Abstract
Background
Avian tuberculosis is a chronic and zoonotic disease that affects a wide variety of birds, mammals, and humans. This study aimed to estimate the frequency of Mycobacterium avium subsp. avium in some domestic birds based on molecular diagnosis, antibiogram profile, and PCR-based detection of inhA, rpoB, rpsL, and otrB antibiotic resistance-related genes.
Methods
A total of 120 fecal samples were collected from small flocks of house-reared domestic birds at Ismailia Governorate, Egypt. The collected samples were processed and subjected to the bacteriological examination. The antimicrobial susceptibility testing of the recovered isolates was performed using the broth microdilution method for the detection of minimum inhibitory concentrations (MICs). The genetic detection of the IS901confirmatory gene, inhA, rpoB, rpsL, and otrB genes was carried out using PCR.
Results
The frequency of M. avium subsp. avium was 4.1% (5/120); 10% (4/40) in ducks, and 2.5% (1/10) in geese. The identification of the recovered isolates was confirmed using PCR, where all the tested isolates were positive for IS901confirmatory gene. The results of the broth microdilution method revealed that most of the recovered isolates exhibited multidrug resistance (MDR) to isoniazid, rifampicin, streptomycin, oxytetracycline, and doxycycline, and harbored the inhA, rpoB, rpsL, and otrB genes.
Conclusion
In brief, to the best of our knowledge this is the first report that emphasized the emergence of avian tuberculosis in house-reared domestic birds in Egypt. The emergence of MDR- M. avium subsp. avium is considered a public health threat. Emerging MDR-M. avium subsp. avium in domestic birds are commonly harbored the IS901, inhA, rpoB, rpsL, and otrB genes. Azithromycin and clofazimine revealed a promising in-vitro antibacterial activity against M. avium subsp. avium.
Similar content being viewed by others
Background
Avian tuberculosis is a chronic, debilitating disease that affects a wide variety of birds including wild and domestic birds [1]. The disease is caused mainly by Mycobacterium avium subsp. avium serotypes 1, 2, and 3, and genotype IS901 segment [2]. Mycobacteria are Gram-positive, aerobic, non-sporulated, non-motile, acid fast-bacilli. Mycobacteria are characterized by their lipid-rich cell wall which constitutes about 60–80% of their cell wall. Mycobacteria are opportunistic, intracellular pathogens that could withstand inside the macrophages and resist the host immune mechanism [3]. The bacilli of M. avium subsp. avium could resist most disinfectants, survive and multiply during adverse environmental conditions such as extreme temperature, low oxygen, and low pH [4].
M. avium subsp. avium transmitted between birds through the ingestion of contaminated food and water, inhalation of contaminated droplets, and from the adult birds to their young during the mouth feeding [5]. The infected birds with M. avium subsp. avium remain alive for a long time carrying the infection and subsequently shedding the pathogen in their dropping that results in the transmission of infection to other birds as well as humans [6, 7]. The infected birds may exhibit certain clinical signs such as diarrhea, emaciation, atrophy of breast muscle, and the development of tuberculous nodules in the last stage of the disease, especially in internal organs such as the intestine, liver, spleen, and lung [6].
Avian tuberculosis has public health importance, especially with immunocompromised persons that handle the infected birds or eat their insufficiently cooked meat [8, 9]. Most of the infected cases with M. avium subsp. avium are keeping domestic or pet birds in their home [10]. M. avium subsp. avium not only infects birds, but also could infect pet animals, pigs, and immunocompromised humans. Avian tuberculosis leads to high economic losses including; 1-High condemnation rates in poultry slaughterhouses, 2-Drop in egg production, 3-Weight loss and emaciation of infected birds, 4-Sudden death, and high mortality rates, and 5-Loss of endangered species of birds [11,12,13]. Avian tuberculosis is considered a serious problem in wild birds, especially the endangered species, so the valuable species should be checked regularly [14].
M. avium subsp. avium has a complex thick cell wall that is responsible for the intrinsic multidrug resistance and virulence [15, 16]. The diagnosis of M. avium complex, especially M. avium subsp. avium is difficult; as there are no specific clinical signs, it depends on;1-The culture technique that is recommended by OIE as the gold-standard technique, 2-The microscopical examination of acid-fast bacilli using Ziehl-Neelsen staining, and 3- PCR-based confirmatory diagnosis [10, 17, 18].
