Abstract
Background
The AP2/ERFs belong to a large family of transcription factors in plants. The AP2/ERF gene family has been identified as a key player involved in both biotic and abiotic stress responses in plants, however, no comprehensive study has yet been carried out on the AP2/ERF gene family in rose (Rosa sp.), the most important ornamental crop worldwide.
Results
The present study comprises a genome-wide analysis of the AP2/ERF family genes (RcERFs) in the rose, involving their identification, gene structure, phylogenetic relationship, chromosome localization, collinearity analysis, as well as their expression patterns. Throughout the phylogenetic analysis, a total of 131 AP2/ERF genes in the rose genome were divided into 5 subgroups. The RcERFs are distributed over all the seven chromosomes of the rose, and genome duplication may have played a key role in their duplication. Furthermore, Ka/Ks analysis indicated that the duplicated RcERF genes often undergo purification selection with limited functional differentiation. Gene expression analysis revealed that 23 RcERFs were induced by infection of the necrotrophic fungal pathogen Botrytis cinerea. Presumably, these RcERFs are candidate genes which can react to the rose’s resistance against Botrytis cinerea infection. By using virus-induced gene silencing, we confirmed that RcERF099 is an important regulator involved in the B.cinerea resistance in the rose petal.
Conclusion
Overall, our results conclude the necessity for further study of the AP2/ERF gene family in rose, and promote their potential application in improving the rose when subjected to biological stress.
Similar content being viewed by others
Background
Transcription factors are important regulators of the expression of various inducible genes in plants, and play an indispensable role in plant growth, development, stress response, as well as pathogen defence [1]. Transcription factors usually comprise a nuclear localization signal, a DNA binding domain, a transactivation domain, as well as an oligomerization site. These domains determine the subcellular localization, cis-regulatory elements binding, and the regulating function of transcription factors [2].
The AP2/ERF superfamily is one of the largest transcription factor gene family in plants, wherein a total of 147 AP2/ERF family members have been identified in Arabidopsis. The AP2/ERF gene family consists of the AP2/ERF domain comprising 60 to 70 amino acids, and recognizes the cis-regulatory element GCC box or DRE elements which regulate the reaction of target genes [3]. The AP2/ERF gene family can be further categorized into five subfamilies, to example ERF, AP2 (APETALA2), DREB (dehydration-responsive element binding), RAV (related to ABI3/VP1) and Soloist [4,5,6]. The AP2/ERFs that regulate growth and development throughout the plant’s life cycle have been detected. The AP2/ERFs also play a very important role when the plant is exposed to abiotic stresses, such as dehydration, salinity, low temperature or heat stress. For example, transgenic Arabidopsis that overexpresses AtERF4 is more sensitive to drought stress and has a lower resistance to Sodium chloride [7]. In addition, overexpressing the RAP2.6 gene (RELATED TO AP2.6, encodes an ERF transcription factor) results in a sensitive phenotype to ABA (Abscisic Acid) and salt/osmotic stress during germination and the early growth stage of Arabidopsis [8].
More importantly, the AP2/ERF gene family is one of the transcription factors considered to be involved in plant defence responses against various phytopathogens [9,10,11,12]. For example, the transcript of ERF1 is induced significantly subsequent to the inoculation of necrotrophic fungi Botrytis cinerea, and overexpression of ERF1 in Arabidopsis enhanced its resistance to both B. cinerea and Plectosphaerella cucumerina [13]. Overexpressing ERF5 or ERF6 also increased resistance to B. cinerea in Arabidopsis, and the erf5 erf6 double mutant showed a significant increase in susceptibility [14].
Rose is the most popular ornamental crop and accounts for over 30% of total cut-flower sales worldwide [15]. However, the flower is a fragile organ and transportation over long distances causes rose flowers to be affected by post-harvest diseases such as gray mold caused by B. cinerea. The function of AP2/ERF transcription factors in disease resistance has been characterized in model plants Arabidopsis as well as many other plant species. However, no rose AP2/ERF family genes involved in disease resistance have yet been identified.
Recently, we performed a de novo RNA-Seq analysis of rose petals infected by B. cinerea. This transcriptome study revealed a large number of rose genes, including AP2/ERF family transcription factors, were significantly up-regulated and implied their involvement of resistance against B. cinerea [16]. In the present study, genome-wide identification and analysis of the AP2/ERF gene family in the rose were carried out. By using virus-induced gene silencing (VIGS), we further confirmed that RcERF099 plays a significant role in B. cinerea resistance in rose flowers.
