Abstract
Background
Methyl-CpG-binding domain (MBD) proteins play important roles in epigenetic gene regulation, and have diverse molecular, cellular, and biological functions in plants. MBD proteins have been functionally characterized in various plant species, including Arabidopsis, wheat, maize, and tomato. In rice, 17 sequences were bioinformatically predicted as putative MBD proteins. However, very little is known regarding the function of MBD proteins in rice.
Results
We explored the expression patterns of the rice OsMBD family genes and identified 13 OsMBDs with active expression in various rice tissues. We further characterized the function of a rice class I MBD protein OsMBD707, and demonstrated that OsMBD707 is constitutively expressed and localized in the nucleus. Transgenic rice overexpressing OsMBD707 displayed larger tiller angles and reduced photoperiod sensitivity—delayed flowering under short day (SD) and early flowering under long day (LD). RNA-seq analysis revealed that overexpression of OsMBD707 led to reduced photoperiod sensitivity in rice and to expression changes in flowering regulator genes in the Ehd1-Hd3a/RFT1 pathway.
Conclusion
The results of this study suggested that OsMBD707 plays important roles in rice growth and development, and should lead to further studies on the functions of OsMBD proteins in growth, development, or other molecular, cellular, and biological processes in rice.
Similar content being viewed by others
Background
DNA methylation, a conserved epigenetic modification in plant and animal genomes, plays an important role in genome stability, genomic imprinting, and gene expression regulation, and exerts effects on various aspects of plant and animal cellular and developmental processes [1, 2]. Methyl-CpG-binding domain (MBD) proteins emerge as important interpreters of DNA methylation marks [3]. MBD proteins can bind to methylated DNA and recruit chromatin remodeling factors, histone deacetylases and methylases to regulate gene expression [4,5,6]. Numerous studies in mammals revealed that mutations or abnormal expression of MBD proteins occur in many neurological diseases and cancers [7]. For example, mutations in the human methyl-CpG-binding protein 2 (MeCP2) cause a postnatal neurodevelopmental disorder Rett syndrome [8], demonstrating that MBD proteins play important roles in maintaining a normal epigenetic and cellular homeostasis.
Plant MBD proteins have been identified based on amino acid sequence similarity with mammalian MBDs [9, 10]. The Arabidopsis genome encodes 13 MBD domain-containing proteins [11]. Studies of the Arabidopsis MBDs showed that plant MBDs can bind to methylated or unmethylated DNA [10, 12], and interact with chromatin-modifying or transcriptional complexes, such as the histone deacetylase AtHDA6 [10], the histone methyltransferase AtPRMT11 [13], the histone acetyltransferase IDM1 and the alpha-crystallin domain protein ROS5/IDM2 in the DNA demethylation pathway [14,15,16], the RNA binding proteins AtRPS2C, AtAGO4 and AtNTF2 which are involved in the RNA-mediated gene silencing pathway [17], or the chromatin remodelers CHR11/CHR17 and chromatin remodeling complex SWR1 subunit PIE1 and ARP6 which are involved in histone H2A.Z deposition [18, 19]. The Arabidopsis MBDs play important roles in diverse molecular and biological processes. For example, knockdown of AtMBD6 or AtMBD10 leads to disturbed nucleolar dominance in Arabidopsis suecica [20]; mutation in AtMBD8 causes late flowering in the C24 ecotype [21]; mutation in AtMBD9 promotes early flowering and enhances shoot-branching in Arabidopsis [22, 23]; and knockdown of AtMBD11 results in Arabidopsis plants with morphological and developmental abnormalities [9].
In addition to the studies in the model plant Arabidopsis, several recent researches have shown the important roles of MBD proteins in different plant species. The wheat TaMBD2 homoeologous genes were significantly induced by salt stress, suggesting their potential functions in stress responses [24]. TaMBD6 was highly responsive to prolonged chilling, suggesting that TaMBD6 was potentially involved in regulating the developmental transition from vegetative to reproductive stages in wheat [25]. The maize ZmMBD101 was identified to be required for maintaining Mutator (Mu) elements chromatin in maize [26]. In tomato, SlMBD5 was identified to interact with the CUL4-DDB1-DET1 complex, which was involved in the transcriptional activation of downstream genes [27]. Overexpressing of the SlMBD5 gene leads to dark fruit color and dwarf phenotype in transgenic plants [27]. A number of MBD genes in four different solanaceae species have been observed to be differentially induced or suppressed during fruit development or abiotic stress responses, suggesting their roles involved in these processes [28].
Rice (Oryza sativa L.) is one of the most important crops worldwide. In the rice genome, 17 genes were bioinformatically predicted to encode putative MBD proteins and were designated as OsMBD701 to OsMBD718 [11]. However, very little is known regarding the functions of OsMBD proteins in rice. In this study, we explored the expression patterns of the predicted OsMBD family genes and detected 13 OsMBDs with active expression in various rice tissues. We further performed functional study of a rice class I MBD protein, OsMBD707, and revealed that overexpression of OsMBD707 resulted in larger tiller angles and reduced photoperiod sensitivity in rice. Our results suggested that OsMBD707 plays biologicalroles in rice growth and development.
Results
Differential expression patterns of 13 OsMBD family genes in rice tissues
A bioinformatics analysis has identified 17 putative OsMBD proteins in rice [11]. Considering the annotation updates of the rice genome over the past decades, in this study, we verified the predicted OsMBD family genes by searching the National Center for Biotechnology Information (NCBI) and MSU Rice Genome Annotation Project (RGAP) databases. Our search revealed 15 genes in both NCBI and RGAP databases matching with previously predicted OsMBD701, OsMBD703, OsMBD704, OsMBD705, OsMBD706, OsMBD707, OsMBD708, OsMBD709, OsMBD710, OsMBD711, OsMBD713, OsMBD714, OsMBD715, OsMBD717, and OsMBD718, respectively (Additional file 1: Table S1). However, no NCBI RAP locus or MSU RGAP locus has been identified that match with OsMBD712 or OsMBD716 (Additional file 1: Table S1). In addition, a RAP locus Os04g0192775 and a MSU RGAP locus LOC_Os04g11510 were retrieved as genes putatively encoding MBD-containing proteins (Additional file 1: Table S1).
