Abstract
Background
HD-Zips (Homeodomain-Leucine Zippers) are a class of plant-specific transcription factors that play multiple roles in plant growth and development. Although some functions of HD-Zip transcription factor have been reported in several plants, it has not been comprehensively studied in peach, especially during adventitious root formation of peach cuttings.
Results
In this study, 23 HD-Zip genes distributed on 6 chromosomes were identified from the peach (Prunus persica) genome, and named PpHDZ01-23 according to their positions on the chromosomes. These 23 PpHDZ transcription factors all contained a homeomorphism box domain and a leucine zipper domain, were divided into 4 subfamilies(I-IV) according to the evolutionary analysis, and their promoters contained many different cis-acting elements. Spatio-temporal expression pattern showed that these genes were expressed in many tissues with different levels, and they had distinct expression pattern during adventitious root formation and development.
Conclusion
Our results showed the roles of PpHDZs on root formation, which is helpful to better understand the classification and function of peach HD-Zip genes.
Similar content being viewed by others
Introduction
HD-Zip transcription factors belong to the Homeodomain (HD) transcription factor superfamily and are only found in plants. HD-Zip protein usually contain two highly conserved domains: homeodomain (HD) and leucine zipper (LZ) [1]. The homeodomain domain is generally composed of about 60 amino acids encoded homeobox, which form three alpha helices. The three helices formed a tight spherical structure with hydrophobic core under the action of two loops and one corner [2]. And the arrangement of the three helices determines the difference in the specific binding of homeodomain to DNA. The leucine zipper domain forms a dimer that is essential for HD-Zip protein recognition of DNA [3]. According to the differences in structure and function, HD-Zip family members can be divided into four subfamilies: I, II, III and IV. The four subfamilies all have HD and LZ domains. In addition to HD and LZ domains, subfamily IV also has a START domain related to sterol binding. Compared with subfamily IV, subfamily III has another C-terminal MEKHLX domain [4].
HD-Zip transcription factors have been identified in a variety of plants, such as Arabidopsis thaliana [5], Zea mays [6] Oryza sativa [7], Populus trichocarpa [8], and so on. As a unique transcription factor in plants, HD-Zip plays an important regulatory role in plant growth and development, environmental response, cell cycle and cell metabolism [9]. For example, NaHD20 could regulate ABA accumulation and activate the expression of dehydration related genes in tobacco leaves under water stress [10]. In Arabidopsis thaliana, overexpression of ATHB17 can increase chlorophyll content and significantly improve photosynthetic capacity [11]. PtrHB7 overexpression enhanced the differentiation of cambium cells into xylem cells and balanced the differentiation of secondary xylem and phloem [12]. ZmOCL1 may regulate the development of corn kernels by regulating gibberellin level [13].
As one of the most important fruits in the world with high nutritional value, peach is widely grown all over the world. In our country, seedling rootstock grafting is the main method of peach propagation. However, due to the obvious separation of traits in the offspring and the poor uniformity of seedlings, it brings many problems to production management. Cutting propagation can preserve the excellent characters of mother plant, and can produce a large number of seedlings in a short period of time, so the cutting propagation of peach has a good application prospect. The formation and growth of adventitious root is the key to cutting propagation, which is regulated by genes. Many studies have found that HD-Zip proteins are involved in the formation and development of roots. Such as MtHB1 that regulate the emergence of lateral roots by mediating LBD1 expression in alfalfa [14], HAT2 that negatively regulates root growth, reducing lateral root length and ATHB10 regulating root hair development by participating in ethylene dependent pathway in Arabidopsis thaliana [15, 16]. The HD-Zip III subfamily in Arabidopsis thaliana regulates root development through plant hormones [17]. However, the roles of HD-Zips in root formation and development of peach has not yet been studied.
In this study, the HD-Zip transcription factor family of peach was analyzed. A total of 23 HD-Zip genes were identified in peach genome, and their chromosome localization, phylogenetic relationship, gene structure, conserved domain and promoter elements were analyzed. Meanwhile, we also investigated the expression patterns of PpHDZ genes in different tissues and in the process of adventitious root development. These results provided a basis for further study on the function of PpHDZ family members in peach.
Results
Identification of HD-Zip gene family members in peach
The HD-Zip gene in peach was identified by whole-genome analysis method, and the HMM model was constructed according to the characteristic domain of HD-Zip protein (PF00046). The HMM model was used to search peach genome to obtain candidate genes, and NCBI was used to further detect whether their sequences contained the HD-Zip domain. Finally, 23 HD-Zip genes were identified, named PpHDZ01-23 according to their location on the chromosomes. These 23 PpHDZ genes were unevenly distributed on 6 chromosomes (Fig. 1). The number of PpHDZ genes on Chr. 3 was up to 6, followed by 5, 4, 3 and 3 PpHDZ genes on Chr. 1, Chr. 2, Chr.5 and Chr. 6, respectively, and there were at least 2 PpHDZ genes on Chr. 4.