The multidrug resistance phenomena have increased globally. The emergence of multidrug-resistant bacterial pathogens from various origins was reported by several recent studies that reflect public health threats [19,20,21,22,23,24,25,26]. Treatment of avian tuberculosis is extremely difficult due to the development of antimicrobial resistance of M. avium complex to most antibiotics. The antimicrobial resistance is attributed to either intrinsic factors such as the waxy lipoid cell wall or mutation in some genes [27]. The most effective drug used in the treatment of avian tuberculosis is azithromycin [28]. Most types of M. avium complex have a weak response to the treatment due to the development of antibiotic resistance [27]. This is the first report concerning the emergence of MDR-M. avium subsp. avium in house-reared domestic birds in Egypt. This study aimed to estimate the frequency of Mycobacterium avium subsp. avium in some domestic birds based on molecular diagnosis, antibiogram profile, and PCR-based detection of inhA, rpoB, rpsL, and otrB antibiotic resistance-related genes.
Materials and methods
Sampling
A total of 120 fecal samples from different domestic birds’ species (chicken (n = 40); 20 apparently healthy and 20 diseased birds, ducks (n = 40); 20 apparently healthy and 20 diseased birds, and geese (n = 40); 20 apparently healthy and 20 diseased birds) were collected from Private house-reared flocks from August 2018 to March 2019 from Ismailia Governorate, Egypt. The diseased birds were suffered from persistent diarrhea and emaciation together with a history of treatment failure with oxytetracycline. The collected samples were prepared according to Parashar et al. [29] and Payeur [30].
Decontamination of fecal samples
The collected specimens were diced by sterile disposable surgical blades and then homogenized in a sterile porcelain mortar and pestle. The homogenized samples were suspended in 10 ml of PBS (Thermo Fisher Scientific, USA). The decontamination was performed as described by Sattar et al. [31]. Briefly, samples were inoculated in 0.9% Hexadecylpryridinium Chloride Monohydrate (HPC) (Sigma-Aldrich, USA), and then were incubated at 37 °C for 24 h. The mixture was centrifuged at 10 °C (3000 xg) for 15 min. The obtained pellets were re-suspended in 1 ml of sterile D.W., then were mixed (using vortex/ 500 rpm for 30 s.) with an equal volume of antibiotic suspension (vancomycin: 100 μg/ml, nalidixic acid: 100 μg/ml, and amphotericin: 50 μg/ml) (Oxoid, UK), followed by incubation at 37 °C for 24 h.
Isolation and identification of M. avium subsp. avium from fecal samples
The processed fecal samples were streaked on Middlebrook 7H10 agar (Oxoid, UK); media was supplemented by a- 5 ml/l Glycerol (Oxoid, UK) and b-Middlebrook OADC Growth Supplement (Oxoid, UK). The inoculated plates were incubated at 37° ± 2 °C under microaerophilic conditions where a CO2 sachet (Oxoid, UK) is placed in a tightly closed anaerobic jar. The incubated plates were examined for bacterial growth at 2, 4, 6, 8, and 10 weeks post-incubation. The suspected colonies were identified according to their culture characters, morphological characters using Ziehl-Neelsen staining, and biochemically using niacin production, nitrate reduction, tween-80 hydrolysis, thermo-stable catalase at 68 °C, and arylsulfatase tests as described by Kubica [32]. Besides, the identification of the recovered isolates was confirmed by PCR-based detection of the IS901 gene (the most conserved gene in M. avium subsp. avium) as described in Table 1.
Antimicrobial susceptibility testing using broth microdilution method
The detection of minimal inhibitory concentrations (MICs) of the tested antimicrobial agents was carried out using the broth microdilution method according to the procedures of CLSI [33]. The test was performed using Middlebrook 7H10 broth (Oxoid, UK). The MIC for each antibiotic for each tested isolate was the mean of two repeated tests. Mycobacterium avium ATCC 700898 was used as a control strain. The MIC breakpoints of the tested antimicrobial agents were expressed as sensitive, intermediate, and resistant as described by the CLSI guidelines. The tested isolates were classified into Multidrug-resistant (MDR: the resistance to at least isoniazid and rifampicin), Extensively drug-resistant (XDR: resistant to isoniazid and rifampicin as well as to fluoroquinolones, and at least one of the second-line drugs such as kanamycin, amikacin, and capreomycin), and Pan-drug resistant (PDR: resistant to all antimicrobial agents listed) according to Prasanna and Niranjan [34].
Molecular detection of IS901, inhA, rpoB, rpsL, and otrB genes
PCR was used to detect the IS901 gene to confirm the diagnosis of the recovered isolates as well as to investigate the presence of inhA, rpoB, rpsL, and otrB genes in the recovered isolates. Extraction of DNA was performed using the QIAamp DNA Mini Kit (Qiagen, GmbH, Germany/ Catalogue No.51305). The PCR reaction performed in a “25- μl” reaction volume containing; “12.5 μl” of Emerald Amp Max PCR Master Mix (Takara, Japan), one μl of each primer of 20 pmol concentration, 4.5 μl of water, and 6 μl of DNA template. The oligonucleotide primers sequences (Metabion International AG, Germany) and their recycling conditions are illustrated in Table 1 [35,36,37,38,39]. Positive control strains (kindly provided by A.H.R.I, Egypt) and negative controls (DNA-free) were involved in each PCR run. Finally, the agar gel electrophoresis was carried out using 1.5% agarose stained with ethidium bromide 0.5 μg/ml (Fermentas, Germany). The gel was visualized by a gel documentation system (Thermo Fisher Scientific, Waltham, MA, USA).