Results
Identifying RcERF genes in the rose genome
In order to identify the potential AP2/ERFs of R. chinensis, we downloaded the AP2/ERF HMM profile (PF00847) from the Pfam database. Using this profile as a query, the HMM search of the rose genome finally lead to the identification of 137 candidate RcERF genes. Conserved Domains Database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and ExPASy (http://web.expasy.org/protparam/) were employed to verify all candidate RcERFs contain a single AP2/ERF motif. We further removed any sequence having less than 150 amino acids, and finally obtained a total of 131 non-redundant RcERF genes. All these 131 ERF family genes can be mapped onto rose chromosomes and we designated the genes RcERF001 to RcERF131 in accordance with their chromosome order.
The length of proteins encoded by RcERF family genes varies from 150 to 832 amino acids, with an average length of 298 amino acids. The longest (RcERF052) contains 832 amino acids, whereas the shortest just has 150 amino acids (RcERF093 and RcERF095). Table 1 summarizes detailed information of all 131 RcERF genes, including their accession numbers, chromosome locations, exon and intron details, protein size and classification.
Chromosomal localization and microsynteny analysis
131 RcERF genes were located on all 7 rose chromosomes, as depicted in Fig. 1. Chromosome 2 contains the largest number of RcERF genes (31), followed by chromosome 7 (26). Chromosomes 3 and 5 contain the least number of chromosomes (11). The RcERF genes were unevenly distributed over 7 chromosomes. 8.40% of RcERFs were located in the long arm of chromosomes 3 and 5, 23.66% of RcERFs were located in chromosome 2, 15.27% of RcERFs were located in chromosome 1, 10.69 and 13.74% of RcERFs were distributed over chromosome 4 and 6. Chromosome 7 contains 19.85% RcERFs, and they were distributed over both the long and short arms.
Furthermore, we studied RcERFs duplication events, and discovered in total 21 gene pairs in the rose genome (Table 2). Only one gene pair was located on the same chromosome (RcERF021 and RcERF042), indicating that they are likely to be tandem repeats. The remaining 20 gene pairs were located on different chromosomes, and indicated that segmental duplication may occur in these regions (Fig. 2).
Microsyntenic analyses of the rose AP2/ERF transcription factors in the Rose chinensis genome. Circular visualization of rose AP2/ERF transcription factors is mapped onto different chromosomes using Circos. The red lines indicate rose AP2/ERF genes having a syntenic relationship. The grey lines represent all syntenic blocks in the genome of R. chinensis
To explore the selective constraints among duplicated RcERF genes, we calculated the ratio of non-synonymous (Ka) to synonymous (Ks) nucleotide substitutions (Ka/Ks ratio) of 21 pairs of duplicated genes (Table 2). A Ka/Ks ratio < 1 indicates a negative or purifying selection of gene pairs, whereas Ka/Ks > 1 depicts a positive selection. Our study revealed that the Ka/Ks ratio for all RcERF gene pairs is < 0.4 (Table 2). These data indicate that RcERF gene pairs had undergone a purifying selection, and functional differentiation is limited.
Phylogenetic and exon-intron structural analysis of RcERF genes
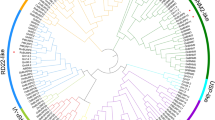
We performed a phylogenetic analysis on all RcERF genes using the neighbor-joining method and established a phylogenetic tree. According to their evolutionary relationships, RcERF genes are further categorized into five subfamilies with supported bootstrap values, including ERF, DREB, AP2, RAV and Soloist, comprising 64, 42, 18, 4 and 3 members, respectively.
Subsequent analysis of the exon-intron structure proved to be consistent with the phylogenetic analysis results. Most of the genes clustered in the same subfamily exhibit a similar exon-intron structure. Members of the RAV subfamily do not comprise intron, however, in contrast, AP2 and Soloist subfamily genes comprise four to twelve introns. Most of the ERF and DREB subfamily members have either no intron or only one, however, some exceptions were also observed; for example, RcERF011 and RcERF045 have two introns and RcERF107 has three (Fig. 3; Table 1). These results demonstrate the presence of highly conserved structures within the subfamilies and diversity among the different subfamilies.