The predicted proteins of PpHDZ genes contain amino acids ranged from 227 (PpHDZ02) to 849 (PpHDZ17), with an average amino acid number of 556. The molecular weight ranged from 25,495.91 Da (PpHDZ02) to 93,018.86 Da (PpHDZ17), and the average molecular weight was 61,445.54 Da. The isoelectric point (pI) is an important physiological indicator of a protein, which mainly depends on the ratio of the number of acidic amino acid to basic amino acid. The theoretical isoelectric point of PpHDZs were between 5.4182 and 9.2656 (Table 1). The isoelectric point of most PpHDZ proteins (87%) was less than 7, which indicates that PpHDZ proteins might be acidic proteins.
Phylogenetic relationships of peach HD-Zip gene family
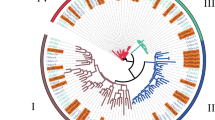
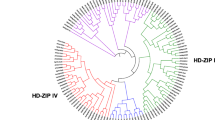
In order to understand the homology of 23 PpHDZ genes in peach and predict their functions, phylogenetic tree was constructed with Arabidopsis HD-Zip gene families. It showed that peach HD-Zip gene family could be divided into four subfamilies (I-IV), which included 6, 5, 4, 8 HD-Zip gene family members respectively, and subfamilies I and II were closely related and in the same branch, (Fig. 2 A). All the PpHDZ proteins contained a HD domain, which were composed of 3 coil-helix structures (Fig. 2B). And the amino acid sequence alignment of HD domains showed that the amino acid number of subfamilies I and II in their characteristic structural domain is the same, with 4 amino acids less than that of subfamily III and 2 amino acids less than that of subfamily IV.
Through phylogenetic tree and amino acid sequence alignment in HD domain, we could infer the function of PpHDZ genes of peach from the function of HD-Zip genes in Arabidopsis thaliana, a model plant with known function.
Analysis of conserved motifs, domains and number of exons
In order to further understand the evolutionary relationship of peach HD-Zip protein, the structure of peach HD-Zip gene family was analyzed by GSDS. There were significant differences in gene structure among the four subfamilies of peach HD-Zip gene family, and the number of exon and intron in the same subfamily was similar. The gene coding sequence of subfamilies I and II was simpler than that of subfamilies III and IV. The number of exon in subfamilies I and II members was 3–4, while that of subfamilies III and IV members was more than 10 (Fig. 3 A). These findings suggested that the exons of the HD-Zip gene family were lost or increased during evolution.
TBtools was used to analyze the protein conserved domain. It was showed that HD-Zip genes from the same subfamily had the same domain distribution and composition, suggesting that members of the same subfamily might have similar functions. All members of peach HD-Zip gene family had HD and LZ domains, that is, Motif 1, 2, 9 together constitute the HD-Zip domain, suggesting that these two domains play an important role in the expression of HD-Zip gene. There were only HD and LZ domains in subfamilies I and II. Both subfamilies III and IV had a START domain related to sterol binding, named Motif 3, 4, 10, 11. Subfamily III also had a special C-terminal MEKHLA domain, named Motif 12 (Fig. 3B). This is consistent with previous reports that the existence of these subfamily-specific domains may be related to subfamily-specific functions [18,19,20]. Therefore, the analysis of conserved motif and exon composition further supported the study of the evolution of HD-Zip gene family.
Gene replication and collinearity analysis of PpHDZ gene
In order to investigate the expansion and evolution mechanism of PpHDZ gene family, gene replication events in peach genome were studied. A total of 5 pairs of homologous genes were found in peach chromosomes, which were PpHDZ01/PpHDZ19, PpHDZ03/PpHDZ23, PpHDZ04/PpHDZ11, PpHDZ05/PpHDZ22 and PpHDZ08/PpHDZ09 (Fig. 4 A). This suggested that segmental duplication was the main cause of PpHDZ gene family amplification. In addition, to further understand the origin and function of the PpHDZ gene family, we also mapped the collinearity of HD-Zip gene in the peach and Arabidopsis genomes. A total of 37 pairs of HD-Zip genes were identified in peach and Arabidopsis (Fig. 4B). This showed that HD-Zip gene in peach and Arabidopsis has high homology, indicating that HD-Zip gene had high conservation.
GO annotation analysis
GO ontology annotation was used to analyze and predict the biological processes, molecular functions and subcellular localization of HD-Zip protein in peach (Fig. 5 A). In molecular function, PpHDZ proteins mainly played a role in DNA binding (46.94%) and nucleic acid binding transcription factor activity (46.94%), and a few proteins played a role in protein binding (6.12%). In biological process, PpHDZ members mainly played roles in biosynthetic process (27.38%), cellular nitrogen compound metabolic process (27.38%),anatomical structure development (11.90%) and cell differentiation (9.52%). In addition, PpHDZ members were involved in response to stress (5.95%), reproduction (3.57%), embryo development (3.57%), anatomical structure formation involved in morphogenesis (3.57%), cell morphogenesis (2.38%), signal transduction (2.38%), pigmentation (1.19%), developmental maturation (1.19%) and so on. In terms of subcellular localization, PpHDZ protein was mainly located in the nuclear (78.57%), and a few proteins were located in the cytoplasmic (10.71%) and plasmamembrane (10.71%).
