Abstract
Background
The study used a set of 407 human extraintestinal pathogenic E. coli strains (ExPEC) isolated from (1) skin and soft tissue infections, (2) respiratory infections, (3) intra-abdominal infections, and (4) genital smears. The set was tested for bacteriocin production, for prevalence of bacteriocin and virulence determinants, and for phylogenetic typing. Results obtained from the group of ExPEC strains were compared to data from our previously published analyses of 1283 fecal commensal E. coli strains.
Results
The frequency of bacteriocinogeny was significantly higher in the set of ExPEC strains (63.1 %), compared to fecal E. coli (54.2 %; p < 0.01). Microcin producers and microcin determinants dominated in ExPEC strains, while colicin producers and colicin determinants were more frequent in fecal E. coli (p < 0.01). Higher production of microcin M and lower production of microcin B17, colicin Ib, and Js was detected in the set of ExPEC strains. ExPEC strains had a significantly higher prevalence of phylogenetic group B2 (52.6 %) compared to fecal E. coli strains (38.3 %; p < 0.01).
Conclusions
Human ExPEC strains were shown to differ from human fecal strains in a number of parameters including bacteriocin production, prevalence of several bacteriocin and virulence determinants, and prevalence of phylogenetic groups. Differences in these parameters were also identified within subgroups of ExPEC strains of diverse origin. While some microcin determinants (mM, mH47) were associated with virulent strains, other bacteriocin types (mB17, Ib, and Js) were associated with fecal flora.
Similar content being viewed by others
Background
Extraintestinal pathogenic E. coli (ExPEC) strains colonize various sites in the human body and cause diverse extraintestinal infections [1, 2]. Compared to commensal E. coli strains, extraintestinal strains have larger genomes and encode more virulence factors [3]. The group of ExPEC strains includes uropathogenic E. coli strains, septicemia-associated E. coli, meningitis-associated E. coli, and other strains [4]. ExPEC strains typically encode i) virulence factors that allow them to bind to human cells (e.g. P-fimbriae, S-fimbriae) [5, 6], ii) factors important for survival in the human body (e.g. siderophores), and iii) factors capable of damaging human cells and tissues (e.g. hemolysin, cytotoxin necrotizing factor) [7].
Another important feature of pathogenic and also commensal E. coli strains is production of bacteriocins [8–12]. Colicins and microcins (bacteriocins) are antibacterial proteins or peptides, respectively, differing in a number of characteristics including molecular mass, presence of post-translational modifications, export from producer cells, etc. Colicin Js is known to share features of both colicins and microcins [9, 10, 13, 14]. Bacteriocin production has been shown to play an important ecological role in bacterial competition [15]. In addition to antimicrobial activity, several bacteriocin types have also been shown to inhibit proliferation of eukaryotic cells [16]. In previous reports, an association of bacteriocin production with E. coli virulence factors was described [10–12, 17] as well as a positive correlation between the frequency of bacteriocinogeny and the number of virulence factors encoded by E. coli strains [17]. In a former study, production of virulence factors typical for the ExPEC pathotype (sfa, pap, aer, iucC, cnf1, α-hly), in the set of fecal E. coli strains, was associated with a higher frequency of bacteriocinogeny, a higher prevalence of bacteriocin multi-producers, and a greater abundance of microcins H47, M, V, B17, and colicins E1, Ia, and S4 [17]. In addition, production of microcin types H47, M, I47, E492, and V, and colicin E1 was associated with the uropathogenic E. coli pathotype [10–12]. Higher production of microcin V was also detected in septicemia-associated ExPEC strains isolated from blood [18].
In this study, we characterized a set of 407 human extraintestinal pathogenic E. coli isolated from different body sites and determined the prevalence of 30 bacteriocin determinants representing most of the known bacteriocin types. In addition, we determined 18 virulence determinants typical of intestinal and extraintestinal pathogenic E. coli strains and also the main phylogroups of E. coli (A, B1, B2, and D). ExPEC strains were isolated from skin and soft tissue infections, as well as from respiratory, intra-abdominal, and genital infections. Results obtained in this study were compared to a previously characterized and published set of 1283 human fecal E. coli strains [19, 20].
Results
Origin of extraintestinal pathogenic E. coli and fecal strains
The ExPEC strains (n = 407) were isolated between 2007 and 2012 from patients attending the University Hospital in Brno, Czech Republic (Table 1 and Additional file 1: Table S1) and characteristics of these strains are shown in Table 1. The ExPEC strains were isolated from patients suffering from skin and/or soft tissue infections (SSTIs) (n = 154), from patients with respiratory infections (n = 111), from patients with intra-abdominal infections (n = 87), and from patients with genital infections (n = 55).