Statistical analyses
The statistical analysis of the obtained findings was carried out using Chi-square (SAS software, version 9.4, SAS Institute, Cary, NC, USA) (significance level; P < 0.05). The correlation analyses were performed between different tested antimicrobial agents as well as the antibiogram results and the presence of resistance genes using R software (version 4.0.2; https//www.r-project.org/).
Results
Phenotypic characteristics of the recovered M. avium subsp. avium from house-reared domestic birds
In the present study, the colonies of the recovered M. avium subsp. avium isolates were small, round, creamy color, and sticky on a Middlebrook 7H10 agar. The colonial growth was obtained within 2 weeks after incubation. The microscopical examination of the retrieved isolates (using Ziehl-Neelsen staining) revealed non-sporulated and non-motile acid-fast bacilli. The recovered isolates were positive for the arylsulfatase test, while were negative to the niacin production, nitrate reduction, thermo-stable catalase, oxidase, and tween − 80 hydrolysis tests (Table 2).
The frequency of M. avium subsp. avium in different species of examined domestic birds
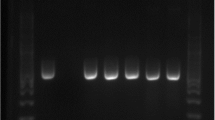
The bacteriological examination revealed that the frequency of M. avium subsp. avium was 10% (4/40) in the examined ducks and 2.5% (1/40) in the examined geese. Moreover, the examined chicken samples were negative to M. avium subsp. avium. The total frequency of M. avium subsp. avium was 4.1% (5/120). All the retrieved isolates were originated from diseased birds suffering from persistent diarrhea and emaciation (Table 3 and Fig. 1). Statistically, there is a significant difference (P < 0.5; P = 0.008783) in the frequency of M. avium subsp. avium among examined samples of different bird species.
Antibiogram of the recovered M. avium subsp. avium isolated from domestic birds
In the present study, the results of the broth microdilution method exhibited harmony with the results of the disc diffusion method. Four isolates were resistant to doxycycline, streptomycin, oxytetracycline rifampicin, and isoniazid with detectable MICs of > 8 μg/ml, > 8 μg/ml, > 8 μg/ml, > 2 μg/ml, and > 0.25 μg/ml, respectively. All the tested isolates (n = 5) were sensitive to azithromycin (MIC ≤2 μg/ml) and clofazimine, while only one isolate was sensitive to rifampicin. Moreover, all the tested isolates were resistant to isoniazid (MIC > 0.25 μg/ml) (Table 4 and Fig. 2). The correlation analyses were conducted among different tested antimicrobial agents. The obtained results showed strong positive correlations between (0.5–1): CFZ and AZM (r = 1); RF, STR, and OT(r = 1); DOX, STR, and OT(r = 1); OT, DOX, RF, and INH (r = 0.97); STR and OT (r = 0.97) and OT, DOX, and STR (r = 0.88) as described in Fig. 3.
PCR-based detection of IS901confirmatory gene and inhA, rpoB, rpsL, and otrB antibiotic resistance-related genes in the recovered M. avium subsp. avium isolates
In the present study, the identification of the recovered isolates is confirmed using PCR, where all the tested isolates were positive for the IS901 confirmatory gene with a specific amplicon size of 577 bp. Moreover, PCR was used to detect the inhA, rpoB, rpsL, and otrB antibiotic resistance-related genes. Our finding revealed that all the tested isolates (100%, 5/5) (four duck isolates and one goose isolate) are harbored inhA genes with a specific amplicon size of 810 bp. Moreover, the rpoB, rpsL, and otrB resistance genes are detected in four recovered isolates (80%, 4/5) (Three of duck origin and one isolate of geese origin) with specific amplicon size of 347 bp, 592 bp, and 947 bp, respectively. The distribution of IS901, inhA, rpoB, rpsL, and otrB genes among the recovered M. avium subsp. avium isolates was illustrated in Table 5 and Fig. 4. Statistically, there is no significant difference in the distribution of IS901, inhA, rpoB, rpsL, and otrB genes in the recovered isolates (P > 0.05, P = 0.9915) as illustrated in Table 5.