Subcellular localization of PpHDZ proteins
Nuclear localization of transcription factors is important for regulating transcription of target genes by binding specific cis-elements in their promoters. Previous studies have shown that HD-Zip protein is mainly located in the nucleus. For example, VvHDZ28 in grape is a nuclear protein [21]. In this study, three cloned PpHDZ genes (PpHDZ2, PpHDZ15 and PpHDZ16) were introduced into the pCAMBIA1300 vector and fused with GFP gene, all under the CaMV35S promoter. The recombinant fusion vector were infiltrated into Nicotiana benthamiana leaves. As shown in Fig. 5B (Fig. 5B), the green fluorescence signals of the fusion proteins PpHDZ2-GFP, PpHDZ15-GFP and PpHDZ16-GFP were heterogeneously distributed in the nucleus. This was consistent with the results of GO analysis. The results showed that PpHDZ2, PpHDZ15 and PpHDZ16 were nuclear proteins.
GO and subcellular localization analyses. A Prediction of biological process, molecular function and subcellular localization of PpHDZ gene family. Figures represent the percentage of each component. B Subcellular localization of three GFP-fused PpHDZ proteins (PpHDZ2-GFP, PpHDZ15-GFP, and PpHDZ16-GFP). The photographs were taken under bright light, in the dark field for the GFP-derived green flourescence and DAPI-derived blue flourescence and merged, respectively
Promoter element analysis
The promoter sequences of 23 PpHDZ genes (2 kb upstream of 5 ‘UTR) were submitted to PlantCARE database for the prediction of promoter elements and a total of 17 cis-elements were identified (Fig. 6A). Except for two conventional promoter elements TATA-box and CAAT-box, the remaining response elements could be divided into three main categories: plant growth and development, stress response and phytohormones response. The phytohormones response elements in HD-Zip gene promoter region mainly included ABA (40%), GA (8%), IAA (13%), JA (13%) and MeJA (26%), indicating that HD-Zip genes might be involved in physiological, biochemical and growth and development processes of plants in response to various upstream hormones. Among the stress-related cis-elements, anaerobic induction (60%), drought induction (13%), low temperature induction (14%) and defense stress response (13%) were detected. In plant growth and development related cis-elements, light response elements accounted for the highest proportion (83%), and each peach HD-Zip family member had light response elements, suggesting that the HD-Zip family would play an important role in the establishment of light morphology. Meristem expression response elements and seed development response elements are mainly distributed in the promoters of members of the subfamily III, perhaps because HD-Zip subfamily III is mainly involved in the development of plant embryos and meristem formation [22, 23]. The HD-Zip subfamily IV is mainly involved in the process of substance accumulation [24], so the endosperm expression response elements related to substance accumulation were mainly distributed in the subfamily IV members (Fig. 6B and C).
Promoters elements analysis of peach PpHDZ genes. A. Cis-elements in the location of the PpHDZ genes promoter sequences. B. Variety of cis-elements of PpHDZ gene. The different colors represent the number of each cis-elements. C. Distribution of cis-elements of PpHDZ gene in plant hormone response, stress response and plant growth and development. Pie charts size represented the percentage of promoter element in each category
Tissue specific expression pattern of PpHDZ genes in peach
The tissue specific expression pattern of 23 PpHDZs was analyzed. Overall, most PpHDZ genes exhibited a relatively wide range of expression patterns in a variety of tissues. However, a small number of gene subfamilies were preferentially expressed in specific tissues and developmental processes. The expression levels of subfamilies I and IV were higher in flower organs, indicating that the members of the two subfamilies played an important role in the development of flower organ, which was consistent with previous studies [25, 26]. Subfamily II members were expressed in stem, leaf and flower organ to varying degrees. Subfamily III genes were highly expressed in stem and leaf (Fig. 7), However, the four subfamily members all had low expression levels in peach fruit. The tissue-specific expression of PpHDZs suggested that different subfamilies played specific regulatory roles in different tissues.
Expression pattern of PpHDZ genes in different stages of rooting in cuttings
In order to explore the role of PpHDZ genes in the formation of adventitious root of cuttings, samples were collected at 0 h, 1 h, 6 h, 2 d, 10 d and 17 d after cuttings respectively, and the expression of the 23 PpHDZ genes were determined by qRT-PCR. The results showed that PpHDZ genes were expressed differently during adventitious root development. The subfamilies I and II showed high expression mainly in the intermediate stage of adventitious root formation. PpHDZ08, PpHDZ09, PpHDZ13, PpHDZ16 and PpHDZ18 of the subfamily IV were strongly expressed in 1 h after cutting, but down-regulated in later cutting period, indicating that they mainly acted in the early stage of adventitious root formation. PpHDZ06, PpHDZ17, and PpHDZ20 were down-regulated in the early stage and up-regulated in the late stage, suggesting that they played a role in the late adventitious root formation (Fig. 8). In addition, the spatio-temporal expression patterns of six homologous genes were analyzed. The expression patterns of PpHDZ08\PpHDZ09, PpHDZ14\PpHDZ15 and PpHDZ17\PpHDZ21 were similar in different adventitious root formation stages. In contrast, the expression patterns of three pairs of homologous genes PpHDZ02\PpHDZ06, PpHDZ01\PpHDZ19 and PpHDZ10\PpHDZ20 were significantly different in different adventitious root formation stages (Fig. 8). This suggests that homologous pairs might have undergone differential differentiation during the evolution of HD-Zip gene.
