Abstract
Background
Hyperuricemia is a major risk for non-alcoholic fatty liver disease. However, the mechanisms for this phenomenon are not fully understood. This study aimed to investigate whether microRNAs mediated the pathogenic effects of uric acid on non-alcoholic fatty liver disease.
Methods
Microarray was used to determine the hepatic miRNA expression profiles of male C57BL/6 mice fed on standard chow diet, high fat diet (HFD), and HFD combined with uric acid-lowering therapy by allopurinol. We validated the expression of the most significant differentially expressed microRNAs and explored its role and downstream target in uric acid-induced hepatocytes lipid accumulation.
Results
Microarray analysis and subsequent validation showed that miR-149-5p was significantly up-regulated in the livers of HFD-fed mice, while the expression was down-regulated by allopurinol therapy. MiR-149-5p expression was also significantly up-regulated in uric acid-stimulated hepatocytes. Over-expression of miR-149-5p significantly aggregated uric acid-induced triglyceride accumulation in hepatocytes, while inhibiting miR-149-5p ameliorated the triglyceride accumulation. Luciferase report assay confirmed that FGF21 is a target gene of miR-149-5p. Silencing FGF21 abolished the ameliorative effects of miR-149-5p inhibitor on uric acid-induced hepatocytes lipid accumulation, while overexpression of FGF21 prevented the lipid accumulation induced by miR-149-5p mimics.
Conclusions
Uric acid significantly up-regulated the expression of miR-149-5p in hepatocytes and induced hepatocytes lipid accumulation via regulation of miR-149-5p/FGF21 axis.
Similar content being viewed by others
Background
Nonalcoholic fatty liver disease (NAFLD) is a group of liver disease characterized by excessive hepatic lipid accumulation without excess alcohol intake [1]. It ranges from simple steatosis to steatohepatitis, fibrosis, cirrhosis, and eventually hepatic carcinoma [2]. NAFLD is the most frequent chronic liver disease worldwide, the prevalence of NAFLD in Asia is increasing, and its prevalence in China has rapidly climbed to 29.2% in 2019 [3, 4]. NAFLD is strongly associated with obesity, type 2 diabetes mellitus and cardiovascular diseases, all of which leads to serious public health issues worldwide [5,6,7]. Despite intensive investigations over past decades, the precise pathogenesis of NAFLD remains poorly understood.
Uric acid is the final enzymatic product of purine metabolism. We previously identified that high serum uric acid level is a major risk factor of NAFLD [8, 9], and uric acid induced hepatic lipid accumulation by activating NLRP3 inflammasome [10]. Uric acid may also induce hepatic lipid accumulation by inducing endoplasmic reticulum stress and mitochondrial oxidative stress [11, 12]. Although increasing researches have emerged to explore the mechanism by which uric acid induced hepatic lipid accumulation, its underlying molecular mechanisms remains not fully clarified. A better understanding of the mechanisms may help for developing novel therapeutic strategy for NAFLD.
MicroRNAs (miRNAs) are members of small non-coding RNAs, which are consist of approximately 18–24 nucleotides. MiRNAs are major in negatively regulate gene expression at the post-transcriptional level by binding to target mRNA followed by silencing or promoting of the mRNA transcription [13]. There are more than 2600 miRNAs have been reported in miRbase and each miRNA can regulate hundreds of gene transcripts [14]. A growing number of studies implicated that miRNAs are participated in the pathogenesis of NAFLD [15]. For instance, miR-122, which is the most abundant miRNA in human liver, was decreased in liver of high fat diet (HFD) fed mice, and miR-122 could prevent hepatic steatosis by decreasing mRNA expression levels of lipogenic genes [16]. Our previous miRNA microarray employed in livers of ob/ob mice and STZ-induced diabetic mice also indicated that many miRNA participate in the pathophysiological processes of NAFLD with hyperglycemia [17]. In addition, another previous study found that miR-34a was elevated in the livers of HFD-fed mice, and miR-34a induced hepatic lipid accumulation through regulating its target gene PPARα [18]. Interestingly, a previous study has reported that a high concentration of uric acid significantly down-regulated miR-92a, which contributed to angiogenesis [19]. However, whether the effects of uric acid on hepatocytes lipid accumulation is mediated by miRNAs remains unclear.
Allopurinol, the prototypical xanthine oxidase (XO) inhibitor, has been considered as the cornerstone of the clinical management of gout or hyperuricemia for several decades [20]. In the previous study, we also have found that lowering the serum uric acid levels with allopurinol significantly alleviated HFD-induced hepatic steatosis in mice [10]. In this study, we aimed to explore the hepatic miRNA expression profiles of HFD-fed mice treated with or without allopurinol-therapy. We found that miR-149-5p is involved in the effects of uric acid on hepatocytes lipid accumulation via regulation of its target gene FGF21.