The correlation between the phenotypic and genotypic resistance patterns of the isolated M. avium subsp. avium
Regarding the correlation between the in-vitro antibiotic resistance and the genotypic resistance patterns, our finding proved that 80% (4/5) of the retrieved M. avium subsp. avium isolates (three isolates of duck origin and one isolate of geese origin) exhibited multidrug resistance patterns to isoniazid, rifampicin, streptomycin, oxytetracycline, and doxycycline, and harbored the inhA rpoB, rpsL, and otrB genes as described in Table 6. The correlation analyses were conducted between various tested antimicrobial agents and the detected antibiotic resistance-related genes in the recovered M. avium subsp. avium isolates. Our results proved strong positive correlations (r = 0.5–1) between: ropB gene and RF (r = 1); inhA gene and INH (r = 0.97); otrB and OT(r = 0.88); rpsL gene and STR (r = 0.88); otrB gene and DOX (r = 0.88) as illustrated in Fig. 5.
Discussion
Regarding the phenotypic characteristics of M. avium subsp. avium, our findings proved no diversity in both morphological and biochemical characteristics among the recovered isolates and revealed a remarkable harmony between different retrieved isolates from duck and geese origins. Our results agreed with those reported by Zhu et al. [35] and Bhalla et al. [40]. The decontamination of fecal samples using 0.9% Hexadecylpryridinium Chloride Monohydrate and the antibiotic mixture is the best method to increase the recovery of M. avium subsp. avium from the fecal samples of examined birds [31]. The culture technique is considered the gold standard method used for the identification of M. avium subsp. avium as it is the most sensitive and specific method (never give false-positive results), but it requires more time [17]. On the other hand, the direct microscopical examination of acid-fast bacilli is not accurate and requires a high load of microorganisms (Approximately, 10.000 bacteria/g) to be effective. Moreover, it may give false-negative results due to its low sensitivity and specificity [41, 42].
In the present study, the frequency of M. avium subsp. avium in the examined house-reared birds agreed with that previously reported by Kindu and Dagnaw [13] who isolated M. avium subsp. avium from domestic birds in Ethiopia with a prevalence of 4.26% (12/282). Moreover, in Bangladesh, Reza et al. [43] reported that the prevalence of M. avium subsp. avium in droppings of different examined birds was 3.75% (3/80). On the other hand, in the Czech Republic, Shitaye et al. [44], and Kazda et al. [45] reported that turkeys are susceptible to M. avium subsp. avium, while ducks, geese, and water birds are resistant to avian tuberculosis. Furthermore, higher prevalence (11.1%) of M. avium subsp. avium was reported in the Czech Republic by Dvorska [46]. The prevalence of infection in birds mainly increased due to the bad hygienic conditions, overcrowding, and multi-aged birds’ populations. Moreover, some birds are highly susceptible to avian tuberculosis due to genetic factors [47, 48]. The prevalence of infection in birds mainly increased due to the bad hygienic conditions, overcrowding, and multi-aged birds’ populations. Moreover, some birds are highly susceptible to avian tuberculosis due to genetic factors [49]. The infection of birds occurs due to the ingestion of contaminated food and water with feces of infected birds, and also may occur via the respiratory route [6]. Besides, avian tuberculosis could be transmitted to the immunocompromised persons through the handling of carrier birds or via the ingestion of insufficiently cooked meat of infected birds [8].
In the current study, PCR was used to confirm the diagnosis of the recovered isolates depending on the detection of the IS901 gene that is specific to M. avium subsp. avium. Our results emphasized the presence of the IS901 gene in all tested isolates. Our findings agreed with those reported by Pavlik et al. [49] who detected the IS901 gene in all tested M. avium subsp. avium strains originated from birds. M. avium subsp. avium are characterized by the existence of the specific insertion sequence IS901 in their genome. PCR could be used as a molecular tool to detect the pathogenic variant of M. avium subsp. avium as well as the screening of elderly birds [50].
Concerning the antibiogram of the retrieved M. avium subsp. avium isolates, our findings emphasized the promising in-vitro antimicrobial activity of azithromycin and clofazimine. M. avium complex commonly exhibits a potential sensitivity to macrolides, especially azithromycin [51]. Moreover, Huang et al. [52] revealed that clofazimine gives optimistic results in the treatment of M. avium-associated infections. Higher frequencies of antimicrobial resistances were identified to doxycycline, streptomycin, oxytetracycline, rifampicin, and isoniazid using the broth microdilution method. In a previous study by Pang et al. [50], among the recovered Mycobacterium avium complex strains, Mycobacterium avium was the most resistant to the tested antimicrobials with a resistance rate of 73.68%. In the present study, most of the retrieved M. avium subsp. avium isolates were resistant to isoniazid, rifampicin, streptomycin, oxytetracycline, and doxycycline, and harbored the inhA, rpoB, rpsL, and otrB genes. The multidrug resistance in Mycobacterium species is defined as the resistance to at least both isoniazid and rifampicin [53]. The complicated cell wall structure, the presence of mycolic acid, and the lipoidal contents are the main factors that control the intrinsic multidrug-resistance of the M. avium complex [15]. The resistance to rifampicin is common in the M. avium complex and mainly occurred due to the mutation of the rpoB gene that encoding for the β-subunit of the DNA-dependent RNA polymerase M. avium complex [54]. Moreover, the resistance to the oxytetracycline is attributed to the presence of the otrB gene encoding for the integral membrane protein that is responsible for the efflux of oxytetracycline from the cell [55]. Aminoglycosides exert their action by ribosomal binding near site A with subsequent inhibition of protein synthesis [56]. The Resistance to streptomycin is common in the M. avium complex and occurred due to mutations in the rpsL gene encoding the ribosomal protein (S12) [57]. The inhA gene encodes for the enoyl acyl reductase which is involved in the mycolic acid synthesis that is responsible for the isoniazid resistance [58]. Briefly, the existence of multidrug resistance in the M. avium complex is attributed to several factors including 1-The intrinsic resistance due to the complex thick cell wall and the presence of mycolic acid, 2-The presence of specific antimicrobial resistance genes, and 3-Mutations that takes place in certain genes such as rpsL and rpoB genes that adversely affect the activity of the antimicrobial agents [59].