Discussion
Plant specific transcription factor HD-Zip plays an important role in plant growth and development and environmental response. HD-Zip transcription factors have been identified in Arabidopsis thaliana [5], Zea mays [6], Oryza sativa [7], Populus trichocarpa [8], and so on. However, there is still no comprehensive and systematic analysis of the HD-Zip gene family of peach, one of the fruit tree crops with high nutritional value in the world. In this study, 23 PpHDZ genes were identified from the whole genome of peach. The phylogenetic relationships, gene structure, conserved domains, promoter elements and the expression patterns of 23 PpHDZ genes in different tissues and adventitious roots were analyzed. It laid a foundation for further study on the function of HD-Zip gene in peach growth and development.
Phylogenetic analysis and multiple sequence alignment revealed that 23 PpHDZ genes were mainly divided into four subfamilies, which was consistent with previous results [19]. Moreover, PpHDZ genes from the same subfamily have similar conserved domain composition, intron and exon composition, which further supports the evolutionary relationship of 23 PpHDZ genes. HD-Zip gene is distributed in various plants. And the number of HD-Zip gene is similar in Arabidopsis thaliana (48 members), Capsicum annuum (42 members) and Carthamus tinctorius (48 members), which is more than in Vitis vinifera (33 members) and Oryza sativa (31 members), but much less than in Zea mays (55 members) and Populus trichocarpa (63 members) [21, 27, 28]. which means that HD-Zip genes are amplified to varying degrees in different species. Evolutionary events such as polyploidy and duplication events could increase the gene family members in plants [29, 30]. Collinearity analysis showed that many HD-Zip genes in Arabidopsis had corresponding genes in peach, but only 5 pairs of homologous HD-Zip genes were also found in peach, According to these results, it concludes that HD-Zip gene family in peach has been more influenced by evolutionary pressures.
More and more studies have shown that the HD-Zip gene family plays an important role in plant growth and development. Such as organ development, environmental response and hormonal response [9]. Based on the annotated information of the Arabidopsis HD-Zip gene family, combined with promoter element analysis and expression analysis of different tissues, the biological function of the peach PpHDZ gene could be inferred. For example, PpHDZ14 was homologous to AtHDZ06 (ATHB1) involved in hypocotyl elongation under short-day condition [31], which indicated that PpHDZ14 might regulate hypocotyl elongation under short-day condition. Both AtHDZ19 (HAT1) and AtHDZ27 (HAT3) belonging to subfamily II of the evolutionary tree were regulated by changes in light quality and induced by shade avoidance response in plants [32, 33]. There were a high abundance of light response elements in the promoters of Subfamilies I and II, suggesting that the two subfamilies genes of peach might be involved in the formation of light morphology. Moreover, the expression level of most members of subfamily I was higher in peach blossom, indicating that the subfamily I might play roles in peach blossom organs. AtHDZ29 (ATHB8) in subfamily III of the evolutionary tree is involved in vein formation and plant hormone signal transduction [34], suggesting that PpHDZ04 and PpHDZ11 belonging to the same subfamily might also have the similar functions. Promoter element analysis revealed that meristem expression response elements were mainly distributed in the genes of subfamily III (Fig. 6B). Expression analysis showed that the genes of subfamily III were highly expressed in stem and leaf (Fig. 7). It could be inferred from these results that subfamily III genes might play a key role in maintaining meristem and be participate in vein formation and hormone transduction. Studies have shown that HD-Zip gene may co-regulate the expression of flavonoid compound synthesis genes [35, 36]. MYB elements involved in the regulation of flavonoid biosynthesis genes were found in promoters of PpHDZ13 and PpHDZ16 genes in subfamily IV. Expression analysis showed that these two genes were also highly expressed in flowers. PpHDZ13 and PpHDZ16 might be involved in the synthesis of flavonoids. AtHDZ04 (GL1), AtHDZ05 (HDG11), and AtHDZ09 (HDG12) are associated with hairy root formation [37, 38], suggesting that subfamily IV might play an important role in trichomes formation. Through evolutionary analysis and tissue expression pattern analysis, the role of the peach HD-Zip gene family could be inferred. Although more experiments would be needed to verify, the study could provide a preliminary theoretical basis for understanding the function of the peach HD-Zip gene family.