Methods
Animals and treatments
Male C57BL/6 mice, 8–10 weeks of age, weighing 18–20 g, were purchased from the Medical Science Institution of Zhejiang Province (Hangzhou, China). Before experiment, mice were allowed to acclimate for 1–2 weeks access to food and water. The mice were randomly divided into 3 groups, including of 6 animals per group and fed with standard chow diet (SCD), HFD, or HFD combined with allopurinol (120 mg/L) in the drinking water (HFD + A) for 8 weeks. The HFD (D12492, Research Diets, New Brunswick, NJ) contains 60% of kcal from fat, 20% from carbohydrates and 20% from protein. Mice were maintained in a 12-h light/dark cycle at controlled room temperature and given free access to food and water.
After intraperitoneal injection of 5% chloral hydrate, mice were sacrificed by cervical dislocation. The liver of each mice was removed and weighted. After rinsing with cold PBS, liver contents were fixed in neutral buffered formalin. All animal experiments were approved by the Animal Care and Use Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University (Reference number: 2019–1096).
Histological analysis
Liver sections were fixed in neutral buffered formalin overnight and then embedded in paraffin blocks. Sections were cut and stained for Hematoxylin and Eosin (H & E). For determination of hepatic lipid accumulation, 8 μm frozen liver sections were sequentially stained with Oil Red O and hematoxylin (Jiancheng Biology, Nanjing, China). Cells grown in glass cover ships in 12-well plates were washed with PBS, and followed by staining with Oil Red O and hematoxylin. Sections were imaged at 200× magnification (Olympus, Tokyo, Japan).
Cell culture and uric acid treatment
The mouse hepatocytes cell line (AML-12) and human hepatoblastoma cell line (HepG2) were obtained from the Chinese Academy of Science (Shanghai, China). AML-12 cells were grown in DMEM/F12 medium supplemented with 10% FBS, and 1% penicillin/streptomycin, ITS Liquid Media Supplement, and Dexamethasone (40 μg/ml). HepG2 cells were grown in DMEM medium supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were incubated at 37 °C in 5% CO2. To established the uric acid-stimulated cell models, AML-12 and HepG2 cells were treated with different concentrations of uric acid (250, 500, and 750 μmol/L) for 48 h.
miRNA array
Total RNA was isolated and the RNA integrity was assessed using Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). The total RNA was transcribed to double strand cDNA, then synthesized into cRNA and labeled with Cyanine-3-CTP. The labeled cRNAs were firstly hybridized onto the microarray, which were scanned by the Agilent Scanner G2505C (Agilent Technologies). Array images were analyzed to get raw data by using feature extraction software (version 10.7.1.1, Agilent Technologies). Genespring (version 14.8, Agilent Technologies) were conducted for the basic analysis of the raw data. The raw data was normalized with the quantile algorithm. The threshold set for up- and down-regulated genes was a fold change ≥1.5.
Cell transfection
Mmu/hsa-miR-149-5p inhibitor (100 nM), mmu/hsa-miR-149-5p (50 nM), and their corresponding negative controls were purchased from RiboBio (Guangzhou, China). The siRNA oligonucleotides and over-expression plasmids of mouse or human FGF21, and their corresponding negative controls were also obtained from RiboBio. The miRNAs, siRNAs and plasmids were transfected into hepatocytes by using Lipofectamine3000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. After 24 h of transfection, the hepatocytes were exposed to uric acid for an additional 48 h.
RNA isolation and real-time PCR
Total RNA was prepared from cells or frozen livers using RNA plus (Takara, Dalian, China). The 2.5 μg total RNA was reversely transcribed with a One Step PrimeScript™ RT-PCR kit (Takara). Real-time PCR analysis was carried out using the SYBR Premix-Ex Tag Kit (Takara) on an ABI prism 7500 sequence Detection System (Applied Biosystems, Foster City, CA). The primer sequences are listed in Table S1.
Western blot analysis
Cells and liver tissues were homogenized by using RIPA buffer (Applygen Technologies Inc., Beijing, China) supplemented with protease and phosphatase inhibitor (Pierce Biotechnology, Rockford, IL). Equal amount of protein was subjected to 12% SDS-PAGE followed by transfer to PVDF membranes (Millipore, Inc., Darmstadt, Germany). Membranes were blocked with 5% non-fat dry milk in TBST, followed by incubation overnight with the following primary antibodies: anti-FGF21 (Abcam, ab17194, 1:1000), anti-β-actin (CST, 3700 s, 1:1000). Bolts were further incubated with HRP-conjugated secondary antibodies: goat anti-rabbit (Sigma, sc-2004) or goat anti-mouse (Sigma, sc-2005). Proteins were visualized with an ECL plus (Fudebio, Hangzhou, China).
Triglyceride analysis
The intrahepatic and intracellular triglyceride contents were detected by using a commercial kit (Applygen Technologies Inc., Beijing, China) according the manufacturer’s instructions. The triglyceride values were normalized by total protein contents.