A set of 1283 E. coli strains of fecal origin had been isolated from patients in the Czech Republic during the same years (i.e. 2007–2012) and previously described in detail [19, 20].
Detection of virulence factors in ExPEC and fecal strains
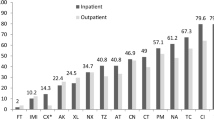
Results of detection of 18 DNA determinants (pCVD432, α-hly, afaI, aer, cnf1, sfa, pap, ial, lt, st, bfpA, eaeA, ipaH, iucC, fimA, stx1, stx2, and ehly) encoding 17 different virulence factors in both ExPEC and fecal strains are shown in Fig. 1 and Additional file 2: Table S2. Virulence genes encoding aerobactin synthesis (iucC, aer), fimbriae type 1 (fimA), S (sfa) and P (pap), afimbrial adhesin I (afaI), cytotoxic necrotizing factor (cnf1), and α-hemolysin (α-hly) were significantly more common in the set of ExPEC strains, compared to fecal E. coli strains (Fig. 1). No virulence determinant was found to be more common among fecal E. coli strains, compared to ExPEC strains.
Significant differences in the prevalence of virulence determinants, E. coli phylogroups, bacteriocin producers, and the prevalence of bacteriocin determinants among ExPEC and fecal strains (for all results see Additional file 3: Table S3). Note: statistically significant results with *0.05 > p > 0.01 or with **p < 0.01; ***bacteriocin producers
Distribution of E. coli phylogenetic groups in ExPEC and fecal strains
Among 407 tested ExPEC strains, the most prevalent phylogenetic group was B2 (n = 214; 52.6 %), followed by phylogroup D (n = 75; 18.4 %), A (n = 75; 18.4 %), and B1 (n = 43; 10.6 %). Compared to fecal E. coli strains, ExPEC strains had a higher prevalence of phylogenetic group B2 (38.3 vs. 52.6 %, respectively; p < 0.01) (Fig. 1 and Additional file 2: Table S2).
Bacteriocinogeny and bacteriocin types in ExPEC and fecal strains
The overall frequency of bacteriocinogeny was significantly higher in the set of ExPEC strains (63.1 %), compared to fecal E. coli strains (54.2 %; p < 0.01) (Fig. 1). While strains producing colicins, but no microcins, were under-represented among ExPEC bacteriocinogenic strains compared to fecal bacteriocinogenic strains (20.2 vs. 28.8 %, respectively; p < 0.01), strains producing only microcins (i.e., no colicin types) were more prevalent (45.1 vs. 30.8 %, respectively; p < 0.01).
The distribution of bacteriocin determinants encoding different bacteriocin types was different between ExPEC and fecal strains. Among ExPEC bacteriocinogenic strains, a higher prevalence of the microcin M determinant (41.2 vs. 23.5 %, respectively; p < 0.01) and a lower prevalence of determinants encoding microcin B17 (5.4 vs. 12.2 %, respectively; p < 0.01), colicins Ib (3.1 vs. 8.9 %; respectively; p < 0.01), and Js (1.6 vs. 4.3 %, respectively; p = 0.05) was detected, compared to fecal E. coli (Fig. 1 and Additional file 2: Table S2).
In general, microcin determinants were more prevalent, while colicin determinants were less prevalent among ExPEC strains compared to fecal strains (Fig. 1 and Additional file 2: Table S2).
Detection of individual bacteriocin determinants in phylogroups of ExPEC and fecal strains
In ExPEC strains, there was a 64.4 % frequency of bacteriocinogeny in the phylogenetic groups A + B1 and 62.6 % in the phylogroups B2 + D. In fecal strains, the frequency of bacteriocinogeny was significantly lower (p = 0.03) in the less pathogenic phylogroups A + B1 (49.9 %), compared to phylogroups B2 + D (56.5 %). Since previous reports showed that several bacteriocin determinants were associated with E. coli phylogroups [20], we tested for the prevalence of bacteriocin determinants within less and more virulent phylogroups (A + B1 vs. B2 + D, respectively, Table 2 and Additional file 3: Table S3). Bacteriocin type mC7 was found to be more common among ExPEC strains of phylogroup A and B1 compared to the same fecal phylogroups. In contrast to microcin C7, the bacteriocin determinant encoding mM was more common in B2 + D phylogroups of ExPEC compared to fecal strains (Table 2). At the same time, microcin B17 was found to be under-represented among ExPEC strains of phylogroups B2 and D.