Phylogenetic and gene structural analysis of rose AP2/ERF transcription factors. The phylogenetic tree is constructed by MEGA6.0 using a Neighbor-joining method. Numbers on the nodes of the branches represent bootstrap values. The gene structure diagram represents UTRs, exons and introns with green boxes, yellow boxes and gray lines, respectively. The scale at the bottom estimated the size of UTRs, exons and introns
There is increasing evidence that AP2/ERF transcription factors play a key role in disease resistance in various plant species (Table 3). In order to evaluate RcERFs’ involvement in rose disease resistance, we generated a composite phylogenetic tree that included defence-related ERFs in other plant species and all RcERFs (Fig. 4). In this composite phylogenetic tree, each subfamily is marked with a different colour, and all plant ERFs that are known to be involved in disease resistance are in bold. ERFs involved in regulating defence responses are distributed in ERF and DREB subfamilies, but not in AP2, RAV, or Soloist.
Phylogenetic analyses of the rose AP2/ERF transcription factors with disease-resistance-related AP2/ERF transcription factors from other plant species. The composite phylogenetic tree that included all rose AP2/ERF transcription factors and disease-resistance-related AP2/ERF transcription factors (in bold) from Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), soybean (Glycine max) and tomato (Solanum lycopersicum) were constructed by MEGA 6.0 with the neighbor-Joining method. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed. The bootstrap values are indicated on the nodes of the branches
The expression of RcERF genes in response to Botrytis cinerea infection
There has been an increasing rise in evidence gained from studying various plant species which indicates that plant AP2/ERF transcription factors play a significant role in pathogen response. In order to study the role of RcERFs in B. cinerea resistance, we analyzed transcriptome data in rose petals at 30 hpi and 48 hpi of this pathogen. The 30 hpi timepoint represents the early response to infection, whereas the 48 hpi timepoint corresponds to the late response [16]. A total of 23 RcERF genes (RhERF004, RhERF005, RhERF015, RhERF019, RhERF023, RhERF024, RhERF054, RhERF063, RhERF064, RhERF066, RhERF068, RhERF070, RhERF072, RhERF080, RhERF088, RhERF089, RhERF092, RhERF093, RhERF095, RhERF099, RhERF114, RhERF123 and RhERF125) were significantly up-regulated, indicating they could be key regulators in resisting B. cinerea infection in rose. Amongst these B. cinerea-induced RcERFs, the expression of 10 RcERF genes was increased significantly at 30 hpi, suggesting that these RcERFs may well be involved in an early response to B. cinerea (Table 4).
In order to further verify the expression profile from RNA-seq, the expression of six RcERFs was analyzed by qPCR. The results of the qPCR analysis proved to be consistent with the expression profile obtained from the transcriptome analysis (Fig. 5).
Validation of RNA-Seq results using qPCR. RcUBI2 was used as a housekeeping gene. Expression profile data of six RcERF genes at 30 hpi and 48 hpi after B. cinerea inoculation were obtained using qPCR. Error bar represent SD in three technical replicates. The primers used are listed in Supplementary Table S1
RcRF099 is required for rose resistance to B. cinerea
In order to further illustrate the potential role of B. cinerea-induced RcERF genes in resistance of this pathogen, we used VIGS to knock down the expression of RcERF099 in rose petals. RcERF099 was selected to conduct this VIGS study because: 1) RcERF099 is up-regulated upon B. cinerea infection (Fig. 5; Table 4); and 2) based on phylogenetic analysis, RcERF099 belongs to the DREB subfamily which comprises many disease-resistant ERFs originating from other plant species, such as AtERF001, AtERF004, AtERF005, AtERF006, AtERF014, and AtERF015 (Fig. 4; Table 3).
In order to silence RcERF099 in rose petals, we cloned a 230 bp fragment of RcERF099 into a pTRV2 vector [27] to generate TRV-RcERF099. Agrobacterium tumefaciens carrying TRV-RcERF099 and TRV1 [27] were co-infiltrated into rose petal discs to generate RcERF099-silenced rose petals. The infiltrated rose petal discs were then inoculated with B. cinerea. Comparing the control petal (TRV-00) inoculated with an empty TRV, the plant inoculated with TRV-RcERF099 showed more serious disease symptoms displaying a significant increase in the size of the disease lesion (Fig. 6a and b). Furthermore, we confirmed the silencing efficiency of VIGS with qPCR (Fig. 6c). These results indicated that RcERF099 is required for rose resistance to B. cinerea.