Luciferase assay
To perform the luciferase reporter, plasmids containing either wild type or mutated 3’UTR sequence were generated (Hanyin, Shanghai, China). The plasmids were transfected into HEK293T cells using Lipofectamine 3000 (Invitrogen, Carlsbad, CA). The cells were also co-transfected with either the negative control or miR-149-5p mimic (50 nM). After 24 h of transfection, the cells were harvested according to the manufacturer’s instructions (Promega Dual Luciferase Assay Kit, Promega). Firefly luciferase values were normalized to those of Renilla luciferase.
Statistical analyses
Statistical analyses were performed by using SPSS 18.0 for windows (SPSS, Chicago, IL). Statistical comparisons were made using t-test or ANOVA where appropriate. All data were expressed as mean ± standard division (SD), with a statistically significant difference defined as P < 0.05.
Results
Uric acid induces lipid accumulation in hepatocytes
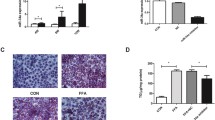
To investigate the regulatory role of uric acid on lipid accumulation in hepatocytes, we treated AML-12 cells with different concentrations of uric acid as previously reported [10]. We found that exposure to 250, 500, and 750 μmol/L uric acid for 48 h induced significant intracellular triglyceride accumulation in AML-12 cells (Fig. 1a). The increased intracellular lipid accumulation was confirmed by Oil Red O staining (Fig. 1b). We observed similar results in HepG2 cells treated with different concentrations of uric acid (Fig. S1). We also investigated the effect of uric acid on hepatic lipid accumulation in mice. Male C57BL/6 mice, 8 weeks of age, weighing 22–23 g, were randomly divided into three groups, consisting of 6 animals per group and fed with SCD, HFD, or HFD combined with allopurinol in drinking water (HFD + A), respectively. We found that the serum uric acid levels were significantly increased about 1.5-fold in HFD-fed mice, while the serum uric acid levels are reduced by about 90% in HFD + A mice than of HFD-mice (Fig. 1c). Consistent with the alterations of serum uric acid levels, we found that the intrahepatic triglyceride contents were reduced approximately by 40% in HFD + A fed mice than of HFD-fed mice (Fig. 1d). Liver sections with H & E and Oil Red O staining confirmed the reduction of hepatic lipid accumulation in HFD + A fed mice (Fig. 1e). These results suggested that uric acid directly induced hepatocytes lipid accumulation, and lowering serum uric acid by allopurinol significantly ameliorated HFD-induced hepatic steatosis in mice.
Uric acid induces hepatocytes lipid accumulation both in vitro and in vivo. a Intracellular triglyceride contents were significantly increased in AML-12 cells after stimulating with uric acid for 48 h; b Representative images of Oil Red O staining of AML-12 cells (200×). c Serum uric acid level was significantly higher in HFD-fed mice than SCD-fed mice, and the level was decreased by allopurinol treatment in HFD + A group. d Intrahepatic triglyceride contents of the mice fed by different diets. f Representative images of liver histology determined by H & E and Oil Red O staining (× 200). Data are presented as the mean ± SD of at least three independent replicates. * P < 0.05, ** P < 0.01, *** P < 0.001 of two-tailed student’s t-test or ANOVA
Differentially expressed miRNA in HFD and HFD + A fed mice
Microarrays were applied to explore the effect of uric acid on hepatic miRNA expressions in mice fed with SCD, HFD and HFD + A. We found that 47 miRNAs were differentially expressed at least 1.5-fold between the HFD and SCD groups, including 28 down-regulated and 19 up-regulated miRNAs. Furthermore, 26 miRNAs were differentially expressed at least 1.5-fold in HFD + A group compared with the HFD groups, including 5 down-regulated and 21 up-regulated miRNAs. We firstly identified that 12 miRNAs were both differentially expressed in HFD vs SCD, and HFD + A vs HFD (Fig. 2a, Table S2), indicating that those 12 miRNAs may both involved in the progress of high uric acid-induced hepatic steatosis and lowering uric acid-ameliorated hepatic steatosis.
The differentially expressed miRNAs in livers of SCD, HFD and HFD + A fed mice. a Hierarchical clustering of the differentially expressed miRNAs in the livers of SCD, HFD and HFD + A fed mice. b Real-time PCR verification of miR-149-5p expression in the livers of SCD, HFD, and HFD + A fed mice. c Real-time PCR verification of miR-149-5p expression in AML-12 cells exposed to different concentrations of uric acid in AML-12 cells. Data are presented as the mean ± SD of at least three independent replicates. * P < 0.05, *** P < 0.001 of two-tailed student’s t-test
Real-time PCR was performed to confirm the expression of differentially expressed miRNAs. Consistent with miRNA array analysis, we found that the expression of miR-149-5p was up-regulated about 2.5-fold in the livers of HFD-fed mice compared with SCD-fed mice, while the expression was down-regulated approximately by 20% in HFD + A mice compared with HFD-fed mice (Fig. 2b). We also found that miR-149-5p was significantly up-regulated about 1.5-fold in AML-12 cells exposed to 500 μmol/L and 750 μmol/L uric acid for 48 h (Fig. 2c). Similar results were observed in HepG2 cells (Fig. S2). These results indicated that the miR-149-5p may be associated with uric acid-induced hepatocytes lipid accumulation.