Virulence factors, E. coli phylogroups and bacteriocinogeny within ExPEC strains
Compared to fecal E. coli strains, ExPEC strains isolated from patients suffering from skin and/or soft tissue infections (SSTIs) (n = 154) showed higher prevalence of determinants encoding aerobactin synthesis and uptake, fimbriae type I, and afimbrial adhesin I (Fig. 2 and Additional file 4: Table S4). The ExPEC strains from patients with respiratory infections (n = 111) showed a higher prevalence of determinants encoding aerobactin synthesis and uptake, fimbriae type I, α-hemolysin, cytotoxic necrotizing factor, and S-fimbriae. ExPEC from patients with intra-abdominal infections (n = 87) had a higher prevalence of determinants encoding aerobactin synthesis and uptake. ExPEC from patients with genital infections (n = 55) showed no detectable differences in encoded virulence factors compared to fecal strains (Fig. 2 and Additional file 4: Table S4).
Significant differences in the distribution of virulence factors, E. coli phylogroups, bacteriocin production, and bacteriocin determinants between subgroups of ExPEC and fecal strains (for all results see Additional file 4: Table S4). Note: statistically significant results with *0.05 > p > 0.01 or with **p < 0.01; ***bacteriocin producers
ExPEC strains from SSTIs, strains from respiratory infections, and strains from genital infections, but not strains from intra-abdominal infections, showed a higher prevalence of phylogroup B2 (p < 0.01, Fig. 2) compared to fecal strains. In addition, the set of strains isolated from SSTIs also had a lower prevalence of phylogenetic group A (p < 0.01) compared to fecal E. coli strains (Fig. 2).
Within ExPEC strains, the highest rate of bacteriocinogeny was found in E. coli strains isolated from genital smears (80.0 %). Compared to fecal strains, strains isolated from SSTIs and strains from genital infections showed a higher prevalence of the mM determinant. Strains from respiratory infections showed a higher prevalence of determinants encoding both mM and mH47 (Fig. 2). Both, ExPEC strains from respiratory infections, and strains from genital infections, showed a higher prevalence of microcin determinants, respectively (p < 0.01). At the same time, these strains showed a lower prevalence of colicin determinants.
Discussion
ExPEC strains characterized in this study were isolated from various sites of infection including skin and/or soft tissue infections, respiratory, intra-abdominal, and genital infections. Compared to fecal strains isolated during the same time period in a similar geographical area, ExPEC strains showed a higher prevalence of seven known virulence factors. These virulence factors comprised adherence factors, aerobactin synthesis and uptake, and cytotoxin synthesis, which is consistent with increased ability to colonize extraintestinal body sites and represents the initial step in the development of extraintestinal infections. These findings are in agreement with previous studies on ExPEC strains [5–7, 21–25].
Phylogenetic analysis revealed that phylogenetic group B2 was dominant in the set of extraintestinal pathogenic E. coli tested in this study. A high prevalence of phylogenetic group B2 is typical of more virulent E. coli strains [26] and a higher prevalence of phylogenetic group B2 (35.0 %) has been described in E. coli strains isolated from bacteremia [27] and in uropathogenic strains (55.0 %) [28]. In a different set of E. coli strains isolated from blood, wound, swab, pus, urine, cerebrospinal fluid, ascitic fluid, and intravascular devices, phylogroups B2 (35.0 %) and D (36.0 %) were found to be most prevalent [29]. Molecular epidemiology, based on MLST (Multilocus sequence typing), also revealed that the phylogroup B2 was correlated with the ExPEC pathotype [23]. ExPEC strains characterized in this study were clearly more virulent compared to fecal strains.
Compared to fecal strains from phylogroup A and B1, microcin type C7 was found to be more common among ExPEC strains of the same phylogroups. These data are consistent with the role of bacteriocin genes regarding increased virulence of E. coli strains. Alternatively to bacteriocin genes, genes encoded on the same plasmids could contribute to the increased virulence of these strains since mC7 plasmids are relatively large with a number of genes of predicted and unknown functions [30–33].