The formation of peach adventitious roots is a complex process and a highly coordinated developmental process. Many studies have shown that HD-Zip gene is involved in root genesis and development [39, 40]. By detecting the expression levels of 23 PpHDZ genes during adventitious root formation, we found that the 23 PpHDZ genes were expressed to varying degrees during the adventitious root formation of peach cuttings, and most of the genes were highly expressed in a specific period of peach adventitious root formation (Fig. 8). For example, PpHDZ08, PpHDZ09, PpHDZ13, PpHDZ16 and PpHDZ18 were highly expressed in the early stage of adventitious root formation. PpHDZ02, PpHDZ03, PpHDZ04 and PpHDZ23 were highly expressed in the medium term. PpHDZ17 and PpHDZ20 were highly expressed in late stage (Fig. 8). These results indicated that HD-Zip gene played an important role in the morphogenesis and development of adventitious roots of peach cuttings. The expression profile of PpHDZ gene is helpful to further understand the functional characteristics of PpHDZ gene family in plant growth and development and adventitious root formation of cuttings.
Conclusion
In this study, a total of 23 PpHDZ genes were identified in peach by genome-wide analysis, distributed on 6 chromosomes. By phylogenetic analysis and conserved domain analysis, PpHDZ gene were divided into four subfamilies. A large number of cis-acting elements were found in promoter of PpHDZ gene, indicating that PpHDZ gene was controlled by a complex regulatory network. The tissue specific expression pattern of PpHDZ gene suggested that PpHDZ gene might play an important role in peach growth and development. PpHDZ gene were expressed variously during the adventitious root formation of peach cuttings, suggesting that PpHDZ would be involved in the formation of peach adventitious roots, and different genes play specific functions in different periods. These results lay a foundation for further exploring the function of PpHDZ gene in peach growth and development.
Materials and methods
Identification of putative PpHDZ in peach
The characteristic domain of HD-Zip transcription factor family protein (PF00046) was obtained from Pfam (Pfam: Home page (xfam.org)). HMMER (Biosequence analysis using profile hidden Markov Models | HMMER (ebi.ac.uk)) was used to construct HMM model to obtain peach HD-Zip transcription factor family candidate protein [41]. NCBI (National Center for Biotechnology Information (nih.gov)) was further used to detect whether the candidate protein contained HD-Zip domain and delete the sequence of the missing domain. Finally, 23 PpHDZ proteins were identified. The molecular weights and isoelectric points of PpHDZ family members were analyzed and identified using the online tool EMBOSS Programs (EMBOSS programs < EMBL-EBI). CELLO2GO (CELLO2GO:Subcellular Localization and Function Analysing System (nctu.edu.tw)) is used to analyze molecular work, biological processes and subcellular localization [42]. And use ChiPlot (bar plot with category (chiplot.online)) to draw GO analysis diagram.
Chromosomal localization analysis
The annotated files of peach genome (Prunus_persica_NCBIv2) and the length information of peach chromosome were obtained from NCBI (National Center for Biotechnology Information (nih.gov)). The chromosome location information of HD-Zip gene family members was extracted from the annotation file of peach genome, and the peach chromosome location map was drawn by TBtools software, and the HD-Zip family members were labeled on the chromosome [43].
Phyloevolutionary analysis and collinearity analysis
ClustaLW was used to analyze the amino acid sequences of Arabidopsis thaliana and peach HD-Zip [44]. The phylogenetic tree is constructed in MEGA11 by ML (maximum likelihood) method. MEGA11 also calculated the nucleotide differences of HD-Zip genes [45]. Genome annotation files of peach (Prunus_persica_NCBIv2) and Arabidopsis (TAIR10.1) were obtained from NCBI (National Center for Biotechnology Information (nih.gov)). Genome-wide collinearity of peach and Arabidopsis was analyzed by MCScanX [46], and genome-wide collinearity was mapped by TBtools.
Gene structure and conserved domain analysis
The exon and intron location information of PpHDZ gene was extracted from peach genome annotation file, and the extracted location information was converted into GSDS2.0 (Gene Structure Display Server 2.0 (gao-lab.org)) readable BED file. The PpHDZ gene structure was mapped by GSDS2.0 [47]. MEME5.4.1 (MEME - Submission form (meme-suite.org)) was used to predict and analyze the conserved protein motifs of PpHDZ protein sequences. The searched motif value was set to 15, and other parameters were set as default [48]. The structure diagram of the conserved protein motif was drawn by TBtools.
Subcellular localization analysis
One PpHDZ gene was randomly selected from each of three subfamilies. The nucleotide sequences of the three genes were obtained through the JGI website (Phytozome (doe.gov)) and amplified by P520 polymerase (Vazyme, Dalian, China). Three cloned PpHDZ genes coding sequence without stop codon were cloned into pCAMBIA1300 vector fused with GFP, all under CaMV 35 S promoter (35 S:PpHDZ2-GFP, 35 S:PpHDZ15-GFP and 35 S:PpHDZ16-GFP), respectively. These constructs were then transferred into Agrobacterium GV3101, and the transformed Agrobacterium cells were activated and infected into Nicotiana benthamiana leaves [49]. After 72 h, GFP was observed using a confocal laser microscope (Zeiss LSM880, Germany) and the nuclei were observed using DAPI-labeled nuclear markers.