Functional analysis of rose AP2/ERF transcription factor gene RcERF099. a Compromised B. cinerea resistance upon silencing of RcERF099 (TRV- RcERF099) was observed at 60 hpi post-inoculation. b. Quantification of B. cinerea disease lesions on TRV-RcERF099- and TRV-00-inoculated rose petal discs. The graph indicates the lesion size of three biological replicates (n = 48) with the standard deviation. c. Expression of RcERF099 relative to that during the control at 6 days of post-silencing. All statistical analyses were performed using Student’s t-test; ** p < 0.01
Discussion
Plant disease resistance-related genes are often induced by the invasion of pathogens, and are regulated at the transcriptional level by specific transcription factors. The AP2/ERFs is a major transcription factor family in plants, and has proved to have important functions in disease resistance in various plant species [28,29,30,31,32]. A genome-wide analysis of the AP2/ERF gene family has been performed in arabidopsis and rice [4]. So far, no comprehensive analysis of the rose AP2/ERF gene family has yet been reported, and the function of most RcERFs is largely generally unknown. In the current study, using the recently available rose genome, we performed a comprehensive analysis of the AP2/ERF gene family, including their gene structure, phylogeny, chromosomal location, gene duplication, as well as expression profiles during infection of gray mold caused by necrotrophic fungal pathogen B. cinerea.
The number of AP2/ERF genes in rose (131) has proved to be lower than those in arabidopsis (147) and rice (164) [4], which indicates that the AP2/ERF gene family in different plants has expanded in various degrees during its evolution. Furthermore, we indicated that gene duplication is involved in the expansion of the RcERF gene family, in which a total of 21 duplication events were identified. Most of the duplicated genes (20) were involved in segmental duplication, whereas only one was involved in tandem duplication. Interestingly, the Ka/Ks ratio of all these 21 RcERF duplicates was < 1, indicating that the RcERF gene family undergoes a purification rather than a positive selection, suggesting a highly conservative evolution of this important transcription factor in the gene family. Previously, it has been demonstrated that the plant immune receptor genes involved in race-specific recognition of an invading pathogen undergo positive selection pressure [15]. It further indicates that the RcERFs generally involved in the basal defence against pathogens, are not race-specific resistance.
Although the role of RcERFs in disease resistance remains unclear, increasing evidence has proved that plant AP2/ERF genes are important players involved in regulating plant disease resistance. It prompts us to search for candidate RcERFs that are involved in the resistance to B. cinerea in roses. Based on their expression in response to gray mold infestation, we identified 23 RcERFs that could well be involved in gray mold resistance in rose petals.
We subsequently added plant ERFs that are known to be involved in disease resistance in the RcERFs phylogenetic tree. We discovered that these disease-related ERFs are mainly distributed within ERF and DREB subfamilies. The RcERF099 belongs to the DREB subfamily, which includes certain members of known disease-related plant ERF genes (Fig. 4). Especially, RcERF099 has a close homolog with Arabidopsis AtERF014, which has proved to play an important role in resistance against both bacterial pathogen Pseudomonas syringae pv. tomato, as well as fungal pathogen Fusarium oxysporum and B. cinerea [22]. More importantly, RcERF099 was induced significantly with B. cinerea. We therefore consider that RcERF099 should be regarded as an important candidate gene involved in the regulation of rose disease resistance. The silencing of RcERF099 in rose petals by VIGS increased its susceptibility to B. cinerea, indicating that it has a positive regulatory function in gray mold resistance.
Conclusion
pt?>In this study, a genome-wide analysis of RcERFs was carried out. A total of 131 non-redundant AP2/ERF family members were identified in the rose genome, and these RcERFs were divided into 5 subfamilies on the basis of phylogeny and conserved domains. Expression analysis indicated that the transcriptional regulation of certain RcERF family genes was induced by B. cinerea infection in rose petals. In addition, plant ERFs involved in disease resistance are usually clustered on the same branch of the phylogenetic tree. Based on these analyses, using VIGS, we further proved that RcERF099 is involved in regulating resistance to B. cinerea in rose petals. The information ensuing from these results may facilitate further research into RcERFs functions and crop improvement.