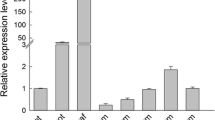
To understand the expression patterns of the OsMBD family genes, we first searched the digital expression values of OsMBDs in the RGAP database (http://rice.plantbiology.msu.edu/). Among the 17 putative OsMBD-encoding genes, OsMBD701, OsMBD704, OsMBD705, OsMBD706, OsMBD707, OsMBD708, OsMBD709, OsMBD710, OsMBD711, OsMBD715, OsMBD717, and OsMBD718 showed differential expression in various tissues. However, there was no detectable expression value of Os04g0192775 and LOC_Os04g11510 in all 13 different tissues, and the predicted OsMBD703, OsMBD713, and MBD714 showed very low FPKM (expected Fragments Per Kilobase of transcript per Million fragments sequenced) values only in callus, seed-5 days after pollination (DAP), or anther, respectively, but no detectable FPKM value in other tissues (Additional file 2: Table S2). We further performed quantitative RT-PCR (qRT-PCR) to validate the expression patterns of the OsMBD family genes in the roots, stems, leaves, spikelets, seeds, and panicle axes of rice plants. qRT-PCR results were nearly consistent with the digital expression database. Among the 17 putative OsMBDs, no transcripts of the predicted OsMBD703, OsMBD713, Os04g0192775, or LOC_Os04g11510 were detected in tested tissues (Additional file 1: Table S1). While OsMBD704 and OsMBD714 were detected to be preferentially expressed in the seeds, OsMBD701, OsMBD705, OsMBD706, OsMBD707, OsMBD708, OsMBD709, OsMBD710, OsMBD711, OsMBD715, OsMBD717, and OsMBD718 were observed to be differentially expressed in various tissues (Fig. 1).
Expression profiles of the OsMBD family genes in various tissues of rice. a. OsMBD701. b. OsMBD704. c. OsMBD705. d. OsMBD706. e. OsMBD707. f. OsMBD708. g. OsMBD709. h. OsMBD710. i. OsMBD711. j. OsMBD714. k. OsMBD715. l. OsMBD717. m. OsMBD718. Relative expression levels of OsMBDs in root, stem, leaf, spikelet, seed, and panicle axis were determined by qRT-PCR. The rice Actin gene was used as an internal control
OsMBD707 is constitutively expressed and localized in the nucleus
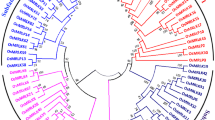
Rice OsMBDs could be divided into six classes [11]. In the present study, we focused on functional analysis of OsMBD707, which belongs to class I and is the only member of class I [11]. Phylogenetic analysis of OsMBD707 and closely related MBDs in various plant species showed that OsMBD707 was clustered together with ObMBD11-like in Oryza brachyantha, hypothetical protein TVU48968.1 in Eragrostis curvula, SiMBD10 and SiMBD11 in Setaria italica, ZmMBD105 and ZmMBD106 in Zea mays, BdMBD11 in Brachypodium distachyon, and SbMBD10 and SbxP2 in Sorghum bicolor (Fig. 2). qRT-PCR analysis showed that the mRNA of OsMBD707 was expressed in various tissues, although the transcript level in the spikelets was relatively low (Fig. 1). A 1931-bp promoter fragment upstream of the translational start of OsMBD707 was cloned and fused with the GUS reporter gene. Histochemical staining of rice plants transformed with the OsMBD707 promoter-GUS fusion construct showed that GUS was expressed throughout the tested tissues, including the roots, stems, leaves, and spikelets, although the expression level was weaker than that of 35S promoter-GUS transgenic plants (Fig. 3a). Overall, these results indicated that OsMBD707 is constitutively expressed in various rice tissues.
Phylogenetic analysis of OsMBD707. Phylogenetic tree was constructed using the neighbor-joining algorithm with 1000 bootstrap replicates. OsMBD707 is indicated by a black triangle. Other OsMBDs are indicated by empty triangles. Only the 13 OsMBDs with active expression detected by qRT-PCR were included in the phylogenetic analysis
Expression pattern of OsMBD707 and subcellular localization of OsMBD707. a. Histochemical GUS staining of the roots, stems, leaves, and spikelets of rice plant transformed with the OsMBD707 promoter-GUS fusion construct. NPB, nontransgenic wild-type Nipponbare; p35S-GUS, rice plant transformed with a CaMV 35S promoter-GUS construct; pMBD707-GUS, riceplant transformed with the OsMBD707 promoter-GUS construct. Scale bar, 2 mm. b. Subcellular localization of the GFP-OsMBD707 fusion in rice protoplasts. 35S-GFP + DsRed-OsH2B, rice protoplast co-transfected with a GFP alone construct and a nucleus marker construct DsRed-OsH2B; GFP-OsMBD707 + DsRed-OsH2B, rice protoplast co-transfected with the GFP-OsMBD707 fusion construct and DsRed-OsH2B; Bright, imagestaken under bright field; GFP, images taken under green fluorescence; RFP, images taken under red fluorescence; Merged, images merged from bright filed and green fluorescence channels. Scale bar, 10 μm
OsMBD707 was predicted to generate two alternative transcripts, XM_015764399.1/LOC_Os12g42550.1, and XM_015764400.2/LOC_Os12g42550.2 (Additional file 3: Figure S1A, B). We performed RT-PCR to clone the cDNA fragment of OsMBD707, but obtained only XM_015764399.1/LOC_Os12g42550.1. An RT-PCR using primers (Additional file 3: Figure S1A) designed to distinguish the two predicted alternative transcripts was further performed. Consistently, only XM_015764399.1/LOC_Os12g42550.1 was detected in all tested tissues, including the roots, stems, leaves, spikelets, seeds, and panicle axes (Additional file 3: Figure S1C, D), suggesting that there is only one isoform, LOC_Os12g42550.1, of OsMBD707. To explore the subcellular localization of OsMBD707, we performed transient expression of a GFP-OsMBD707 fusion construct in rice protoplasts. Microscopy revealed that the fluorescence signal of GFP-OsMBD707 was inside the nucleus region, which was merged with the signal of the nucleus DsRed-OsH2B marker [29] (Fig. 3b). This result demonstrated that OsMBD707 is a nuclear-localized protein which is consistent with a function as a methyl-CpG-binding protein.