Promoter element analysis
The promoter sequences of each PpHDZ gene, 2 kb sequence upstream of 5’UTR, were retrieved from the JGI website (Phytozome (doe.gov)). All the promoter sequences were submitted to the PlantCARE (PlantCARE, a database of plant promoters and their cis-acting regulatory elements (ugent.be)) database for the prediction of promoter elements, and then the distribution map and heat map of promoter elements were drawn using TBtools software [50].
Plant material acquisition, RNA extraction and qRT-PCR
The peach rootstock ‘GF677’ is planted in the experimental base of Shandong Agricultural University. The tissue samples of ‘GF677’ leaf, stem, flower and fruit were frozen in liquid nitrogen and stored at -80℃. The peach cutting were hardwood shoot of ‘GF677’. The work was carried out in a greenhouse of experimental base of Shandong Agricultural University, on November 12, 2021. The hardwood shoots were collected and cut in to pieces with 4–5 buds, the leaves of base were remove. The upper end of cuttings were cut flat, and the lower end of cutting were cut a 2 cm bevel with a scalpel. The cuttings were put into 1 mg/L IBA solution soaked for 2 min and then inserted into the sand pool. After completion, the sand was watered thoroughly. The sand kept moist by spraying during the whole expreiment. Samples were taken at 0 h, 1 h, 6 h, 2 d, 10 d and 17 d after cutting, and the samples were frozen with liquid nitrogen and stored at − 80 ℃.Total RNA was extracted from the samples using the RNA Plant Plus Reagent Kit (TIANGEN, China) and the first-strand cDNA was synthesized using the PrimeScript First-strand cDNA Synthesis Kit (Takara, Dalian, China). Real-time quantitative polymerase chain reaction (qRT-PCR) was performed in an ABI7500 system using SYBR premix ExTaq (Takara). qRT-PCR was calculated by 2−ΔΔCT. At least 3 replicates per sample were used for qPCR [51]. Primers used for qRT-PCR were listed in Table S1.
Availability of data and materials
All the data generated or analyzed during this study are included in this published article and its supplementary information files. The peach sequences in this article can be found from phytozome (Phytozome (doe.gov)). The Arabidopsis thaliana sequences in this article were downloaded from TAIR (TAIR - Home Page (arabidopsis.org)). All plant materials were selected from peach provided by the F. Peng lab, Shandong Agricultural University, Taian, China.
Change history
06 March 2023
The email addresses of the two corresponding authors were interchanged. This has been corrected.
References
Ruberti I, Sessa G, Luccetti S, Morelli G. A novel class of plant proteins containing a homeodomain with a closely linked leucine zipper motif. Embo J. 1991;10:1781–91.
Viola IL, Gonzalez DH. Structure and evolution of plant homeobox genes.Plant transcripton factor. 2016;Pages101–112.
Tron AE, Welchen E, Gonzalez DH. Engineering the loop region of a homeodomain-leucine zipper protein promotes efficient binding to a monomeric DNA binding site. Biochemistry. 2004;43(50):15845–51.
Ariel FD, Manavella PA, Dezar CA, Chan RL. The true story of the HD-Zip family. Trends Plant Sci. 2007;12(9):419–26.
Henriksson E, Olsson AS, Johannesson H, et al. Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol. 2005;139(1):509–18.
Zhao Y, Zhou Y, Jiang H, Li X, Gan D, Peng X, Zhu S, Cheng B. Systematic analysis of sequences and expression patterns of drought-responsive members of the HD-Zip gene family in maize. PLoS ONE. 2011;6(12):e28488.
Agalou A, Purwantomo S, Overnäs E, Johannesson H, Zhu X, Estiati A, de Kam RJ, Engström P, Slamet-Loedin IH, Zhu Z, Wang M, Xiong L, Meijer AH, Ouwerkerk PB. A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol Biol. 2008;66(1–2):87–103.
Hu R, Chi X, Chai G, Kong Y, He G, Wang X, Shi D, Zhang D, Zhou G. Genome-wide identification, evolutionary expansion, and expression profile of homeodomain-leucine zipper gene family in poplar (Populus trichocarpa). PLoS ONE. 2012;7(2):e31149.
Sharif R, Raza A, Chen P, et al. HD-ZIP Gene Family: potential roles in improving Plant Growth and regulating stress-responsive mechanisms in plants. Genes (Basel). 2021;12(8):1256.
Ré DA, Dezar CA, Chan RL, Baldwin IT, Bonaventure G. Nicotiana attenuata NaHD20 plays a role in leaf ABA accumulation during water stress, benzylacetone emission from flowers, and the timing of bolting and flower transitions. J Exp Bot. 2011;62(1):155–66.