Methods
Identification of the rose AP2/ERF family gene
The genome sequences and CDS sequences of rose were downloaded from the website (https://lipm-browsers.toulouse.inra.fr/pub/RchiOBHm-V2/) to construct a local genome database. Based on AP2/ERF HMM (Hidden Markov model) from Pfam (PF00847, http://pfam.xfam.org), we initially identified AP2/ERF candidate genes in the rose genome with E-value <1e− 3. Finally, all candidate AP2/ERF sequences were verified that they contain at least one AP2/ERF domain through the CDD (Conserved Domains Database; https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and ExPASy (http://web.expasy.org/protparam/). Sequences without relevant domains or conserved motifs were removed. Chromosomal distribution of each AP2/ERF gene was mapped using Mapchart 2.2 software [33].
Gene structure and phylogenetic analysis of RcERFs
The map of exon-intron structures of the RcERF genes was carried out using TBtools software [34] by comparing the coding sequences (CDS) with their corresponding protein sequences. Furthermore, the phylogenetic analysis of RcERFs in the rose was conducted using the NJ method in MEGA 6.0 software and the bootstrap test was carried out with 1000 replicates.
In addition, 17 ERFs were previously reported that involved in disease resistance. These ERFs originate from various plant species, including tomato (Solanum lycopersicum), rice (Oryza sativa), soybean (Glycine max), and Arabidopsis thaliana. Amino acid sequences of these disease resistance-related ERFs and rose AP2/ERFs were then aligned using ClustalW. The alignment of protein sequences which resulted was subsequently used for phylogenetic analysis. A phylogenetic analysis was conducted using the NJ method in MEGA 6.0 software [35] and the bootstrap test was carried out with 1000 replicates. On the phylogenetic dendrograms, the percentage of replicated trees in which the associated taxa clustered together in the bootstrap test is indicated alongside the branches.
Collinearity analyses
For the purpose of identifying the collinearity of RcERFs, we downloaded the genome sequence of rose on a local server, and a Multiple Collinearity Scan toolkit [36] was used to determine microsyntenic relationships between RcERF genes. The resultant microsynteny relationships were further evaluated by CollinearScan set at an E-value of <1e− 10.
Calculation of non-synonymous (Ka) to synonymous (Ks) substitution rates
TBtools was used to calculate the synonymous (Ks) and non-synonymous (Ka) nucleotide substitution rates. The Ka/Ks ratios of duplicated gene pairs were calculated to determine the selection mode driving the evolution of RcERFs.
Expression of RcERFs in response to B. cinerea
RNA-Seq data (accession number PRJNA414570) of rose petals undergoing B. cinerea infection was downloaded from the National Center for Biotechnology Information (NCBI) database. The clean sequencing reads were mapped to the Rosa chinensis ‘Old Blush’ reference genome. Gene expression levels of RcERFs were calculated by Reads per kb per million reads (RPKM). And differentially expressed gene based on Log2 fold change was performed by DEseq2. In order to verify the RNA-Seq results, the expression of 6 RcERF genes was analyzed using quantitative PCR (qPCR). To this end, total RNA was extracted from rose petals at 30 h and 48 h post-inoculation (hpi) respectively with B. cinerea using the hot borate method as previously described [37]. One microgram of DNase-treated RNA was used to synthesize the first-strand cDNA by using HiScript II Q Select RT SuperMix (Vazyme) in a 20-μL reaction volume. An qPCR reaction was performed using the SYBR Green Master Mix (Takara), and detection was achieved in StepOnePlus Real-Time PCR System (Thermo Fisher Scientific). RcUBI2 was used as an internal control. A delta-delta-Ct method calculation method was used for expression analysis. All primers that were used as qPCR are listed in Supplementary Table S1.
VIGS and B. cinerea inoculation assays
The rose plants (Rosa hybrida) used in this study were grown in soil in a greenhouse in Yunnan, China. In order to obtain the constructs for silencing, a 230 bp sequence of RcERF099 was amplified using primers TRV-RcERF099-F (5′- GGGGACAAGTTTGTACAAAAAAGCAGGCTGCTCATTTGGGTCCTATACT − 3′) and TRV-RcERF099-R (5′- GGGGACCACTTTGTACAAGAAAGCTGGGTAGTAATATCTTCAAGCAATT − 3′). The fragment generated was subsequently cloned into TRV2 vectors [27]. The VIGS of detached rose petal discs has been described previously [38]. In brief, detached petals are obtained from the outermost whorls of the rose, and 15-mm petal discs were punched. Agrobacterium consisting of TRV1 [27] and TRV2 constructs were mixed at a ratio of 1: 1 and vacuum infiltrated into petal discs. Petal discs were then inoculated with B. cinerea at 6 days after TRV infection. At least three biological repeats were performed, using at least 16 discs for each repeat. The disease lesion was estimated at 60 h post-inoculation, and a Student’s t-test conducted to determine the significance. All primers used for this study are listed in Supplementary Table S1.