Effect of miR-149-5p on lipid accumulation in hepatocytes
To verify whether miR-149-5p mediates the regulatory effects of uric acid on lipid accumulation in hepatocytes, loss- or gain-of-function of miR-149-5p on uric acid-stimulated hepatocytes was analyzed. miR-149-5p was overexpressed in AML-12 cells by transfection of miR-149-5p mimic (Fig. 3a). We found that overexpression of miR-149-5p significantly aggregated uric acid-induced intracellular triglyceride accumulation in AML-12 cells (Fig. 3b). On the other hand, inhibiting miR-149-5p expression by miR-149-5p inhibitor significantly ameliorated uric acid-induced triglyceride accumulation in AML-12 cells (Fig. 3c and d). Oil Red O staining of hepatocytes confirm the altered intracellular lipid accumulation (Fig. 3e). Similar results were observed in uric acid-stimulated HepG2 cells treated with miR-149-5p mimic or inhibitor (Fig. S3). These results suggested that miR-149-5p was involved in the regulatory effects of uric acid on lipid accumulation in hepatocytes.
MiR-149-5p mediated the regulatory effects of uric acid on lipid accumulation in hepatocytes. a Real-time PCR confirmed miR-149-5p mimic increased miR-149-5p expression in AML-12 cells. b Overexpression of miR-149-5p aggregated uric acid-induced intracellular triglyceride accumulation in AML-12 cells. c Real-time PCR confirmed that miR-149-5p inhibitor decreased miR-149-5p expression in AML-12 cells. d Inhibition of miR-149-5p alleviated uric acid-induced intracellular triglyceride accumulation in AML-12 cells. e Oil Red O staining conformed the regulatory roles of miR-149-5p on uric acid-induced hepatocytes lipid accumulation (× 200). Data are presented as the mean ± SD of at least three independent replicates. * P < 0.05, ** P < 0.01, *** P < 0.001 of two-tailed student’s t-test
FGF21 is a target gene of miR-149-5p involved in uric acid-induced hepatocytes lipid accumulation
By searching miRwalk prediction database, we found that FGF21 was predicted to be a target gene of miR-149-5p. FGF21 is a well-known protein that plays a key role in lipid metabolism [21, 22]. In order to determine whether FGF21 was a target gene of miR-149-5p, renilla luciferase reporter plasmids was constructed with the insertion of predicted miR-149-5p binding sites inside of human FGF21 (Fig. 4a). Co-transfection of miR-149-5p mimic led to a 30% reduction of the luciferase activity from the FGF21 WT 3’UTR in 293 T cells (Fig. 4b). However, mutation of predicted binding sites recovered the loss of luciferase activities mediated by excess miR-149-5p (Fig. 4b). Moreover, we also examine whether miR-149-5p regulated FGF21 endogenously. We found that transfecting with miR-149-5p mimic markedly suppressed the protein levels of FGF21 in AML-12 cells (Fig. 4c). Uric acid stimulation also suppressed the protein levels of FGF21 in AML-12 cells, whereas miR-149-5p inhibitor restored the expression of FGF21 in uric acid-stimulated hepatocytes (Fig. 4d). Similar results were observed in uric acid-stimulated HepG2 cells (Fig. S4a and b). These results suggested that miR-149-5p target regulated FGF21 through translation inhibition.
FGF21 is a downstream target of miR-149-5p that mediated uric acid-induced lipid accumulation in hepatocytes. a Diagram showing miR-149-5p binding sites predicted by miRwalk in the FGF21 3’UTR region. b Luciferase activity assays showed that miR-149-5p mimic significantly reduced the luciferase activity of wild type but not mutant 3’UTR of FGF21. c Western blot confirmed that miR-149-5p mimic significantly inhibited FGF21 expression. d Uric acid stimulation significantly down-regulated FGF21 expression, while miR-149-5p inhibitor restored the expression of FGF21 in uric acid-stimulated AML-12 cells. e Inhibiting FGF21 abolished the ameliorative effect of miR-149-5p inhibitor on uric acid-induced triglyceride accumulation in AML-12 cells. f Overexpression of FGF21 prevented the triglyceride accumulation induced by miR-149-5p mimic in AML-12 cells. Data are presented as the mean ± SD of at least three independent replicates. * P < 0.05, ** P < 0.01, *** P < 0.001 of two-tailed student’s t-test
We further applied FGF21 siRNA and overexpression plasmids to determine the effect of FGF21 on lipid accumulation in hepatocytes. We found that silencing FGF21 abolished the ameliorative effect of miR-149-5p inhibitor on uric acid-induced triglyceride accumulation in hepatocytes (Fig. 4e). On the contrary, overexpression of FGF21 significantly prevented the triglyceride accumulation induced by miR-149-5p mimic in AML-12 cells (Fig. 4f). Similar results were observed in HepG2 cells treated with FGF21 siRNA and overexpression plasmids (Fig. S4c and d). These results suggested that FGF21 is the downstream target of miR-149-5p and mediates the regulatory effects of uric acid on lipid accumulation in hepatocytes.