Study limitations
Gene sequencing of the antibiotic-resistance related genes of isolates (rpoB and rpsL) should be carried out for the detection of mutations that adversely affect the activity of the antimicrobial agents.
Conclusion
In conclusion, to the best of our knowledge, this is the first report that emphasized the emergence of MDR-M.avium subsp. avium in house-reared domestic birds (duck and geese) in Egypt. The emergence of MDR-M. avium subsp. avium is considered a public health threat. The combination of phenotypic and genotypic characterization is an effective epidemiological tool for the identification of M. avium subsp. avium. Emerging MDR-M. avium subsp. avium in house-reared domestic birds are commonly harbored the IS901, inhA, rpoB, rpsL, and otrB genes. Azithromycin and clofazimine revealed a promising in-vitro antimicrobial activity against M. avium subsp. avium. The continuous monitoring of bird flocks is essential to reduce the prevalence of avian tuberculosis. The regular application of antimicrobial susceptibility testing is necessary to determine the most effective antimicrobial agent and detect the emerging MDR-strains.
Availability of data and materials
All the data has been included in the manuscript.
Abbreviations
- OIE:
-
World Organization for Animal Health
- CLSI:
-
Clinical and Laboratory Standards Institute
- PDR:
-
Pan-drug resistant.
- XDR:
-
Extensive drug-resistant
- MDR:
-
Multi-drug resistant
References
Dhama K, Mahendran M, Tiwari R, Dayal Singh S, Kumar D, Singh S, et al. Tuberculosis in birds: insights into the Mycobacterium avium infections. Vet Med Int. 2011;2011:712369.
Dhama K, Mahendran M, Tomar S. Avian tuberculosis: an overview. Poultry Punch. 2007;24(3):38–52.
Schorey JS, Sweet L. The mycobacterial glycopeptidolipids: structure, function, and their role in pathogenesis. Glycobiology. 2008;18(11):832–41. https://doi.org/10.1093/glycob/cwn076.
Coelho AC, de Lurdes Pinto M, Matos A, Matos M, dos Anjos Pires M: Mycobacterium avium complex in domestic and wildanimals: InTechOpen; 2013.
Leite CQF, Souza CWOd, Leite SRdA: Identification of mycobacteria by thin layer chromatographic analysis of mycolic acidsand conventional biochemical method: four years of experience. Mem Inst Oswaldo Cruz. 1998;93(6):801–5. . https://doi.org/10.1590/S0074-02761998000600019.
Tell LA, Woods L, Cromie R. Mycobacteriosis in birds. Revue scientifique et technique (International Office of Epizootics). 2001;20(1):180–203. https://doi.org/10.20506/rst.20.1.1273.
Tell LA, Woods L, Foley J, Needham ML, Walker RL. A model of avian mycobacteriosis: clinical and histopathologic findings in Japanese quail (Coturnix coturnix japonica) intravenously inoculated with Mycobacterium avium. Avian Dis. 2003;47(2):433–43. https://doi.org/10.1637/0005-2086(2003)047[0433:AMOAMC]2.0.CO;2.
Moravkova M, Lamka J, Kriz P, Pavlik I. The presence of Mycobacterium avium subsp. avium in common pheasants (Phasianus colchicus) living in captivity and in other birds, vertebrates, non-vertebrates and the environment. Vet Med (Praha). 2011;56(56):333–43. https://doi.org/10.17221/1588-VETMED.
Broomand V, Namazizadeh M: The relationship between the bacteria Mycobacterium tuberculosis Avium in human city ofArdabil. In: Biological Forum: 2015: Research Trend; 2015:61.
Babacan O, Bülent B, SAREYYÜPOĞLU B. PCR detection of Mycobacterium genavense DNA in fecal samples of caged birds. Ankara Üniversitesi Veteriner Fakültesi Dergisi. 2020;67(2):201–4.