Overexpression of OsMBD707 causes larger tiller angles and reduced photoperiod sensitivity in rice
To explore the function of OsMBD707 in rice, we generated overexpression and RNAi knockdown transgenic plants. For each type, more than 40 independent transgenic T0 plants were generated, and five independent plants were chosen for initial analysis. qRT-PCR analysis showed that the transcription levels of OsMBD707 were significantly higher in plants transformed with OsMBD707-overexpression construct (about 12- to 43- fold) and lower in plants transformed with OsMBD707-RNAi construct (about 11 to 27%), as compared to wild-type plants (Fig. 4a, b), confirming the overexpression and knock-down of OsMBD707, respectively, in the transgenic plants. In addition, we generated CRISPR/Cas9 knockout plants of OsMBD707. Genotyping of the CRISPR/Cas9 transgenic plants identified nine independent T0 plants with homozygous mutations in at least one of the single guide RNA (sgRNA) targeting sites of OsMBD707,among which seven carried homozygous frameshift mutations: mbd707-#6, mbd707-#9, mbd707-#12, mbd707-#15, mbd707-#22, mbd707-#25, and mbd707-#28 (Fig. 4c). Initial phenotypic observation showed that the OsMBD707-overexpression plants (referred to as OX707) displayed a larger tiller angle after tillering stage compared to wild-type. In contrast, no obvious morphological differences were observed among wild-type, the OsMBD707-RNAi plants (referred to as 707i) and the CRISPR/Cas9 edited frameshift plants.
Molecular characterization of OsMBD707-overexpression, −knockdown and -frameshiftplants. a, b. qRT-PCR analysis of the transcript levels of OsMBD707 in rice plants transformed with OsMBD707-overexpression construct (OX707-#8, OX707-#14, OX707-#20, OX707-#21, OX707-#40) and OsMBD707-RNAi construct (707i-#11, 707i-#22, 707i-#26, 707i-#30, 707i-#39), respectively. NPB, nontransgenic wild-type Nipponbare. The rice Actingene was used as an internal control. c. Schematic illustration of the OsMBD707 gene structure and mutations of OsMBD707 in CRISPR/Cas9-mediatedmutated plants. Black rectangles represent the 2 exons of OsMBD707; Red characters indicate the sequences of the target sites; PAM sequences are underlined; Deletions are indicated by dashes; Numbers on the right side indicate the numbers of deletion nucleotides; WT indicates wild-type sequence
OsMBD707-overexpression, -knockdown, and -frameshiftlines were generated up to T3 to T4 generations, and two independent overexpression lines, one knockdown line, and one frameshift mutant line were chosen for further analysis. Consistent with initial phenotypic observation, the two OsMBD707-overexpression lines OX707-#20 and OX707-#21 displayed larger tiller angles, compared to wild-type, the knockdown line 707i-#30, and the frameshift line mbd707-#6 (Fig. 5a). In addition, we observed significant delays in flowering of the two overexpression lines grown under short day (SD) condition (Fig. 5a). We further investigated the heading dates of the OsMBD707-overexpression, −knockdown, and -frameshift lines in growth chambers under SD and long day (LD) conditions. As showed in Fig. 5b, under SD, the flowering times of OX707-#20 and OX707-#21 were significantly delayed (about 15–17 days) compared with that of wild-type, 707i-#30 or mbd707-#6(Fig. 5b). In contrast, under LD, the flowering times of OX707-#20 and OX707-#21 were significantly earlier (about 10.5–12.5 days) compared with that of wild-type, 707i-#30 or mbd707-#6 (Fig. 5c), indicating that overexpression of OsMBD707 caused reduced photoperiod sensitivity in transgenic rice plants.
Overexpression of OsMBD707 causes larger tiller angles and reduced photoperiod sensitivity. a. Morphological phenotypes of OsMBD707-overexpression (OX707-#20, OX707-#21), -knockdown (707i-#30) and -frameshift (mbd707-#6) plants. NPB, nontransgenic wild-type Nipponbare. b. Flowering times of OsMBD707-overexpression (OX707-#20, OX707-#21), -knockdown (707i-#30) and -frameshift (mbd707-#6) plants under short day (9.5-h light, 28 °C/14.5-h dark, 26 °C). c. Flowering times of OsMBD707-overexpression (OX707-#20, OX707-#21), -knockdown (707i-#30) and -frameshift (mbd707-#6) plants under long day (14.5-h light, 28 °C/9.5-h dark, 26 °C). The letters A and B indicate significant differences according to LSD multiple range test at P ≤ 0.01
Global transcriptome analysis reveals transcriptional changes in key flowering regulator genes induced by overexpression of MBD707
RNA-seq-based transcriptome analysis was performed to investigate global transcript changes in the OsMBD707-overexpression transgenic line OX707-#21under both SD and LD conditions. Under SD, about 1026 genes were identified that were differentially expressed between OX707-#21 and wild-type, and of these, 616 genes were up-regulated, whereas 410 genes were down-regulated in OX707-#21 (Additional file 4: Figure S2A, Additional file 5: Table S3). Under LD, about 1653 differentially expressed genes (DEGs) were identified between OX707-#21 and wild-type, including 997 up-regulated genes and 656 down-regulated genes in OX707-#21 (Additional file 4: Figure S2A, Additional file 6: Table S4). In total, about 2353 DEGs were identified under SD and/or LD (Additional file 4: Figure S2B).
Gene Ontology (GO) analysis of these 2353 DEGs showed that the significant biological process categories were associated with translation, peptide biosynthetic, peptide metabolic, amide biosynthetic, and cellular amide metabolic processes; the significant cellular component categories were ribosome, ribonucleoprotein complex, non-membrane-bounded organelle, intracellular non-membrane-bounded organelle, and cytoplasmic part; and the significant molecular function categories were involved in structural constituent of ribosome, structural molecule activity, serine-type endopeptidase inhibitor activity, endopeptidase inhibitor activity, peptidase inhibitor activity, peptidase regulator activity, endopeptidase regulator activity, transferase activity/transferring glycosyl groups, N-acetyltransferase activity, N-acyltransferase activity (Additional file 7: Figure S3,Additional file 8: Table S5). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that the identified DEGs were significantly enriched in ribosome and phenylpropanoid biosynthesis pathways (Additional file 9: Figure S4, Additional file 10: Table S6).