Discussion
In this study, we provide novel evidence that uric acid-induced hepatocytes lipid accumulation through miR-149-5p/FGF21 dependent mechanism. First, uric acid stimulation significantly up-regulated miR-149-5p expression in hepatocytes, while lowering uric acid by allopurinol significantly down-regulated hepatic miR-149-5p expression in HFD-fed mice. Second, loss- or gain-of-function studies found that miR-149-5p was responsible for pathogenic effect of uric acid on hepatocytes lipid accumulation. Third, we confirmed that FGF21 is a downstream target of miR-149-5p and FGF21 mediated the regulatory effects of uric acid on hepatocytes lipid accumulation.
Our previous studies have shown that SUA levels are significantly elevated in NAFLD patients, and that elevated SUA levels are associated with increased risk of incident NAFLD [9, 23]. Other large population-based cohort studies also found that high SUA levels are positively associated with risk of NAFLD [24, 25]. Histological studies have demonstrated that the SUA levels were significantly associated with histological disease severity of NAFLD [26,27,28]. Moreover, uric acid-lowering therapy by allopurinol significantly alleviated HFD-induced hepatic steatosis in an animal model of NAFLD [29]. These studies suggested that high uric acid may be a cause for the development of NAFLD, while its underlying molecular mechanism remains unclear.
MiRNA has been implicated as a crucial regulator in metabolic diseases by silencing genes at the post-transcriptional level. Growing evidence demonstrated that a large number of miRNAs, such as miR-122, miR-378 and miR-34a, are involved in the pathogenesis of NAFLD [30, 31]. However, whether miRNAs are involved in the uric acid-induced hepatocytes lipid accumulation is unclear. In this study, we aimed to explore the hepatic miRNA expression profiles of HFD-fed mice treated with or without allopurinol-therapy. In this study, we analyzed the miRNA profiles altered in response to HFD, and HFD combined with lowering uric acid therapy. We found that 47 miRNAs were differentially expressed between the HFD and SCD groups, including 19 up-regulated miRNAs and 28 down-regulated miRNAs. This finding is partially consistent with the previously observed in ob/ob mice [17, 32], MCD-fed mice [33], and NASH patients [34]. Our study confirmed that miRNAs are participated in the pathogenesis of NFALD.
We also found that 12 miRNAs were both differentially expressed in HFD vs SCD, and HFD + A vs HFD, suggesting potential involvement of those miRNAs in the progress of uric acid-induced hepatocyte lipid accumulation. Specifically, miR-149-5p was 5.31-fold up-regulated in HFD vs SCD, and 2.29-fold down- regulated in HFD + A vs HFD. Because miR-149-5p was the most obvious up-regulated miRNAs in response to HFD, we focused on this miRNA and explored its underlying mechanism on uric acid-induced hepatocytes lipid accumulation. MiR-149 was reported to be both a tumor suppressor and an onco-miRNA in various types of cancers, and has distinct functions including proliferation inhibition, apoptosis induction, cell-cycle regulation, and metastasis promotion separately [35,36,37]. A recent study reported that deficient of miR-149 increased mice whole-body energy expenditure, combined with enhanced thermogenesis of inguinal fat [38]. This finding suggested that miR-149 may play a potential role in the development of metabolic diseases. However, the role of miR-149-5p in NAFLD remains unclear. Here, we found that the expression of miR-149-5p was significantly elevated in response to uric acid stimulation in hepatocytes. Overexpression of miR-149-5p aggravated uric acid-induced hepatocytes lipid accumulation, whereas inhibition of miR-149-5p had the opposite effects. These results suggested that miR-149-5p mediated the regulatory of uric acid on hepatocytes lipid accumulation.
Furthermore, we also explored the down-stream mechanism of miR-149-5p on hepatocytes lipid accumulation. FGF21 is a member of the endocrine FGF family, mainly expressed and secreted in the liver and adipose tissue. FGF21 is an important regulator of lipid and glucose metabolism and plays a key role in the development in NAFLD [39, 40]. Some clinical studies reported that serum FGF21 level is increased in NAFLD patients and is considered to be a predictor of NAFLD [41]. However, recent studies reported different results that deficient of FGF21 induced hepatic steatosis [42], and overexpression of FGF21 was able to ameliorate liver fat accumulation [43]. In our study, we confirmed that FGF21 is a down-stream target of miR-149-5p in the process of uric acid-induced hepatocytes lipid accumulation. Our results suggest a new mechanism of uric acid-induced hepatocytes lipid accumulation.