Radkowski M, Uradziński J, Szteyn J. The occurrence of infectious and parasitic diseases in poultry slaughtered in the district of Olsztyn, Poland, 1986-91. Avian Dis. 1996;40(2):285–9. https://doi.org/10.2307/1592222.
Kahn CM, Line S, Aiello S. The merck veterinary manual. Whitehouse Station: Merck & Co. Inc; 2005.
Kindu A, Getaneh G. Prevalence of avian tuberculosis in domestic chickens in selected sites of Ethiopia. J Vet Sci Technol. 2016;7(377):2.
Hoenerhoff M, Kiupel M, Sikarskie J, Bolin C, Simmons H, Fitzgerald S. Mycobacteriosis in an American bald eagle (Haliaeetus leucocephalus). Avian Dis. 2004;48(2):437–41. https://doi.org/10.1637/7133.
Gygli SM, Borrell S, Trauner A, Gagneux S. Antimicrobial resistance in Mycobacterium tuberculosis: mechanistic and evolutionary perspectives. FEMS Microbiol Rev. 2017;41(3):354–73. https://doi.org/10.1093/femsre/fux011.
Rocco J, Irani V. Mycobacterium avium and modulation of the host macrophage immune mechanisms. Int J Tuberculosis Lung Dis. 2011;15(4):447–52. https://doi.org/10.5588/ijtld.09.0695.
Manual OOT. OIE; Paris, France: 2014. Chapter. 2014;2(12):1–15.
Algammal AM, Wahdan A, Elhaig MM. Potential efficiency of conventional and advanced approaches used to detect Mycobacterium bovis in cattle. Microb Pathog. 2019;134:103574. https://doi.org/10.1016/j.micpath.2019.103574.
Algammal AM, Mohamed MF, Tawfiek BA, Hozzein WN, El Kazzaz WM, Mabrok M. Molecular typing, antibiogram and PCR-RFLP based detection of Aeromonas hydrophila complex isolated from Oreochromis niloticus. Pathogens. 2020;9(3):238. https://doi.org/10.3390/pathogens9030238.
Algammal AM, Hetta HF, Elkelish A, Alkhalifah DHH, Hozzein WN, Batiha GE-S, et al. Methicillin-resistant Staphylococcus aureus (MRSA): one health perspective approach to the bacterium epidemiology, virulence factors, antibiotic-resistance, and zoonotic impact. Infect Drug Resistance. 2020;13:3255–65. https://doi.org/10.2147/IDR.S272733.
El-Sayed M, Algammal A, Abouel-Atta M, Mabrok M, Emam A. Pathogenicity, genetic typing, and antibiotic sensitivity of vibrio alginolyticus isolated from Oreochromis niloticus and Tilapia zillii. Rev Med Vet. 2019;170:80–6.
Algammal AM, Enany ME, El-Tarabili RM, Ghobashy MO, Helmy YA. Prevalence, antimicrobial resistance profiles, virulence and enterotoxin-determinant genes of MRSA isolated from subclinical bovine mastitis samples in Egypt. Pathogens. 2020;9(5):362. https://doi.org/10.3390/pathogens9050362.
Algammal AM, El-Kholy AW, Riad EM, Mohamed HE, Elhaig MM, Yousef SAA, et al. Genes encoding the virulence and the antimicrobial resistance in enterotoxigenic and shiga-toxigenic E. coli isolated from diarrheic calves. Toxins. 2020;12(6):383.
Algammal AM, El-Sayed ME, Youssef FM, Saad SA, Elhaig MM, Batiha GE, et al. Prevalence, the antibiogram and the frequency of virulence genes of the most predominant bacterial pathogens incriminated in calf pneumonia. AMB Express. 2020;10(1):1–8.
Enany ME, Algammal AM, Nasef SA, Abo-Eillil SA, Bin-Jumah M, Taha AE, et al. The occurrence of the multidrug resistance (MDR) and the prevalence of virulence genes and QACs resistance genes in E. coli isolated from environmental and avian sources. AMB Express. 2019;9(1):1–9.
Abolghait SK, Fathi AG, Youssef FM, Algammal AM. Methicillin-resistant Staphylococcus aureus (MRSA) isolated from chicken meat and giblets often produces staphylococcal enterotoxin B (SEB) in non-refrigerated raw chicken livers. Int J Food Microbiol. 2020;328:108669. https://doi.org/10.1016/j.ijfoodmicro.2020.108669.
Nasiri MJ, Haeili M, Ghazi M, Goudarzi H, Pormohammad A, Imani Fooladi AA, et al. New insights in to the intrinsic and acquired drug resistance mechanisms in mycobacteria. Front Microbiol. 2017;8:681. https://doi.org/10.3389/fmicb.2017.00681.
