexidine eradicates gram-positive bacteria from the skin [32–37]. In the MOSAR trial, patients were washed from the neck down. Since we mainly used swabs taken from the nose for MRSA, one could expect that chlorhexidine would not influence colonization at this site. However, chlorhexidine may remove colonization or temporal contamination with MRSA at other body sites. Hence, chlorhexidine may lower the colonization pressure of gram-positive bacteria and could be effective in prevention of transmission. Little is known about the influence of chlorhexidine on colonization with gram-negative bacteria [33]. However, eradication seems unlikely given that the gut, the main reservoir of gram-negative bacteria, is not targeted with this decolonization therapy. To prevent a potential influence of chlorhexidine washings on colonization duration, we only included patients with at least one positive culture in the last two cultures of their ‘first’ admission. In this way, patients in whom carriage was eradicated were excluded. Furthermore, when we analyzed the different study phases separately, no significant differences in colonization duration were found.
A limitation of this study is that the reasons for patient readmissions were not known. We had no information on whether these reasons were correlated with the colonization status of the patients. Therefore, we assumed admission was non-informative with regard to colonization status. We did have data on the ‘most specific reason for ICU admission’ and there was no difference in the prevalence of sepsis or in the type of sepsis (urinary tract or other origin) in patients admitted to the ICU for the first time in the study period or readmitted patients (data not shown). Furthermore, there were no structured follow-up cultures taken of discharged patients. Therefore, we do not know exactly when colonization disappeared.
The prevalence and incidence of AMRB are different per hospital and country, but this did not seem to influence the results. The maximal contribution of one hospital to all cases was 18.7 % for AMRB, 21.8 % for HRE, 26.2 % for E. coli, 25.9 % for KESC, 62.5 % for PPM (note: there were only eight cases), 21.0 % for MRSA, and 29.4 % for VRE.
To our knowledge, this is the first study to assess the duration of colonization of ICU patients with HRE. Further studies with more readmitted colonized patients could be of help to assess the time to clearance more precisely.
References
Cooper BS, Medley GF, Stone SP, Kibbler CC, Cookson BD, Roberts JA, Duckworth G, Lai R, Ebrahim S (2004) Methicillin-resistant Staphylococcus aureus in hospitals and the community: stealth dynamics and control catastrophes. Proc Natl Acad Sci USA 101:10223–10228
Robicsek A, Beaumont JL, Peterson LR (2009) Duration of colonization with methicillin-resistant Staphylococcus aureus. Clin Infect Dis 48:910–913
MacKinnon MM, Allen KD (2000) Long-term MRSA carriage in hospital patients. J Hosp Infect 46:216–221
Sanford MD, Widmer AF, Bale MJ, Jones RN, Wenzel RP (1994) Efficient detection and long-term persistence of the carriage of methicillin-resistant Staphylococcus aureus. Clin Infect Dis 19:1123–1128
Scanvic A, Denic L, Gaillon S, Giry P, Andremont A, Lucet JC (2001) Duration of colonization by methicillin-resistant Staphylococcus aureus after hospital discharge and risk factors for prolonged carriage. Clin Infect Dis 32:1393–1398
Larsson AK, Gustafsson E, Nilsson AC, Odenholt I, Ringberg H, Melander E (2011) Duration of methicillin-resistant Staphylococcus aureus colonization after diagnosis: a 4-year experience from southern Sweden. Scand J Infect Dis 43:456–462
Mattner F, Biertz F, Ziesing S, Gastmeier P, Chaberny IF (2010) Long-term persistence of MRSA in re-admitted patients. Infection 38:363–371
Marschall J, Muhlemann K (2006) Duration of methicillin-resistant Staphylococcus aureus carriage, according to risk factors for acquisition. Infect Control Hosp Epidemiol 27:1206–1212