We further surveyed the DEGs with known or putative functions involved in tiller angle or flowering time regulation. The phytochrome-interacting factor-like protein gene OsPIL15 (Os01g0286100) that negatively regulates tiller angle [30] was significantly down-regulated in OX707-#21 under both SD and LD (Additional file 5: Table S3, Additional file 6: Table S4). However, no other known tiller angle regulator genes were identified as DEGs. Notably, a number of genes with functions in controlling flowering time were identified among the DEGs, including FLAVIN-BINDING, KELCH REPEAT, F-Box 1(OsFKF1) [31], Early heading date1 (Ehd1) [32, 33], Days to heading on chromosome 2 (DTH2) [34], Heading date3a (Hd3a) and RICE FLOWERING LOCUS T1 (RFT1) [35, 36], OsMADS14 and OsMADS15 [37], and Flowering Locus T gene homologs FT-L7, FT-L8 and FT-L12 that promote flowering, and Grain number, plant height, and heading date2 (GHd2) [38] that inhibits flowering. Under SD, Hd3a, RFT1, FT-L7, OsMADS14, and OsMADS15 were down-regulated in OX707-#21. In contrast, OsFKF1, Ehd1, Hd3a, RFT1, FT-L8, FT-L12, and OsMADS14 were up-regulated, whereas Ghd2 was down-regulated in OX707-#21 under LD (Fig. 6a, b). The transcriptional changes of these flowering regulator genes in OX707-#21 were consistent with the delayed flowering and early flowering phenotypes of the MBD707-overexpression line under SD and LD, respectively, except that the minor-effect heading promoting gene DTH2 was paradoxically down-regulated under LD (Fig. 6a, b). The transcriptional profiles of five key flowering regulator genes, Ehd1, Hd3a, RFT1, OsMADS14, and OsMADS15 were verified by qRT-PCR, and the results were consistent with the RNA-seq data (Fig. 6c). The frameshift mutant line mbd707-#6 was also included for qRT-PCR, and the expression patterns of the five genes in mbd707-#6 were similar to that in wild-type plant (Fig. 6c).
Transcriptional changes of key flowering regulator genes in the Ehd1-Hd3a/RFT1 pathway induced by overexpression of MBD707. a. Heatmap showing the differential expression levels of flowering genes in the OsMBD707-overexpression line (OX707-#21) and wild-type (NPB) revealed by RNA-seq. Grey blocks indicate that the genes were not detected as differentially expressed genes (DEGs) by RNA-seq. b. Schematic model showing the differentially expressed flowering genes involved in the Ehd1-Hd3a/RFT1 regulatory network. Red and green backgrounds indicate that the genes were down-regulated and up-regulated, respectively, in OX707-#21. Yellow background represents that the down-regulation of DTH2 was paradoxical to the early flowering phenotype of OX707-#21. Grey backgrounds represent that the genes were not detected as DEGs by RNA-seq. Black arrows indicate a promoting effect, bars indicate a repressive effect, and dotted lines indicate an unknown pathway. c. qRT-PCR validation of five key flowering regulator genes Ehd1, Hd3a, RFT1, OsMADS14, and OsMADS15 that were identified as DEGs between OX707-#21 and NPB by RNA-seq. The frameshift mutant line mbd707-#6 was also included in the experiment. The rice Actin gene was used as an internal control.
Discussion
MBD family proteins have been functionally characterized in various plant species, including Arabidopsis [9,10,11,12,13,14,15,16,17,18,19,20,21,22,23], wheat [24, 25], maize [26], and tomato [27, 28]. MBD proteins play pivotal roles in plant growth, development, and stress responses. In the present study, we characterized OsMBD707 in rice and demonstrated its roles in regulating tiller architecture and flowering time.
Seventeen sequences were predicted as putative MBD proteins from the rice genome (Additional file 1: Table S1). In the present study, 13 predicted OsMBD genes were detected to be actively expressed in the roots, stems, leaves, spikeletes, seeds, or panicle axes (Fig. 1). However, no transcripts were detected in any tested tissues for the rest four predicted sequences (Additional file 1: Table S1). The digital expression values of these four sequences were undetectable or almost undetectable in the RGAP database (Additional file 2: Table S2). Whether these four sequences are pseudogenes, or their expression is restricted to other specific tissues or developmental stages, remains to be further elucidated. The plant MBDs were grouped into eight classes, but among them, class IV and class VI were present only in dicots [11]. OsMBD707 belongs to the class I MBD proteins [11]. Previous studies have demonstrated that the Arabidopsis class I MBD proteins AtMBD10 and AtMBD11 had important functions in regulating nucleolar dominance [20], and morphological development [9]. RNAi-mediated knockdown of AtMBD10 revealed that AtMBD10 was required for rRNA gene silencing in nucleolar dominance [20], and knockdown of AtMBD11 led to aerial rosettes, serrated leaves, abnormal flower position, late flowering, and reduced fertility in Arabidopsis [9]. In the present study, we observed that overexpression of OsMBD707 caused larger tiller angles and reduced photoperiod sensitivity in rice. On the contrary, both knockdown and frameshift mutation of OsMBD707 did not result in observable changes in the morphology of rice plants. One possible reason might be the functional redundancy between OsMBD707 and other OsMBDs, although they could be bioinformatically grouped into different classes.