Hymus GJ, Cai S, Kohl EA, Holtan HE, Marion CM, Tiwari S, Maszle DR, Lundgren MR, Hong MC, Channa N, Loida P, Thompson R, Taylor JP, Rice E, Repetti PP, Ratcliffe OJ, Reuber TL, Creelman RA. Application of HB17, an Arabidopsis class II homeodomain-leucine zipper transcription factor, to regulate chloroplast number and photosynthetic capacity. J Exp Bot. 2013;64(14):4479–90.
Zhu Y, Song D, Sun J, Wang X, Li L. PtrHB7, a class III HD-Zip gene, plays a critical role in regulation of vascular cambium differentiation in Populus. Mol Plant. 2013;6(4):1331–43.
Khaled AS, Vernoud V, Ingram GC, Perez P, Sarda X, Rogowsky PM. Engrailed-ZmOCL1 fusions cause a transient reduction of kernel size in maize. Plant Mol Biol. 2005;58(1):123–39.
Ariel F, Diet A, Verdenaud M, Gruber V, Frugier F, Chan R, Crespi M. Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1. Plant Cell. 2010;22(7):2171–83.
Sawa S, Ohgishi M, Goda H, Higuchi K, Shimada Y, Yoshida S, Koshiba T. The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J. 2002;32(6):1011–22.
Di Cristina M, Sessa G, Dolan L, Linstead P, Baima S, Ruberti I, Morelli G. The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 1996;10(3):393–402.
Singh A, Roy S, Singh S, Das SS, Gautam V, Yadav S, Kumar A, Singh A, Samantha S, Sarkar AK. Phytohormonal crosstalk modulates the expression of miR166/165s, target class III HD-ZIPs, and KANADI genes during root growth in Arabidopsis thaliana. Sci Rep. 2017;7(1):3408.
Chen D, Chen Z, Wu M, Wang Y, Wang Y, Yan H, Xiang Y. Genome-wide identification and expression analysis of the HD-Zip Gene Family in Moso Bamboo (Phyllostachys edulis). J Plant Growth Regul. 2017;36(2):323–37.
Li Y, Xiong H, Cuo D, Wu X, Duan R. Genome-wide characterization and expression profiling of the relation of the HD-Zip gene family to abiotic stress in barley (Hordeum vulgare L.). Plant Physiol Biochem. 2019;141:250–8.
Li Z, Gao Z, Li R, Xu Y, Kong Y, Zhou G, Meng C, Hu R. Genome-wide identification and expression profiling of HD-ZIP gene family in Medicago truncatula. Genomics. 2020;112(5):3624–35.
Li Z, Zhang C, Guo Y, Niu W, Wang Y, Xu Y. Evolution and expression analysis reveal the potential role of the HD-Zip gene family in regulation of embryo abortion in grapes (Vitis vinifera L.). BMC Genomics. 2017;18(1):744.
Izhaki A, Bowman JL. KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell. 2007;19(2):495–508.
Kim YS, Kim SG, Lee M, Lee I, Park HY, Seo PJ, Jung JH, Kwon EJ, Suh SW, Paek KH, Park CM. HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell. 2008;20(4):920–33.
Wang Z, Wang S, Xiao Y, Li Z, Wu M, Xie X, Li H, Mu W, Li F, Liu P, Wang R, Yang J. Functional characterization of a HD-ZIP IV transcription factor NtHDG2 in regulating flavonols biosynthesis in Nicotiana tabacum. Plant Physiol Biochem. 2020;146:259–68.
Ma YJ, Li PT, Sun LM, Zhou H, Zeng RF, Ai XY, Zhang JZ, Hu CG. HD-ZIP I transcription factor (PtHB13) negatively regulates Citrus flowering through binding to FLOWERING LOCUS C promoter. Plants (Basel). 2020;9(1):114.
Cai Y, Bartholomew ES, Dong M, Zhai X, Yin S, Zhang Y, Feng Z, Wu L, Liu W, Shan N, Zhang X, Ren H, Liu X. The HD-ZIP IV transcription factor GL2-LIKE regulates male flowering time and fertility in cucumber. J Exp Bot. 2020;71(18):5425–37.
Shao C, Huang Z, Bai X, Wang Y, Duan W, Identification. Systematic evolution and expression analysis of HD-Zip Gene Family in Capsicum annuum. Scientia Agricultura Sinica. 2020;53(05):1004–17.
Peng S, Zhao L, Wang T, Dong C, Zhu Y. Genome-wide Identification Bioinformatics and Expression Analysis of HD-Zip Gene Family in Carthamus tinctorius.Molecular Plant Breeding. 2022;1–23.
Faraji S, Heidari P, Amouei H, Filiz E, Abdullah, Poczai P. Investigation and Computational Analysis of the Sulfotransferase (SOT) Gene Family in Potato (Solanum tuberosum): Insights into Sulfur Adjustment for Proper Development and Stimuli Responses. Plants (Basel). 2021;10(12):2597.