A higher frequency of bacteriocin determinant mM among B2 + D phylogroups suggest that microcin M and/or genes in the same linkage group contribute to the virulence of ExPEC strains. Previous reports have shown that the mM and mH47 locus is abundant in uropathogenic isolates, while the cluster seldom appeared in intestinal or in other extraintestinal E. coli isolates [10–12, 17, 34]. Our study showed that the percentage of strains coding for mM and mH47 differed among ExPEC strains of diverse origin suggesting that the source of extraintestinal E. coli isolates affects the prevalence of mM and mH47 determinants. Despite the fact that microcins H47 and M are typically produced together, sequence analyses have revealed frequent rearrangements at this locus [7, 17, 20, 27, 34]. The incomplete linkage of the microcin H47 and M locus is also supported by our results.
E. coli strains isolated from intra-abdominal infections were not phylogenetically distinct from fecal flora suggesting that any E. coli strain having access to the abdominal cavity can cause an infection. This was also true for all other tested parameters with just one exception, which related to increased synthesis and uptake of siderophore aerobactin. Access to iron thus appears to be of importance to E. coli strains causing infections in intra-abdominal environment
E. coli strains isolated from respiratory infections showed a lower number of colicin producers and a higher number of microcin producers, and also a higher representation of phylogroup B2. Unlike other tested ExPEC strains, these strains also showed a higher prevalence of microcin H47, in addition to a higher prevalence of microcin M determinants, suggesting selection for a complete chromosomal region encoding both microcins H47 and M [31].
Strains isolated from genital infections showed the highest rate of bacteriocinogeny (reaching 80 %) and also of microcin production, even though they produced only six different colicin types. Based on the prevalence of phylogroup B2, E. coli strains isolated from genital smears were the most virulent in our set of ExPEC strains. Although, there was no evidence for a direct association between ExPEC strains and bacteriocinogeny, previous studies have found an association between virulence factors and bacteriocin determinants, suggesting a role for bacteriocin in bacterial virulence [10–12, 17]. It is widely accepted that strains causing genital infections and uropathogenic strains originate from fecal flora, which represents a reservoir of these strains [35, 36]. Genital infections appear to be caused by only the most virulent and bacteriocinogenic subset of fecal E. coli strains.
E. coli strains isolated from skin and soft tissue infections, i.e., decubiti, abscesses, and surgical wounds, showed a higher prevalence of microcin M and a higher prevalence of phylogenetic group B2. Results published by Petkovšek et al. (2009) revealed a similarly higher incidence of phylogroup B2 in a group of 102 SSTIs strains [22]. However, in general, E. coli strains isolated from skin and soft tissue infections showed a bacteriocinogeny frequency comparable to fecal E. coli. Bacteriocin synthesis, therefore, in these skin and soft tissue infections does not appear to be of selective advantage. At the same time, skin and soft tissue infections including decubiti, abscesses, and surgical wounds are typical for hospitalized patients and therefore likely hospital-acquired [22]. It has been previously reported that uropathogenic ESBL-producing hospital-acquired strains showed a low frequency of bacteriocinogeny [37]. Since hospital-acquired strains are often of clonal origin, we have tested genetic heterogeneity among the SSTIs strains used in this study. Detection of four E. coli phylogroups, 18 different virulence, and 30 different bacteriocin determinants among the 154 strains analyzed in this study revealed 103 distinct phenotypes (data not shown) suggesting that the strains causing SSTIs are not of clonal origin.
Conclusions
In summary, we have described a higher frequency of bacteriocinogeny in a set of ExPEC strains isolated from various human extraintestinal infections relative to fecal E. coli. Production of bacteriocins is important for most extraintestinal strains isolated from various locations on and in the human body. In general, the extraintestinal environment appears to select for strains with chromosomally encoded microcins (predominantly of mH47 and mM), while plasmid-encoded microcin and colicin types (e.g. mB17, Ia, M) appear to be contra-selected among ExPEC strains suggesting their contribution to fitness in fecal E. coli strains.
Methods
Bacterial strains
Human extraintestinal E. coli strains were collected between 2007 and 2012 from patients attending the University Hospital in Brno (n = 407) (Table 1 and Additional file 1: Table S1). From each patient, a single E. coli strain was isolated and the ENTEROtest16 (Erba Lachema, Czech Republic) was used for biochemical identification. ExPEC strains used in the study were isolated from various extraintestinal infections and included 1) E. coli strains isolated from skin and/or soft tissue infections (SSTIs), 2) E. coli strains isolated from respiratory infections (nasal, oral, and throat smears and/or sputum), 3) E. coli strains isolated from intra-abdominal infections, and 4) E. coli strains isolated from genital smears. ExPEC strains were defined as strains isolated from extraintestinal environment causing an extraintestinal infection (Additional file 1: Table S1).