There were several limitations in this study. First, the function and regulation of miRNA are complex, FGF21 may not be the only and the master target gene regulated by uric acid, further investigations are required to clarify the detailed downstream mechanisms of miR-149-5p on uric acid-induced hepatic steatosis. Second, although our in vitro cell line results clearly showed that uric acid regulates hepatocyte lipid accumulation through miR-149-5p/FGF21 dependent mechanism, primary hepatocytes experiments and in vivo experiments are needed to further confirm the regulatory mechanisms.
Conclusions
In conclusion, our study provided evidence for the first time that uric acid induced hepatocytes lipid accumulation through miR-149-5p/FGF21 dependent mechanism.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Abbreviations
- DMEM:
-
Dulbecco’s modified eagle medium
- FBS:
-
Fetal bovine serum
- HFD + A:
-
HFD combined with allopurinol therapy
- HFD:
-
High fat diet
- miRNA:
-
Microrna
- NAFLD:
-
Non-alcoholic fatty liver disease
- NLRP3:
-
NACHT, LRR and PYD domains-containing protein 3
- PPARα:
-
Peroxisome proliferator activated receptor alpha
- SCD:
-
Standard chow diet
- SD:
-
Standard division
- siRNA:
-
Small interfering RNA
- UTR:
-
Untranslated region
References
Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, Harrison SA, Brunt EM, Sanyal AJ. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018;67(1):328–57.
Fan JG, Wei L, Zhuang H, National Workshop on Fatty Liver and Alcoholic Liver Disease, Chinese Society of Hepatology, Chinese Medical Association, Fatty Liver Disease Expert Committee, Chinese Medical Doctor Association. Guidelines of prevention and treatment of nonalcoholic fatty liver disease (2018, China). J Dig Dis. 2019;20(4):163–73.
Li J, Zou B, Yeo YH, et al. Prevalence, incidence, and outcome of non-alcoholic fatty liver disease in Asia, 1999-2019: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2019;4(5):389–98.
Zhou F, Zhou J, Wang W, Zhang XJ, Ji YX, Zhang P, She ZG, Zhu L, Cai J, Li H. Unexpected rapid increase in the burden of nonalcoholic fatty liver disease in China from 2008 to 2018: a systematic review and meta-analysis. Hepatology. 2019;70(4):1119–33.
Xiao J, Bei Y, Liu J, et al. miR-212 downregulation contributes to the protective effect of exercise against non-alcoholic fatty liver via targeting FGF-21. J Cell Mol Med. 2016;20(2):204–16.
Mantovani A, Byrne CD, Bonora E, Targher G. Nonalcoholic fatty liver disease and risk of incident type 2 diabetes: a meta-analysis. Diabetes Care. 2018;41(2):372–82.
Targher G, Day CP, Bonora E. Risk of cardiovascular disease in patients with nonalcoholic fatty liver disease. N Engl J Med. 2010;363(14):1341–50.
Xu C, Yu C, Ma H, Xu L, Miao M, Li Y. Prevalence and risk factors for the development of nonalcoholic fatty liver disease in a nonobese Chinese population: the Zhejiang Zhenhai study. Am J Gastroenterol. 2013;108(8):1299–304.
Li Y, Xu C, Yu C, Xu L, Miao M. Association of serum uric acid level with non-alcoholic fatty liver disease: a cross-sectional study. J Hepatol. 2009;50(5):1029–34.
Wan X, Xu C, Lin Y, Lu C, Li D, Sang J, He H, Liu X, Li Y, Yu C. Uric acid regulates hepatic steatosis and insulin resistance through the NLRP3 inflammasome-dependent mechanism. J Hepatol. 2016;64(4):925–32.
Choi YJ, Shin HS, Choi HS, Park JW, Jo I, Oh ES, Lee KY, Lee BH, Johnson RJ, Kang DH. Uric acid induces fat accumulation via generation of endoplasmic reticulum stress and SREBP-1c activation in hepatocytes. Lab Investig. 2014;94(10):1114–25.
Lanaspa MA, Sanchez-Lozada LG, Choi YJ, et al. Uric acid induces hepatic steatosis by generation of mitochondrial oxidative stress: potential role in fructose-dependent and -independent fatty liver. J Biol Chem. 2012;287(48):40732–44.
Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4(9):1179–84.
Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–62.
Su Q, Kumar V, Sud N, Mahato RI. MicroRNAs in the pathogenesis and treatment of progressive liver injury in NAFLD and liver fibrosis. Adv Drug Deliv Rev. 2018;129:54–63.
Wu G, Rui C, Chen J, Sho E, Zhan S, Yuan X, Ding Y. MicroRNA-122 inhibits lipid droplet formation and hepatic triglyceride accumulation via yin Yang 1. Cell Physiol Biochem. 2017;44(4):1651–64.
Li S, Chen X, Zhang H, Liang X, Xiang Y, Yu C, Zen K, Li Y, Zhang CY. Differential expression of microRNAs in mouse liver under aberrant energy metabolic status. J Lipid Res. 2009;50(9):1756–65.