Availability of data and materials
The datasets used and/or analyzed during the current study have been included within supplemental data. The Raw data of RNA-Seq of rose petals undergoing B. cinerea infection can be found in the BioProject database (accession nr. PRJNA414570). The plant materials are available from the corresponding author on request.
Abbreviations
- hpi:
-
Hours post-inoculation
- NJ:
-
Neighbor-joining
- HMM:
-
Hidden Markov Model
- CDD:
-
Conserved Domains Database
- VIGS:
-
Virus-induced gene silencing
References
Dou L, Zhang X, Pang C, Song M, Wei H, Fan S, et al. Genome-wide analysis of the WRKY gene family in cotton. Mol Gen Genomics. 2014;289(6):1103–21.
Cao Y, Han Y, Li D, Lin Y, Cai Y. MYB transcription factors in Chinese pear (Pyrus bretschneideri Rehd.): genome-wide identification, classification, and expression profiling during fruit development. Front Plant Sci. 2016;7:577.
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290(3):998–1009.
Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006;140(2):411–32.
Zhang G, Chen M, Chen X, Xu Z, Guan S, Li LC, et al. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.). J Exp Bot. 2008;59(15):4095–107.
Zhuang J, Cai B, Peng RH, Zhu B, Jin XF, Xue Y, et al. Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem Biophys Res Commun. 2008;371(3):468–74.
Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol. 2005;58(4):585–96.
Zhu Q, Zhang J, Gao X, Tong J, Xiao L, Li W, et al. The Arabidopsis AP2/ERF transcription factor RAP2.6 participates in ABA, salt and osmotic stress responses. Gene. 2010;457(1–2):1–12.
Shin R, Park JM, An JM, Paek KH. Ectopic expression of Tsi1 in transgenic hot pepper plants enhances host resistance to viral, bacterial, and Oomycete pathogens. Mol Plant-Microbe Interact. 2002;15(10):983–9.
Zuo K, Qin J, Zhao J, Ling H, Zhang L, Cao Y, et al. Over-expression GbERF2 transcription factor in tobacco enhances brown spots disease resistance by activating expression of downstream genes. Gene. 2007;391(1–2):80–90.
Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH. Overexpression of the tobacco Tsi1 gene encoding an EREBPAP2–type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell. 2001;13(5):1035–46.
Gutterson N, Reuber TL. Regulation of disease resistance pathways by AP2ERF transcription factors. Curr Opin Plant Biol. 2004;7(4):465–71.
Berrocal-Lobo M, Molina A, Solano R. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 2002;29(1):23–32.
Moffat CS, Ingle RA, Wathugala DL, Saunders NJ, Knight H, Knight MR. ERF5 and ERF6 play redundant roles as positive regulators of JA/et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS One. 2012;7(4):e35995.
Liorzou M, Pernet A, Li S, Chastellier A, Thouroude T, Michel G, et al. Nineteenth century French rose (Rosa sp.) germplasm shows a shift over time from a European to an Asian genetic background. J Exp Bot. 2016;67(15):4711–25.
Liu X, Cao X, Shi S, Zhao N, Li D, Fang P, et al. Comparative RNA-Seq analysis reveals a critical role for brassinosteroids in rose (Rosa hybrida) petal defense against Botrytis cinerea infection. BMC Genet. 2018;19(1):62.
Liu D, Chen X, Liu J, Ye J, Guo Z. The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J Exp Bot. 2012;63(10):3899–912.
Zhang G, Chen M, Li L, Xu Z, Chen X, Guo J, et al. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot. 2009;60(13):3781–96.
Zhao Y, Chang X, Qi D, Dong L, Wang G, Fan S, et al. A novel soybean ERF transcription factor, GmERF113, increases resistance to Phytophthora sojae infection in soybean. Front Plant Sci. 2017;8:299.
Dong L, Cheng Y, Wu J, Cheng Q, Li W, Fan S, et al. Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J Exp Bot. 2015;66(9):2635–47.