Both tiller architecture and flowering time are important traits for rice cultivation. Over the past decades, several key/major regulators of the tiller angle have been identified, such as LAZY1 [39], Tiller Angle Control 1 (TAC1) [40], PROSTRATE GROWTH 1 (PROG1) [41, 42], Loose Plant Architecture 1 (LPA1) [43], PLANT ARCHITECTURE AND YIELD 1 (PAY1) [44], TAC3 and D2 [45], HEAT STRESS TRANSCRIPTION FACTOR 2D (HSFA2D) [46], TILLER INCLINED GROWTH 1 (TIG1) [47], Tiller Angle Control 4 (TAC4) [48], etc. However, RNA-seq analysis did not identify any of these tiller angle regulating genes as DEGs between the OsMBD707-overexpression line OX707-#21 and wild-type, except that an OsPIL15 gene negatively regulating tiller angle [30] was found to be down-regulated in OX707-#21. Since RNA-seq was conducted using RNAs extracted from fully expanded leaves, one reason for rare detection of the key/major tiller angle regulating genes among the DEGs could be that thesegenes were expressed abundantly in tissues involved in tillering (e.g., stems, tiller base, and tiller node), but not expressed or expressed at very low levels in mature leaves [39, 40]. To test this, we performed qRT-PCR to investigate the expression patterns of these known tiller angle regulator genes in mature leaves of OX707-#21 and wild-type. While HSFA2D was detected to be expressed highly in mature leaves, no transcripts of LAZY1, PROG1, LPA1 and TIG1, and very low expression of TAC1, PAY1, TAC3, D2, and TAC4 were detected (Additional file 11: Figure S5). Overall, the expression patterns in mature leaves of these genes, except OsPIL15 (Additional file 11: Figure S5B), were not correlated with the larger tiller angle phenotype in OsMBD707-overexpression plant. Nevertheless, the mechanism by which OsMBD707 regulates tiller angle remains to be further explored.
Rice is an SD plant whose flowering is promoted under SD, but postponed under LD. Rice flowering time is largely determined by photoperiod sensitivity, which is controlled by an intricate genetic network of an antagonistic regulation of flowering promotion under SD, and repression under LD [49]. In the present study, a number of flowering time genes were identified as DEGs between OX707-#21 and wild-type (Fig. 6a, b). Among these differentially expressed flowering time genes, Hd3a, RFT1, and Ehd1 are primary integrators of photoperiodic signals. Hd3a and RFT1 are two florigens of rice and Ehd1 promotes the expression of Hd3a and RFT1 to induce heading [32, 35, 36]. Via RNA-seq, Hd3a and RFT1 were found to be down-regulated in OX707-#21 under SD, but up-regulated under LD (Fig. 6). Ehd1 was identified to be up-regulated in OX707-#21 under LD (Fig. 6a), and although RNA-seq did not detect Ehd1 as a DEG under SD, qRT-PCR verified that Ehd1 was down-regulated in OX707-#21 under SD (Fig. 6b). Besides, RNA-seq revealed that three flowering-promoting genes/homologs involved in the Ehd1-Hd3a/RFT1 pathway, OsMADS14, OsMADS15, and FTL7 were down-regulated in OX707-#21 under SD; and four genes/homologs OsFKF1, OsMADS14, FTL8, and FTL12 were up-regulated in OX707-#21 under LD (Fig. 6a, b). Although a DTH2 gene having a function in promoting heading by inducing Hd3a and RFT1 was paradoxically down-regulated in OX707-#21 under LD (Fig. 6a, b), its effect did not seem to change the up-regulation of Hd3a and RFT1. Overall, RNA-seq results indicated that overexpression of OsMBD707 led to transcriptional changes in key flowering regulator genes in the Ehd1-Hd3a/RFT1 pathway. In the present study, we observed that OsMBD707 itself is expressed at similar levels under SD or LD (Additional file 11: Figure S5C). Our results suggested that overexpression of OsMBD707 may cause certain epigenetic changes related to the expression of these key flowering regulator genes. However, we have not explored the detailed connections between OsMBD707 and the flowering regulator genes, the mechanism underlying the role of OsMBD707 in regulating photoperiod sensitivity remains to be further investigated.
Conclusion
In this study, the bioinformatically predicted OsMBD family genes were verified and 13 OsMBDs were identified to be actively expressed in various rice tissues. We further performed functional study of OsMBD707, and demonstrated that OsMBD707 is constitutively expressed and localized in the nucleus. Overexpression of OsMBD707 causes larger tiller angles, delayed flowering under SD and early flowering under LD in rice. RNA-seq analysis revealed that overexpression of OsMBD707 led to reduced photoperiod sensitivity in rice by down-regulating flowering-promoting genes under SD and up-regulating flowering-promoting genes under LD. Our results suggested the biological roles of OsMBD707 in rice growth and development, and lay the foundation for future studies on the function of OsMBD proteinsin molecular, cellular, and biological processes in rice.
Methods
Plant materials and growth conditions
Oryza sativa L. ssp. japonica (cv. Nipponbare) (maintained in Biotechnology Research Institute, Fujian Academy of Agricultural Sciences) was used in this study. Rice plants were cultured in a greenhouse under partially regulated conditions of 26–32 °C with a 14-h light/10-h dark cycle. For flowering time investigation, rice plants were grown in environmentally controlled growth chambers under SD (9.5-h light, 28 °C/14.5-h dark, 26 °C) and LD (14.5-h light, 28 °C/9.5-h dark, 26 °C) conditions, respectively.
Gene expression analysis by qRT-PCR
Total RNAs were extracted from rice tissues using TRIzol reagent (Invitrogen, USA). The RNA samples were treated with DNase I (Takara, Dalian, China). The first-strand complementary DNA was generated from 0.5 μg RNA using a ReverTra Ace qPCR RT Kit (TOYOBO, Japan). qRT-PCR was performed on an ABI QuantStudio 6 Flex System (Applied Biosystems, USA) using a FastStart Universal SYBR green Master (ROX) (Roche, Germany). Three replications were conducted for each sample. The primers used for analysis are listed in Additional file 12: Table S7.
Phylogenetic analysis
The full-length amino acid of OsMBD707 was used as a query to search against the PLAZA database (https://bioinformatics.psb.ugent.be/plaza/). The retrieved sequences were verified against the NCBI non-redundant (NR) protein database (http://blast.ncbi.nlm.nih.gov/). The homologs were aligned using Clustal X program [50], and the phylogenetic tree was constructed using the neighbor-joining algorithm with 1000 bootstrap replicates in MEGA X [51].