Heidari P, Faraji S, Ahmadizadeh M, Ahmar S, Mora-Poblete F. New Insights Into Structure and Function of TIFY Genes in Zea mays and Solanum lycopersicum: A Genome-Wide Comprehensive Analysis. Front Genet. 2021;12:657970.
Capella M, Ribone PA, Arce AL, Chan RL. Arabidopsis thaliana HomeoBox 1 (AtHB1), a homedomain-leucine Zipper I (HD-Zip I) transcription factor, is regulated by PHYTOCHROME-INTERACTING FACTOR 1 to promote hypocotyl elongation. New Phytol. 2015;207(3):669–82.
Bou-Torrent J, Salla-Martret M, Brandt R, Musielak T, Palauqui JC, Martínez-García JF, Wenkel S. ATHB4 and HAT3, two class II HD-ZIP transcription factors, control leaf development in Arabidopsis. Plant Signal Behav. 2012;7(11):1382–7.
Carabelli M, Turchi L, Ruzza V, Morelli G, Ruberti I. Homeodomain-leucine Zipper II family of transcription factors to the limelight: central regulators of plant development. Plant Signal Behav. 2013;8(9):e25447.
Donner TJ, Sherr I, Scarpella E. Auxin signal transduction in Arabidopsis vein formation. Plant Signal Behav. 2010;5(1):70–2.
Elhiti M, Stasolla C. Structure and function of homodomain-leucine zipper (HD-Zip) proteins. Plant Signal Behav. 2009;4(2):86–8.
Li X, Li Y, Zhao M, Hu Y, Meng F, Song X, Tigabu M, Chiang VL, Sederoff R, Ma W, Zhao X. Molecular and metabolic insights into anthocyanin biosynthesis for Leaf Color Change in Chokecherry (Padus virginiana). Int J Mol Sci. 2021;22(19):10697.
Hülskamp M, Misŕa S, Jürgens G. Genetic dissection of trichome cell development in Arabidopsis. Cell. 1994;76(3):555–66.
Nakamura M, Katsumata H, Abe M, Yabe N, Komeda Y, Yamamoto KT, Takahashi T. Characterization of the class IV homeodomain-leucine Zipper gene family in Arabidopsis. Plant Physiol. 2006;141(4):1363–75.
Perotti MF, Arce AL, Chan RL. The underground life of homeodomain-leucine zipper transcription factors. J Exp Bot. 2021;72(11):4005–21.
Li X, Shen F, Xu X, Zheng Q, Wang Y, Wu T, Li W, Qiu C, Xu X, Han Z, Zhang X. An HD-ZIP transcription factor, MxHB13, integrates auxin-regulated and juvenility-determined control of adventitious rooting in Malus xiaojinensis. Plant J. 2021;107(6):1663–80.
El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. The pfam protein families database in 2019. Nucleic Acids Res. 2019;47(D1):D427–32.
Sabir IA, Manzoor MA, Shah IH, Liu X, Zahid MS, Jiu S, Wang J, Abdullah M, Zhang C. MYB transcription factor family in sweet cherry (Prunus avium L.): genome-wide investigation, evolution, structure, characterization and expression patterns. BMC Plant Biol. 2022;22(1):2.
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative Toolkit developed for interactive analyses of big Biological Data. Mol Plant. 2020;13(8):1194–202.
Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX.Curr Protoc Bioinformatics. 2002;Chap. 2:Unit 2.3.
Tamura K, Stecher G, Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol. 2021;38(7):3022–7.
Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49.
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–7.
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Clementi L, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202-8.
Sparkes IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc. 2006;1(4):2019–25.
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–7.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–8.
Acknowledgements
We would like to thank the Rural Revitalization Science and Technology Innovation Action Plan Project of Shandong (2021TZXD013), and China Agriculture Research System Fund (No. CARS-30-2-02) for their financial support.
Funding
This research was supported by Rural Revitalization Science and Technology Innovation Action Plan Project of Shandong (2021TZXD013), and China Agriculture Research System Fund (No. CARS-30-2-02).
Author information
Authors and Affiliations
Contributions
QJC designed the experiment. ZW, XLW, BBZ and JL performed experiments, material sampling, laboratory data measurements and analyzed the data, and ZW wrote the first draft of the manuscript. QJC, FTP, YSX and JG edited the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Study complied with local and national regulations for using plants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1
. The primers sequences of PpHDZ genes for qRT-PCR and gene cloning. Table S2. The Gene IDs of AtHDZ genes. Figure S1. The conserved protein motifs in the PpHDZ proteins. The x-axis indicates the conserved sequences of the domain. The height of each letter indicates the conservation of each residue across all proteins. The y-axis is a scale of the relative entropy, which reflects the conservation rateof each amino acid.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, Z., Wu, X., Zhang, B. et al. Genome-wide identification, bioinformatics and expression analysis of HD-Zip gene family in peach. BMC Plant Biol 23, 122 (2023). https://doi.org/10.1186/s12870-023-04061-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-023-04061-w