Pacio GA, Visintainer P, Maguire G, Wormser GP, Raffalli J, Montecalvo MA (2003) Natural history of colonization with vancomycin-resistant enterococci, methicillin-resistant Staphylococcus aureus, and resistant gram-negative bacilli among long-term-care facility residents. Infect Control Hosp Epidemiol 24:246–250
Apisarnthanarak A, Bailey TC, Fraser VJ (2008) Duration of stool colonization in patients infected with extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Clin Infect Dis 46:1322–1323
Alsterlund R, Carlsson B, Gezelius L, Haeggman S, Olsson-Liljequist B (2009) Multiresistant CTX-M-15 ESBL-producing Escherichia coli in southern Sweden: description of an outbreak. Scand J Infect Dis 41:410–415
Alsterlund R, Axelsson C, Olsson-Liljequist B (2012) Long-term carriage of extended-spectrum beta-lactamase-producing Escherichia coli. Scand J Infect Dis 44:51–54
Tande D, Boisrame-Gastrin S, Munck MR, Hery-Arnaud G, Gouriou S, Jallot N, Nordmann P, Naas T (2010) Intrafamilial transmission of extended-spectrum-beta-lactamase-producing Escherichia coli and Salmonella enterica Babelsberg among the families of internationally adopted children. J Antimicrob Chemother 65:859–865
Mascini EM, Jalink KP, Kamp-Hopmans TE, Blok HE, Verhoef J, Bonten MJ, Troelstra A (2003) Acquisition and duration of vancomycin-resistant enterococcal carriage in relation to strain type. J Clin Microbiol 41:5377–5383
Montecalvo MA, de Lencastre H, Carraher M, Gedris C, Chung M, VanHorn K, Wormser GP (1995) Natural history of colonization with vancomycin-resistant Enterococcus faecium. Infect Control Hosp Epidemiol 16:680–685
Byers KE, Anglim AM, Anneski CJ, Farr BM (2002) Duration of colonization with vancomycin-resistant Enterococcus. Infect Control Hosp Epidemiol 23:207–211
O’Fallon E, Gautam S, D’Agata EM (2009) Colonization with multidrug-resistant gram-negative bacteria: prolonged duration and frequent colonization. Clin Infect Dis 48:1375–1381
Birgand G, Armand-Lefevre L, Lolom I, Ruppe E, Andremont A, Lucet JC (2012) Duration of colonization by extended-spectrum beta-lactamase-producing Enterobacteriaceae after hospital discharge. Am J Infect Control. doi:10.1016/j.ajic.2012.05.015
Bonten MJ, Weinstein RA (1996) The role of colonization in the pathogenesis of nosocomial infections. Infect Control Hosp Epidemiol 17:193–200
Haverkate MR, Derde LPG, Brun-Buisson C, Bonten MJM, Bootsma MCJ, On behalf of the MOSAR research consortium (2012) Duration of colonisation with antimicrobial-resistant bacteria after ICU discharge. 22nd ECCMID, London, Abstract O126
Derde LP, Cooper BS, Goossens H, Malhotra-Kumar S, Willems RJ et al (2013) Interventions to reduce colonisation and transmission of antimicrobial-resistant bacteria in intensive care units: an interrupted time series study and cluster randomised trial. Lancet Infect Dis. doi:10.1016/S1473-3099(13)70295-0
Goggins WB, Finkelstein DM (2000) A proportional hazards model for multivariate interval-censored failure time data. Biometrics 56:940–943
Andersen PK, Ronn BB (1995) A nonparametric test for comparing two samples where all observations are either left- or right-censored. Biometrics 51:323–329
Brusselaers N, Vogelaers D, Blot S (2011) The rising problem of antimicrobial resistance in the intensive care unit. Ann Intensive Care 1:47
Weinstein JW, Tallapragada S, Farrel P, Dembry LM (1996) Comparison of rectal and perirectal swabs for detection of colonization with vancomycin-resistant enterococci. J Clin Microbiol 34:210–212
Sewell DL, Potter SA, Jacobson CM, Strausbaugh LJ, Ward TT (1993) Sensitivity of surveillance cultures for the detection of methicillin-resistant Staphylococcus aureus in a nursing-home-care unit. Diagn Microbiol Infect Dis 17:53–56
Lautenbach E, Harris AD, Perencevich EN, Nachamkin I, Tolomeo P, Metlay JP (2005) Test characteristics of perirectal and rectal swab compared to stool sample for detection of fluoroquinolone-resistant Escherichia coli in the gastrointestinal tract. Antimicrob Agents Chemother 49:798–800