Plasmids construction
A 1935-bp DNA sequence upstream of the ATG start codon of OsMBD707 was amplified by PCR from Nipponbare genomic DNA. The amplified fragment was cloned into pCXGUS-P [52], resulting in a binary vector containing a fusion of the OsMBD707 promoter and a GUS reporter gene. The open reading frame (ORF) of OsMBD707 was amplified by using specific primers and was cloned into pCXUN [52], resulting in a binary vector pCXUN-OsMBD707 in which OsMBD707 was driven by the maize ubiquitin-1 promoter. A 213-bp DNA fragment of OsMBD707 and a 388-bp stuffer DNA fragment from the GUS gene were amplified by using specific primers. Overlapping PCR was performed using the two amplified fragments as templates. The resultant hairpin RNAi fragment was cloned into pCXUN [52], resulting in a binary RNAi vector pCXUN-OsMBD707-RNAi. Two sgRNA sequences targeting the coding region of OsMBD707 were designed according to Shan’s program [53]. Construction of the CRISPR/Cas9 vector was carried out as previously described [54]. Tow DNA oligos corresponding to the designed sgRNAs were synthesized and the dimer was cloned into pYLgRNA-OsU6a and pYLgRNA-OsU6b, respectively. The resultant sgRNA expression cassettes were thus cloned into pYLCRISPR/Cas9Pubi-H [54]. To generate subcellular localization construct for OsMBD707, the ORF of a GFP gene was amplified and cloned into pCXSN [52], resulting in a binary vector pCS-NGFP. The OsMBD707 ORF digested from pCXUN-OsMBD707 by BamHI was then cloned into the BamHI-digested pCS-NGFP to fused in-frame with the GFP gene. All primers or oligos used for plasmid construction are listed in Additional file 12: Table S7.
Rice protoplast transfection and stable transformation
Rice protoplasts were prepared from the sheath and stem tissues of 2-week-old etiolated seedlings. The GFP alone and GFP-OsMBD707 fusion constructs were, respectively, co-transfected with a nucleus marker construct DsRed-OsH2B [29] into rice protoplasts via a PEG-mediated method as previously described [55]. Rice stable transformation was conducted as previously described [56]. The binary vectors were introduced into Agrobacterium tumefaciens EHA105, and the transformant strains were used to transform rice calli of cv. Nipponbare. Homozygous transgenic plants were screened in T1 generation derived from self-pollination of T0 plants, and were maintained up to T3 to T4 generations.
GUS staining and GFP detection
GUS histochemical staining was performed following the procedure described by [57]. Rice tissues were immersed in X-Gluc (Thermo Fisher Scientific, USA) staining solution at 37 °C overnight and were subsequently rinsed in 70% ethanol at room temperature for 1 or 3 days. Pictures were taken with an Olympus SZX12 stereo microscope. The transfected rice protoplasts were incubated at room temperature for 16–20 h. Fluorescence microscopy was carried out on a Leica DMi8 Laser Scanning Confocal microscope (Leica, Germany) with Excitation/emission wavelengths 488/535 nm for green fluorescence.
RNA-seq analysis
Fully expanded leaves of the overexpression line OX707-#21 and wild-type were collected at about 50 days after sowing. Total RNAs extracted from leaf tissues were subjected to RNA-seq analysis at Novogene (Beijing, China). Three to four biological replicates of each sample were used for RNA-seq analysis. Briefly, sequencing libraries were generated using NEB Next UltraTM RNA Library Prep Kit for Illumina (NEB, USA) following manufacturer’s instructions. The libraries were sequenced on an Illumina Novaseq platform. Raw reads were processed through in-house perl scripts, and the filtered clean reads were mapped to the reference genome using Hisat2 v2.0.5. FPKM value of each gene was calculated for estimating gene expression levels [58]. All gene count and FPKM values are available in Additional file 13: Table S8. Gene expression difference between OX707-#21 and wild-type was performed using the DESeq2 [59]. Genes with padj< 0.05 and log2FoldChange > 1 were assigned as differentially expressed. GO and KEGG enrichment analysis of DEGs were implemented by the cluster Profiler [60].
Availability of data and materials
Raw data was deposited in NCBI database under SRA accession: SRP303927 (https://www.ncbi.nlm.nih.gov/sra/SRP303927).
Abbreviations
- MBD:
-
Methyl-CpG-binding domain
- NCBI:
-
National Center for Biotechnology Information
- RGAP:
-
Rice Genome Annotation Project
- FPKM:
-
Expected Fragments PerKilobase of transcript per Million fragments sequenced
- DAP:
-
Days after pollination
- qRT-PCR:
-
Quantitative RT-PCR
- SD:
-
Short day
- LD:
-
Long day
- DEGs:
-
Differentially expressed genes
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- ORF:
-
Open reading frame
- sgRNA:
-
Single guide RNA
References
He X-J, Chen T, Zhu J-K. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011;21:442–65.
Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–6.
Zemach A, Grafi G. Methyl-CpG-binding domain proteins in plants: interpreters of DNA methylation. Trends Plant Sci. 2007;12:80–5.
Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–91.
Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61.
Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–9.
Du Q, Luu PL, Stirzaker C, Clark SJ. Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics. 2015;7:1051–73.
Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–8.
Berg A, Meza TJ, Mahic M, Thorstensen T, Kristiansen K, Aalen RB. Ten members of the Arabidopsis gene family encoding methyl-CpG-binding domain proteins are transcriptionally active and at least one, AtMBD11, is crucial for normal development. Nucleic Acids Res. 2003;31:5291–304.
Zemach A, Grafi G. Characterization of Arabidopsis thaliana methyl-CpG-binding domain (MBD) proteins. Plant J. 2003;34:565–72.
Springer NM, Kaeppler SM. Evolutionary divergence of monocot and dicot methyl-CpG-binding domain proteins. Plant Physiol. 2005;138:92–104.
Ito M, Koike A, Koizumi N, Sano H. Methylated DNA-binding proteins from Arabidopsis. Plant Physiol. 2003;133:1747–54.
Scebba F, De Bastiani M, Bernacchia G, Andreucci A, Galli A, Pitto L. PRMT11: a new Arabidopsis MBD7 protein partner with arginine methyltransferase activity. Plant J. 2007;52:210–22.
Lang Z, Lei M, Wang X, Tang K, Miki D, Zhang H, Mangrauthia SK, Liu W, Nie W, Ma G, Yan J, Duan CG, Hsu CC, Wang C, Tao WA, Gong Z, Zhu JK. The methyl-CpG-binding protein MBD7 facilitates active DNA demethylation to limit DNA hyper-methylation and transcriptional gene silencing. Mol Cell. 2015;57:971–83.