Ding J, Li M, Wan X, Jin X, Chen S, Yu C, Li Y. Effect of miR-34a in regulating steatosis by targeting PPARalpha expression in nonalcoholic fatty liver disease. Sci Rep. 2015;5:13729.
Yu S, Hong Q, Wang Y, et al. High concentrations of uric acid inhibit angiogenesis via regulation of the Kruppel-like factor 2-vascular endothelial growth factor-a axis by miR-92a. Circ J. 2015;79(11):2487–98.
Takir M, Kostek O, Ozkok A, et al. Lowering uric acid with allopurinol improves insulin resistance and systemic inflammation in asymptomatic Hyperuricemia. J Investig Med. 2015;63(8):924–9.
Maratos-Flier E. Fatty liver and FGF21 physiology. Exp Cell Res. 2017;360(1):2–5.
Rusli F, Deelen J, Andriyani E, Boekschoten MV, Lute C, van den Akker EB, Muller M, Beekman M, Steegenga WT. Fibroblast growth factor 21 reflects liver fat accumulation and dysregulation of signalling pathways in the liver of C57BL/6J mice. Sci Rep. 2016;6:30484.
Xu C, Yu C, Xu L, Miao M, Li Y. High serum uric acid increases the risk for nonalcoholic fatty liver disease: a prospective observational study. PLoS One. 2010;5(7):e11578.
Afzali A, Weiss NS, Boyko EJ, Ioannou GN. Association between serum uric acid level and chronic liver disease in the United States. Hepatology. 2010;52(2):578–89.
Sirota JC, McFann K, Targher G, Johnson RJ, Chonchol M, Jalal DI. Elevated serum uric acid levels are associated with non-alcoholic fatty liver disease independently of metabolic syndrome features in the United States: liver ultrasound data from the National Health and nutrition examination survey. Metab Clin Exp. 2013;62(3):392–9.
Huang Q, Yu J, Zhang X, Liu S, Ge Y. Association of the serum uric acid level with liver histology in biopsy-proven non-alcoholic fatty liver disease. Biomed Rep. 2016;5(2):188–92.
Petta S, Cammà C, Cabibi D, Di Marco V, Craxì A. Hyperuricemia is associated with histological liver damage in patients with non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2011;34(7):757–66.
Fernández Rodríguez CM, Aller R, Gutiérrez García ML, et al. Higher levels of serum uric acid influences hepatic damage in patients with non-alcoholic fatty liver disease (NAFLD). Rev Esp Enferm Dig. 2019;111(4):264–9.
Xu CF, Yu CH, Xu L, Sa XY, Li YM. Hypouricemic therapy: a novel potential therapeutic option for nonalcoholic fatty liver disease. Hepatology. 2010;52(5):1865–6.
Tsai WC, Hsu SD, Hsu CS, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122(8):2884–97.
Zhang T, Duan J, Zhang L, Li Z, Steer CJ, Yan G, Song G. LXRalpha promotes Hepatosteatosis in part through activation of MicroRNA-378 transcription and inhibition of Ppargc1beta expression. Hepatology. 2019;69(4):1488–503.
Zhang M, Sun W, Zhou M, Tang Y. MicroRNA-27a regulates hepatic lipid metabolism and alleviates NAFLD via repressing FAS and SCD1. Sci Rep. 2017;7(1):14493.
Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, Patel T, Schmittgen TD, Croce C, Ghoshal K, Jacob ST. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50(4):1152–61.
Estep M, Armistead D, Hossain N, Elarainy H, Goodman Z, Baranova A, Chandhoke V, Younossi ZM. Differential expression of miRNAs in the visceral adipose tissue of patients with non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2010;32(3):487–97.
Zhang Y, Guo X, Xiong L, Yu L, Li Z, Guo Q, Li Z, Li B, Lin N. Comprehensive analysis of microRNA-regulated protein interaction network reveals the tumor suppressive role of microRNA-149 in human hepatocellular carcinoma via targeting AKT-mTOR pathway. Mol Cancer. 2014;13:253.
Jin L, Hu WL, Jiang CC, et al. MicroRNA-149*, a p53-responsive microRNA, functions as an oncogenic regulator in human melanoma. Proc Natl Acad Sci U S A. 2011;108(38):15840–5.
Zhi Y, Zhou H, Mubalake A, Chen Y, Zhang B, Zhang K, Chu X, Wang R. Regulation and functions of MicroRNA-149 in human cancers. Cell Prolif. 2018;51(5):e12465.
Ding H, Zheng S, Garcia-Ruiz D, et al. Fasting induces a subcutaneous-to-visceral fat switch mediated by microRNA-149-3p and suppression of PRDM16. Nat Commun. 2016;7:11533.
Gao M, Ma Y, Cui R, Liu D. Hydrodynamic delivery of FGF21 gene alleviates obesity and fatty liver in mice fed a high-fat diet. J Control Release. 2014;185:1–11.