Zhang H, Huang L, Dai Y, Liu S, Hong Y, Tian L, et al. Arabidopsis AtERF15 positively regulates immunity against pseudomonas syringae pv. Tomato DC3000 and Botrytis cinerea. Front Plant Sci. 2015;6:686.
Onate-Sanchez L, Anderson JP, Young J, Singh KB. AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol. 2007;143(1):400–9.
Berrocal-Lobo M, Molina A, Solano R. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1in Arabidopsis confers resistance to several necrotrophicfungi. Plant J. 2002;9(1):23–32.
Pre M, Atallah M, Champion A, De Vos M, Pieterse CM, Memelink J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008;147(3):1347–57.
He P, Warren RF, Zhao T, Shan L, Zhu L, Tang X, et al. Overexpression of Pti5 in tomato potentiates pathogen-induced defense gene expression and enhances disease resistance to pseudomonas syringae pv. Tomato. Mol Plant-Microbe Interact. 2001;14(12):1453–7.
Zhang H, Zhang D, Chen J, Yang Y, Huang Z, Huang D, et al. Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum. Plant Mol Biol. 2004;55(6):825–34.
Liu YL, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. Plant J. 2002;31(6):777–86.
Li Z, Tian Y, Xu J, Fu X, Gao J, Wang B, et al. A tomato ERF transcription factor, SlERF84, confers enhanced tolerance to drought and salt stress but negatively regulates immunity against pseudomonas syringae pv. Tomato DC3000. Plant Physiol Biochem. 2018;132:683–95.
Lu X, Zhang L, Zhang F, Jiang W, Shen Q, Zhang L, et al. AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea. New Phytol. 2013;198(4):1191–202.
Jin JH, Zhang HX, Ali M, Wei AM, Luo DX, Gong ZH. The CaAP2/ERF064 regulates dual functions in pepper: plant cell death and resistance to Phytophthora capsici. Genes (Basel). 2019;10:7.
Tian Z, He Q, Wang H, Liu Y, Zhang Y, Shao F, et al. The potato ERF transcription factor StERF3 negatively regulates resistance to Phytophthora infestans and salt tolerance in potato. Plant Cell Physiol. 2015;56(5):992–1005.
Maruyama Y, Yamoto N, Suzuki Y, Chiba Y, Yamazaki K, Sato T, et al. The Arabidopsis transcriptional repressor ERF9 participates in resistance against necrotrophic fungi. Plant Sci. 2013;213:79–87.
Voorrips RE. MapChart software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93(1):77–8.
Chen C, Xia R, Chen H, He Y. TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv. 2018;1:289660.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9.
Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49.
Wu L, Ma N, Jia Y, Zhang Y, Feng M, Jiang CZ, et al. An ethylene-induced regulatory module delays flower senescence by regulating Cytokinin content. Plant Physiol. 2017;173(1):853–62.
Cao X, Yan H, Liu X, Li D, Sui M, Wu J, et al. A detached petal disc assay and virus-induced gene silencing facilitate the study of Botrytis cinerea resistance in rose flowers. Hortic Res. 2019;6:136.
Acknowledgements
Not Applicable.
Funding
Financial support for this study was provided by the National Natural Science Foundation of China (grant number 31772344 and 31972444) to Zhao Zhang. It was also financially supported by the Natural Science Foundation of Shaanxi province (grant number 2019JQ645), the Fundamental Research Funds for the Central Universities (grant number 2452019112), and the Scientific Research Startup Fund for Talents, Northwest A&F University (grant number 2452019040) to Yin Song. The funding agents were not involved in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Z.Z., Y.S., D.L. and X.L. conceived and designed the experiments. D.L., X.L., L.S., S.Z. and Y.S. carried out the experiments and analyzed the data. Z.Z., Y.S., H.Z. and D.L. have written the paper. All the authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable. Our research did not involve any human or animal subjects, material, or data. The plant materials used in this study were provided by the China Agricultural University and are freely available for research purposes following institutional, national and international guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
List of primers used in this study.
Additional file 2: Figure S1.
Melting curves for qPCR.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, D., Liu, X., Shu, L. et al. Global analysis of the AP2/ERF gene family in rose (Rosa chinensis) genome unveils the role of RcERF099 in Botrytis resistance. BMC Plant Biol 20, 533 (2020). https://doi.org/10.1186/s12870-020-02740-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-020-02740-6