Li Q, Wang X, Sun H, Zeng J, Cao Z, Li Y, Qian W. Regulation of active DNA demethylation by a methyl-CpG-binding domain protein in Arabidopsis thaliana. PLoS Genet. 2015;11:e1005210.
Wang C, Dong X, Jin D, Zhao Y, Xie S, Li X, He X, Lang Z, Lai J, Zhu JK, Gong Z. Methyl-CpG-binding domain protein MBD7 is required for active DNA demethylation in Arabidopsis. Plant Physiol. 2015;167:905–14.
Parida AP, Sharma A, Sharma AK. AtMBD6, a methyl CpG binding domain protein, maintains gene silencing in Arabidopsis by interacting with RNA binding proteins. J Biosci. 2017;42:57–68.
Potok ME, Wang Y, Xu L, Zhong Z, Liu W, Feng S, Naranbaatar B, Rayatpisheh S, Wang Z, Wohlschlegel JA, Ausin I, Jacobsen SE. Arabidopsis SWR1-associated protein methyl-CpG-binding domain 9 is required for histone H2A.Z deposition. Nat Commun. 2019;10:3352.
Luo YX, Hou XM, Zhang CJ, Tan LM, Shao CR, Lin RN, Su YN, Cai XW, Li L, Chen S, He XJ. A plant-specific SWR1 chromatin-remodeling complex couples histone H2A.Z deposition with nucleosome sliding. EMBO J. 2020;39:e102008.
Stangeland B, Rosenhave EM, Winge P, Berg A, Amundsen SS, Karabeg M, Mandal A, Bones AM, Grini PE, Aalen RB. AtMBD8 is involved in control of flowering time in the C24 ecotype of Arabidopsis thaliana. Physiol Plant. 2009;136:110–26.
Peng M, Cui Y, Bi Y-M, Rothstein SJ. AtMBD9: a protein with a methyl-CpG-binding domain regulates flowering time and shoot branching in Arabidopsis. Plant J. 2006;46:282–96.
Yaish MWF, Peng M, Rothstein SJ. AtMBD9 modulates Arabidopsis development through the dual epigenetic pathways of DNA methylation and histone acetylation. Plant J. 2009;59:123–35.
Preuss SB, Costa-Nunes P, Tucker S, Pontes O, Lawrence RJ, Mosher R, Kasschau KD, Carrington JC, Baulcombe DC, Viegas W, Pikaard CS. Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol Cell. 2008;32:673–84.
Hu Z, Yu Y, Wang R, Yao Y, Peng H, Ni Z, Sun Q. Expression divergence of TaMBD2homoeologous genes encoding methyl CpG-binding domain proteins in wheat (Triticumaestivum L.). Gene. 2011;471:13–8.
Shi R, Zhang J, Li J, Wang K, Jia H, Zhang L, Wang P, Yin J, Meng F, Li Y. Cloning and characterization of TaMBD6homeologues encoding methyl-CpG-binding domain proteins in wheat. Plant Physiol Biochem. 2016;109:1–8.
Questa JI, Rius SP, Casadevall R, Casati P. ZmMBD101 is a DNA-binding protein that maintains Mutator elements chromatin in a repressive state in maize. Plant Cell Environ. 2016;39:174–84.
Li Y, Deng H, Miao M, Li H, Huang S, Wang S, Liu Y. Tomato MBD5, a methyl CpG binding domain protein, physically interacting with UV-damaged DNA binding protein-1, functions in multiple processes. New Phytol. 2016;210:208–26.
Parida AP, Raghuvanshi U, Pareek A, Singh V, Kumar R, Sharma AK. Genome-wide analysis of genes encoding MBD domain-containing proteins from tomato suggest their role in fruit development and abiotic stress responses. Mol Biol Rep. 2018;45:2653–69.
Chen Z, Zheng W, Chen L, Li C, Liang T, Chen Z, Xu H, Han Y, Kong L, Zhao X, Wang F, Wang Z, Chen S. Green fluorescent protein- and Discosomasp red fluorescent protein-tagged organelle marker lines for protein subcellular localization in rice. Front Plant Sci. 2019;10:1421.
Xie C, Zhang G, An L, Chen X, Fang R. Phytochrome-interacting factor-like protein OsPIL15 integrates light and gravitropism to regulate tiller angle in rice. Planta. 2019;250:105–14.
Han SH, Yoo SC, Lee BD, An G, Paek NC. Rice FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (OsFKF1) promotes flowering independent of photoperiod. Plant Cell Environ. 2015;38:2527–40.
Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004;18:926–36.
Wu M, Liu H, Lin Y, Chen J, Fu Y, Luo J, Zhang Z, Liang K, Chen S, Wang F. In-frame and frame-shift editing of the Ehd1gene to develop Japonicarice with prolonged basic vegetative growth periods. Front Plant Sci. 2020;11:307.
Wu W, Zheng XM, Lu G, Zhong Z, Gao H, Chen L, Wu C, Wang HJ, Wang Q, Zhou K, Wang JL, Wu F, Zhang X, Guo X, Cheng Z, Lei C, Lin Q, Jiang L, Wang H, Ge S, Wan J. Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc Natl AcadSci USA. 2013;110:2775–80.
Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. Hd3a, a rice ortholog of the ArabidopsisFT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002;43:1096–105.
Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. Hd3a and RFT1 are essential for flowering in rice. Development. 2008;135:767–74.
Kobayashi K, Yasuno N, Sato Y, Yoda M, Yamazaki R, Kimizu M, Yoshida H, Nagamura Y, Kyozuka J. Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell. 2012;24:1848–59.
Liu J, Shen J, Xu Y, Li X, Xiao J, Xiong L. Ghd2,aCONSTANS-like gene, confers drought sensitivity through regulation of senescence in rice. J Exp Bot. 2016;67:5785–98.
Li P, Wang Y, Qian Q, Fu Z, Wang M, Zeng D, Li B, Wang X, Li J. LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 2007;17:402–10.
Yu B, Lin Z, Li H, Li X, Li J, Wang Y, Zhang X, Zhu Z, Zhai W, Wang X, Xie D, Sun C. TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 2007;52:891–8.