D’Agata EM, Gautam S, Green WK, Tang YW (2002) High rate of false-negative results of the rectal swab culture method in detection of gastrointestinal colonization with vancomycin-resistant enterococci. Clin Infect Dis 34:167–172
Green HP, Johnson JA, Furuno JP, Strauss JM, Perencevich EN, Lautenbach E, Lee D, Harris AD (2007) Impact of freezing on the future utility of archived surveillance culture specimens. Infect Control Hosp Epidemiol 28:886–888
Veguilla W, Peak KK, Luna VA, Roberts JC, Davis CR, Cannons AC, Amuso P, Cattani J (2008) Two-year study evaluating the potential loss of methicillin resistance in a methicillin-resistant Staphylococcus aureus culture collection. J Clin Microbiol 46:3494–3497
Bonten MJ, Nathan C, Weinstein RA (1997) Recovery of nosocomial fecal flora from frozen stool specimens and rectal swabs: comparison of preservatives for epidemiological studies. Diagn Microbiol Infect Dis 27:103–106
Climo MW, Yokoe DS, Warren DK, Perl TM, Bolon M, Herwaldt LA, Weinstein RA, Sepkowitz KA, Jernigan JA, Sanogo K, Wong ES (2013) Effect of daily chlorhexidine bathing on hospital-acquired infection. N Engl J Med 368:533–542
Derde LP, Dautzenberg MJ, Bonten MJ (2012) Chlorhexidine body washing to control antimicrobial-resistant bacteria in intensive care units: a systematic review. Intensive Care Med 38:931–939
Batra R, Cooper BS, Whiteley C, Patel AK, Wyncoll D, Edgeworth JD (2010) Efficacy and limitation of a chlorhexidine-based decolonization strategy in preventing transmission of methicillin-resistant Staphylococcus aureus in an intensive care unit. Clin Infect Dis 50:210–217
Climo MW, Sepkowitz KA, Zuccotti G, Fraser VJ, Warren DK, Perl TM, Speck K, Jernigan JA, Robles JR, Wong ES (2009) The effect of daily bathing with chlorhexidine on the acquisition of methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus, and health care-associated bloodstream infections: results of a quasi-experimental multicenter trial. Crit Care Med 37:1858–1865
Bleasdale SC, Trick WE, Gonzalez IM, Lyles RD, Hayden MK, Weinstein RA (2007) Effectiveness of chlorhexidine bathing to reduce catheter-associated bloodstream infections in medical intensive care unit patients. Arch Intern Med 167:2073–2079
Milstone AM, Passaretti CL, Perl TM (2008) Chlorhexidine: expanding the armamentarium for infection control and prevention. Clin Infect Dis 46:274–281
Acknowledgments
This study was supported by the European Commission under the Life Science Health Priority of the 6th Framework Program (MOSAR network contract LSHP-CT-2007-037941). M.J.M.B. and M.R.H. were supported by the Netherlands Organization of Scientific Research (VICI NWO Grant 918.76.611 and Priority Medicines Antimicrobial Resistance Grant 205100013). M.C.J.B., M.J.M.B, and C.B-B. were supported by funding from the European Community [R-GNOSIS Integrated project (FP7/2007-2013) under grant agreement no. 282512].
Conflicts of interest
None declared.
Author information
Authors and Affiliations
Corresponding author
Additional information
On behalf of the MOSAR Work Packages 3 and 7.
Take-home message: To gain insight into the role of readmitted patients in the nosocomial dynamics of antimicrobial-resistant bacteria, we used colonization and readmission data from the MOSAR intensive care study, a study on intensive care units in eight European countries, to estimate the duration of colonization with highly resistant Enterobacteriaceae, methicillin resistant Staphylococcus aureus (MRSA), and vancomycin-resistant enterococci (VRE). We found that 50 % of the patients had lost colonization when readmitted 2 or more months after previous ICU discharge.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Haverkate, M.R., Derde, L.P.G., Brun-Buisson, C. et al. Duration of colonization with antimicrobial-resistant bacteria after ICU discharge. Intensive Care Med 40, 564–571 (2014). https://doi.org/10.1007/s00134-014-3225-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00134-014-3225-8