Jimenez V, Jambrina C, Casana E, et al. FGF21 gene therapy as treatment for obesity and insulin resistance. EMBO Mol Med. 2018;10(8):e8791.
Li H, Dong K, Fang Q, Hou X, Zhou M, Bao Y, Xiang K, Xu A, Jia W. High serum level of fibroblast growth factor 21 is an independent predictor of non-alcoholic fatty liver disease: a 3-year prospective study in China. J Hepatol. 2013;58(3):557–63.
Singhal G, Kumar G, Chan S, Fisher FM, Ma Y, Vardeh HG, Nasser IA, Flier JS, Maratos-Flier E. Deficiency of fibroblast growth factor 21 (FGF21) promotes hepatocellular carcinoma (HCC) in mice on a long term obesogenic diet. Mol Metab. 2018;13:56–66.
Ye D, Wang Y, Li H, Jia W, Man K, Lo CM, Wang Y, Lam KS, Xu A. Fibroblast growth factor 21 protects against acetaminophen-induced hepatotoxicity by potentiating peroxisome proliferator-activated receptor coactivator protein-1alpha-mediated antioxidant capacity in mice. Hepatology. 2014;60(3):977–89.
Acknowledgements
Not applicable.
Funding
This work was supported by National Natural Science Foundation of China (Nos. 81722009, 81770573, and 81870400), Key Research and Development Program of Zhejiang Province (No. 2020C03033), and the Fundamental Research Funds for the Central Universities (No. 2018XZZX002–10, and 2019QNA7034).
Author information
Authors and Affiliations
Contributions
SC, and CX contributed to the conception and design of the study. SC, DC, HY, XW, JW, and CX performed and collected the data. SC, DC, HY, and CX analyzed and interpreted the data, and drafted the manuscript. SC, and CX prepared the manuscript. SC, DC, HY, XW, JW, and CX reviewed the manuscript. SC, DC, HY, XW, JW and CX approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All animal experiments were approved by the Animal Care and Use Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University (Reference number: 2019–1096).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Fig. S1.
Uric acid stimulation induced significant lipid accumulation in HepG2 cells. (a) Intracellular triglyceride contents in HepG2 cells exposed to different concentrations of uric acid for 48 h. (b) Representative image of Oil Red O staining of HepG2 cells (× 200). Data are presented as the mean ± SD of at least three independent replicates. * P < 0.05, ** P < 0.01 of two-tailed student’s t-test.
Additional file 2: Fig. S2.
Real-time PCR verification of miR-149-5p expression in HepG2 cells exposed to different concentrations of uric acid for 48 h. Data are presented as the mean ± SD of at least three independent replicates. * P < 0.05 of two-tailed student’s t-test.
Additional file 3: Fig. S3.
miR-149-5p mediated the regulatory effects of uric acid on intracellular lipid accumulation in HepG2 cells. (a) miR-149-5p mimic increased miR-149-5p expression levels in HepG2 cells. (b) miR-149-5p mimic enhanced uric acid-induced intracellular triglyceride accumulation in HepG2 cells. (c) miR-149-5p inhibitor decreased miR-149-5p expression levels in HepG2 cells. (d) miR-149-5p inhibitor ameliorated uric acid-induced intracellular triglyceride accumulation in HepG2 cells. (e) Oil Red O staining conformed the regulatory roles of miR-149-5p on uric acid-induced intracellular lipid accumulation in HepG2 cells (× 200). Data are presented as the mean ± SD of at least three independent replicates. * P < 0.05, ** P < 0.01 of two-tailed student’s t-test.
Additional file 4: Fig. S4.
FGF21 is a target gene of miR-149-5p. (a) Western blot confirmed that miR-149-5p mimic significantly inhibited FGF21 expression in HepG2 cells. (b) Uric acid stimulation significantly down-regulated FGF21 expression, while miR-149-5p inhibitor restored the FGF21 expression in uric acid-stimulated HepG2 cells. (c) Silencing FGF21 abolished the ameliorative effect of miR-149-5p inhibitor on uric acid-induced intracellular triglyceride accumulation in HepG2 cells. (d) Overexpression of FGF21 decreased intracellular triglyceride contents induced by miR-149-5p mimic in HepG2 cells. Data are presented as the mean ± SD of at least three independent replicates. * P < 0.05 of two-tailed student’s t-test.
Additional file 5: Table S1.
Primer sequences of genes analyzed by Real-time PCR.
Additional file 6: Table S2.
Differential expressed miRNAs identified by microarray analysis of liver samples from SCD, HFD and HFD + A fed mice.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Chen, S., Chen, D., Yang, H. et al. Uric acid induced hepatocytes lipid accumulation through regulation of miR-149-5p/FGF21 axis. BMC Gastroenterol 20, 39 (2020). https://doi.org/10.1186/s12876-020-01189-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12876-020-01189-z