Jin J, Huang W, Gao JP, Yang J, Shi M, Zhu MZ, Luo D, Lin HX. Genetic control of rice plant architecture under domestication. Nat Genet. 2008;40:1365–9.
Tan L, Li X, Liu F, Sun X, Li C, Zhu Z, Fu Y, Cai H, Wang X, Xie D, Sun C. Control of a key transition from prostrate to erect growth in rice domestication. Nat Genet. 2008;40:1360–4.
Wu X, Tang D, Li M, Wang K, Cheng Z. Loose plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol. 2013;161:317–29.
Zhao L, Tan L, Zhu Z, Xiao L, Xie D, Sun C. PAY1 improves plant architecture and enhances grain yield in rice. Plant J. 2015;83:528–36.
Dong H, Zhao H, Xie W, Han Z, Li G, Yao W, Bai X, Hu Y, Guo Z, Lu K, Yang L, Xing Y. A novel tiller angle gene, TAC3, together with TAC1 and D2largely determine the natural variation of tiller angle in rice cultivars. PLoS Genet. 2016;12:e1006412.
Zhang N, Yu H, Yu H, Cai Y, Huang L, Xu C, Xiong G, Meng X, Wang J, Chen H, Liu G, Jing Y, Yuan Y, Liang Y, Li S, Smith SM, Li J, Wang Y. A core regulatory pathway controlling rice tiller angle mediated by the LAZY1-dependent asymmetric distribution of auxin. Plant Cell. 2018;30:1461–75.
Zhang W, Tan L, Sun H, Zhao X, Liu F, Cai H, Fu Y, Sun X, Gu P, Zhu Z, Sun C. Natural variations at TIG1encoding a TCP transcription factor contribute to plant architecture domestication in rice. Mol Plant. 2019;12:1075–89.
Li H, Sun H, Jiang J, Sun X, Tan L, Sun C. TAC4 controls tiller angle by regulating the endogenous auxin content and distribution in rice. Plant Biotechnol J. 2020. https://doi.org/10.1111/pbi.13440.
Hori K, Matsubara K, Yano M. Genetic control of flowering time in rice: integration of Mendelian genetics and genomics. Theor Appl Genet. 2016;129:2241–52.
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9.
Chen S, Songkumarn P, Liu J, Wang GL. A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 2009;150:1111–21.
Shan Q, Wang Y, Li J, Gao C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc. 2014;9:2395–410.
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu YG. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8:1274–84.
Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL. A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol. 2006;7:417–27.
Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryzasativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–82.
Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBOJ. 1987;6:3901–7.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Yu G, Wang LG, Han Y, He QY. ClusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16:284–7.
Acknowledgements
Not applicable.
Funding
National Key Research and Development Program of China (2016YFD0102103) to SZ and Technological Innovation Projects of Fujian Academy of Agricultural Sciences (ZYTS2019014) to MW. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
SZ and SC conceived and designed the experiments. MQ, ZZ, TL, PN, and ZQC performed the experiments. MQ, MW, WC, and SC analyzed the data. MW and ZJC contributed reagents and tools. MQ and SC wrote the manuscript. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1
. Methyl-CpG-binding domain protein genes predicted in rice genome.
Additional file 2: Table S2.
Digital expression profiles of rice Methyl-CpG-binding domain protein genes.
Additional file 3: Figure S1
. Detection of OsMBD707 transcript.(A) Schematic diagrams of predicted OsMBD707 splicing variants. (B) Sequence alignment of predicted OsMBD707 splicing variants. Sequences for primers designed to distinguish the two predicted alternative transcripts (predicted amplification sizes of 106 bp and 94 bp for XM_015764399.1/LOC_Os12g42550.1 and XM_015764400.2/LOC_Os12g42550.2, respectively.) are indicated by arrow. (C) RT-PCR analysis for detecting predicted OsMBD707 splicing variants. Only an amplification size of 106 bp for XM_015764399.1/LOC_Os12g42550.1 was detected in the roots, stems, leaves, spikelets, seeds, and panicle axes. (D) Sequencing confirmation of the 106 bp-amplified product of XM_015764399.1/LOC_Os12g42550.1. (PPT 2129 kb)
Additional file 4: Figure S2
. Summary of differentially expressed genes (DEGs) identified between the OsMBD707-overexpression line OX707-#21 and wild-type. (A) Number of DEGs between OX707-#21 and wild-type under short day (SD) and long day (LD) conditions, respectively. Up, up-regulated in OX707-#21; Down, down-regulated in OX707-#21. (B) Venn diagrams of DEGs between OX707-#21 and wild-type under SD and LD. (PPT 128 kb)
Additional file 5: Table S3
. DEGs between the OsMBD707-overexpression line OX707-#21 and wild-type under short day condition. (XLS 314 kb)
Additional file 6: Table S4
. DEGs between the OsMBD707-overexpression line OX707-#21 and wild-type under long day condition. (XLS 486 kb)
Additional file 7: Figure S3
. GO analysis of DEGs between the OsMBD707-overexpression line OX707-#21 and wild-type. (PPT 758 kb)
Additional file 8: Table S5
. Significant categories identified by GO analysis of the 2353 DEGs between the OsMBD707-overexpression line OX707-#21 and wild-type. (XLS 42 kb)
Additional file 9: Figure S4
. KEGG analysis of DEGs between the OsMBD707-overexpression line OX707-#21 and wild-type. (PPT 374 kb)
Additional file 10: Table S6
. Significant pathways identified by KEGG analysis of the 2353 DEGs between the OsMBD707-overexpression line OX707-#21 and wild-type. (XLS 29 kb)
Additional file 11: Figure S5
. Expression profiles of tiller angle regulator genes in OsMBD707-overexpression plant, and of OsMBD707 under short day (SD) and long day (LD). (PPT 2596 kb)
Additional file 12: Table S7
. Primers used in this study.
Additional file 13: Table S8
. Gene count and gene FPKM values from the RNA-seq in this study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Qu, M., Zhang, Z., Liang, T. et al. Overexpression of a methyl-CpG-binding protein gene OsMBD707 leads to larger tiller angles and reduced photoperiod sensitivity in rice. BMC Plant Biol 21, 100 (2021). https://doi.org/10.1186/s12870-021-02880-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-021-02880-3