Brown-Elliott BA, Philley JV, Benwill JL, Wallace RJ. Current opinions in the treatment of pulmonary nontuberculous mycobacteria in non-cystic fibrosis patients: Mycobacterium abscessus group, Mycobacterium avium complex, and Mycobacterium kansasii. Curr Treat Options Infect Dis. 2014;6(4):392–408. https://doi.org/10.1007/s40506-014-0032-2.
Parashar D, Das R, Chauhan D, Sharma V, Lavania M, Yadav V, et al. Identification of environmental mycobacteria isolated from Agra, North India by conventional & molecular approaches. Indian J Med Res. 2009;129(4):424–31.
Payeur, J.B.: Current culture methods for Mycobacterium avium subspecies paratuberculosis. Proceedings of 81CP 2005, 2005:352–358.
Sattar A, Zakaria Z, Abu J, Aziz SA, Gabriel R-P. Evaluation of six decontamination procedures for isolation of Mycobacterium avium complex from avian feces. PLoS One. 2018;13(8):e0202034. https://doi.org/10.1371/journal.pone.0202034.
Kubica GP. Differential identification of mycobacteria: VII. Key features for identification of clinically significant mycobacteria. Am Rev Respir Dis. 1973;107(1):9–21.
CLSI: Clinical and Laboratory Standards Institute. "Performance standards for susceptibility testing of mycobacteria,Nocardia spp., and other aerobic actinomycetes." CLSI supplement M62 (2018).
Prasanna A, Niranjan V. Classification of Mycobacterium tuberculosis DR, MDR, XDR isolates and identification of signature mutationpattern of drug resistance. Bioinformation. 2019;15(4):261–8. https://doi.org/10.6026/97320630015261.
Zhu L, Peng Y, Ye J, Wang T, Bian Z, Qin Y, et al. Isolation, identification, and characterization of a new highly Pathogenic Field isolate of Mycobacterium avium spp. avium. Front Vet Sci. 2018;4:243.
Kongpetchsatit O, Phatihattakorn W, Mahakunkijcharoen Y, Eampokalarp B, Boonyasopun J, Ramasoota P. Mutation in the rpoB gene of the rifampicin resistant M avium complex strains from Thailand. Southeast Asian J Trop Med Public Health. 2006;37:165.
Pang H, Wan K, Wei L. Single-nucleotide polymorphisms related to fluoroquinolone and aminoglycoside resistance in Mycobacterium avium isolates. Infect Drug Resistance. 2018;11:515–21. https://doi.org/10.2147/IDR.S160899.
Kyselková M, Chroňáková A, Volná L, Nĕmec J, Ulmann V, Scharfen J, Elhottová D: Tetracycline resistance and presenceof tetracycline resistance determinants tet (V) and tap in rapidly growing mycobacteria from agricultural soils and clinical isolates.Microbes Environments 2012:ME12028.
Tseng S-T, Tai C-H, Li C-R, Lin C-F, Shi Z-Y. The mutations of katG and inhA genes of isoniazid-resistant Mycobacterium tuberculosis isolates in Taiwan. J Microbiol Immunol Infect. 2015;48(3):249–55. https://doi.org/10.1016/j.jmii.2013.08.018.
Bhalla GS, Sarao MS, Kalra D, Bandyopadhyay K, John AR. Methods of phenotypic identification of non-tuberculous mycobacteria. Practical Laboratory Med. 2018;12:e00107. https://doi.org/10.1016/j.plabm.2018.e00107.
Cudahy P, Shenoi SV. Diagnostics for pulmonary tuberculosis. Postgrad Med J. 2016;92(1086):187–93. https://doi.org/10.1136/postgradmedj-2015-133278.
Gerlach H. Bacteria. Avian medicine: principles and application. 1994;142:949–83.
Reza MR, Lijon MB, Khatun MM, Islam MA. Prevalence and antibiogram profile of Mycobacterium spp. in poultry and its environments. J Adv Vet Animal Res. 2015;2(4):458–63. https://doi.org/10.5455/javar.2015.b118.
Shitaye J, Matlova L, Horvathova A, Moravkova M, Dvorska-Bartosova L, Treml F, et al. Mycobacterium avium subsp. avium distribution studied in a naturally infected hen flock and in the environment by culture, serotyping and IS901 RFLP methods. Vet Microbiol. 2008;127(1–2):155–64. https://doi.org/10.1016/j.vetmic.2007.07.026.
Kazda J, Pavlik I, Falkinham JO III, Hruska K. The ecology of mycobacteria: impact on animal's and human's health: Springer Science & Business Media; 2010.
Dvorska L, Matlova L, Ayele W, Fischer O, Amemori T, Weston R, et al. Avian tuberculosis in naturally infected captive water birds of the Ardeideae and Threskiornithidae families studied by serotyping, IS901 RFLP typing, and virulence for poultry. Vet Microbiol. 2007;119(2–4):366–74. https://doi.org/10.1016/j.vetmic.2006.09.010.
Saggese MD, Tizard I, Phalen DN. Comparison of sampling methods, culture, acid-fast stain, and polymerase chain reaction assay for the diagnosis of mycobacteriosis in ring-neck doves (Streptopelia risoria). Journal of avian medicine and surgery. 2010;24(4):263–71. https://doi.org/10.1647/2008-009.1.
Saggese MD, Riggs G, Tizard I, Bratton G, Taylor R, Phalen DN. Gross and microscopic findings and investigation of the aetiopathogenesis of mycobacteriosis in a captive population of white-winged ducks (Cairina scutulata). Avian Pathology. 2007;36(5):415–22. https://doi.org/10.1080/03079450701595909.
Pavlik I, Svastova P, Bartl J, Dvorska L, Rychlik I. Relationship between IS901 in theMycobacterium avium complex strains isolated from birds, animals, humans, and the environment and virulence for poultry. Clin Diagn Lab Immunol. 2000;7(2):212–7. https://doi.org/10.1128/CDLI.7.2.212-217.2000.
Haque M, Rima U, Hossain M, Islam M, Chowdhury S, Hossain M, et al. Standardize polymerase chain reaction (PCR) technique for the detection of pathogenic serovars of Mycobacterium a. avium infection in layer chicken. Asian J Poultry Sci. 2016;10(4):175–83. https://doi.org/10.3923/ajpsaj.2016.175.183.
Dovriki E, Papaioannou A, Gourgouliannis K: Identification and Drug Treatment of Nontuberculous Mycobacteria Isolated from Patients Lived in Greece during 2000–2017. of 2018, 9:25–50.
Huang C-C, Wu M-F, Chen H-C, Huang W-C. In vitro activity of aminoglycosides, clofazimine, d-cycloserine and dapsone against 83 Mycobacterium avium complex clinical isolates. J Microbiol Immunol Infect. 2018;51(5):636–43. https://doi.org/10.1016/j.jmii.2017.05.001.
Organization WH: Multidrug and extensively drug-resistant TB (M. In: World Health Organization; 2010.
Salah IB, Adekambi T, Raoult D, Drancourt M. rpoB sequence-based identification of Mycobacterium avium complex species. Microbiology. 2008;154(12):3715–23. https://doi.org/10.1099/mic.0.2008/020164-0.
Pang Y, Brown B, Steingrube V, Wallace R, Roberts M. Tetracycline resistance determinants in Mycobacterium and Streptomyces species. Antimicrob Agents Chemother. 1994;38(6):1408–12. https://doi.org/10.1128/AAC.38.6.1408.
Sanz-García F, Anoz-Carbonell E, Pérez-Herrán E, Martín C, Lucía A, Rodrigues L, et al. Mycobacterial aminoglycoside acetyltransferases: a little of drug resistance, and a lot of other roles. Front Microbiol. 2019;10:46. https://doi.org/10.3389/fmicb.2019.00046.
Springer B, Kidan YG, Prammananan T, Ellrott K, Böttger EC, Sander P. Mechanisms of streptomycin resistance: selection of mutations in the 16S rRNA gene conferring resistance. Antimicrob Agents Chemother. 2001;45(10):2877–84. https://doi.org/10.1128/AAC.45.10.2877-2884.2001.
Sharma BK, Bhandari S, Maharjan B, Shrestha B, Banjara MR. Rapid detection of rifampicin and isoniazid resistant mycobacterium tuberculosis using genotype mtbdrplus assay in Nepal. Int Scholarly Res Notices. 2014;2014.
Parker H, Lorenc R, Ruelas Castillo J, Karakousis PC. Mechanisms of antibiotic tolerance in Mycobacterium avium complex: lessons from related mycobacteria. Front Microbiol. 2020;11:2348.
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
A.M.A. conception and study design. A.M.A, E.M.E, A.W.E, H.R.H, H.F.H, K.J.A, E. M, A.S.A, H. R, and R.M.E; conducted the experiments. A.M.A. and E.M.E. drafted the manuscript. A.M.A, K.J.A, E.M.E, E. M, A.S.A, H.R.H, H.F.H, A.W.E, H. R, and R.M.E; acquisition of data, statistical analysis, and interpretation of data. AMA; critically reviewing the article. All authors have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The handling of birds and all used protocols were approved by the Animal Ethics Review Committee of Suez Canal University (AERC-SCU), Egypt. The written informed consent is not applicable. We confirmed that all methods were performed in accordance with the relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Algammal, A.M., Hashem, H.R., Al-otaibi, A.S. et al. Emerging MDR-Mycobacterium avium subsp. avium in house-reared domestic birds as the first report in Egypt. BMC Microbiol 21, 237 (2021). https://doi.org/10.1186/s12866-021-02287-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-021-02287-y