A set of 1283 E. coli strains of fecal origin had been isolated from patients at two university hospitals in Brno (n = 1181) and one in Hradec Králové in the Czech Republic during the same years (i.e. 2007–2012) and previously described in detail [19, 20].
For identification of colicin and microcin determinants among tested strains, known bacteriocin producers were used as positive controls: E. coli BZB2101pColA - CA31, BZB2102 pColB - K260, BZB2103 pColD - CA23, BZB2107 pColE4 - CT9, BZB2108 pColE5 - 099, BZB2150 pColE6 - CT14, BZB2120 pColE7 - K317, BZB2279 pColIa - CA53, BZB2202 ColIb - P9, BZB2116 pColK - K235, PAP1 pColM - BZBNC22, BZB2123 pColN - 284 (original source: A. P. Pugsley), E. coli 189BM pColE2 - P9 (B. A. D. Stocker), E. coli 385/80 pColE1, pColV (H. Lhotová), E. coli 185 M4 pColE3 - CA38 (P. Fredericq), E. coli W3110 pColE8, W3110 pColE9 (J. R. James), E. coli K-12 pColS4 (D. Šmajs), S. boydii M592 (serovar 8) pColU (V. Horák), E. coli K339 pColY (D. Friedman), Shigella sonnei (colicinotype 7) pColJs (J. Šmarda), E. coli pCol5, E. coli pCol10 (H. Pilsl), E. coli 449/82 pColX (microcin B17), E. coli 313/66 pColG (microcin H47), E. coli 363/79 pColV (microcin V, original source: H. Lhotová), E. coli TOP10F' pDS601 (microcin C7), E. coli D55/1 (microcin J25), and E. coli B1239 (microcin L, D. Šmajs) [11, 17].
Positive controls for detection of virulence determinants were taken from our laboratory stock and comprised the following strains: E. coli B2917 (pCVD432), E. coli B3428 (α-hly), E. coli B3406 (afaI), E. coli B3427 (aer), E. coli B3410 (cnf1), E. coli B3418 (sfa), E. coli B3406 (pap), E. coli B3430 (ial), E. coli B2541 (st), E. coli B2802 (lt), E. coli B1804 (bfpA), E. coli B2905 (eaeA), E. coli B2987 (ipaH), E. coli B3411 (iucC), E. coli B3404 (aer), E. coli B3423 (fimA), and E. coli B2871 (ehly).
Detection of colicin and microcin determinants
A previously described method [11, 17] was used for detection of bacteriocin producers. Briefly, bacteriocinogeny of ExPEC strains (n = 407) was tested on agar plates against six indicator strains E. coli K12-Row, C6 (ϕ), B1, P400, S40, and Shigella sonnei 17. Identification of genetic determinants encoding 23 colicin (A, B, D, E1, E2-9, Ia, Ib, Js, K, L, M, N, S4, U, Y, and 5/10) and 7 microcin (H47, M, B17, C7, J25, L, and V) types, in bacteriocin producers, was performed using the DNA-PCR and colony PCR method. The list of primer pairs and the length of PCR products are in a separate file (Additional file 5: Table S5). The PCR protocol was as follows: 94 °C (2 min for DNA-PCR method; 5 min for colony PCR); 94 °C (30 s), 60 °C (30 s), 72 °C (1 min), 30 cycles; 72 °C (7 min). Because of the sensitivity of microcin types H47 and M to chloroform vapors, all investigated ExPEC and fecal E. coli strains were tested for the presence of microcin encoding genes using PCR [38]. PCR products of related bacteriocin types (colicins E2-9, Ia-Ib, and U-Y) were sequenced using dideoxy chain terminator sequencing with amplification primers (Additional file 5: Table S5) and sequences were analyzed using Lasergene software (DNASTAR, Inc., Madison, WI).
Detection of virulence factors
The presence of 18 virulence determinants (pCVD432, α-hly, afaI, aer, cnf1, sfa, pap, ial, lt, st, bfpA, eaeA, ipaH, iucC, fimA, stx1, stx2, and ehly) encoding 17 virulence factors was screened in ExPEC strains. The primer pair sequences, PCR product lengths and PCR protocols used, were previously described [39–46].
Phylogenetic group analysis
The phylogenetic groups (A, B1, B2, and D) of 407 extraintestinal E. coli strains were determined using the triplex PCR protocol (detection of chuA, yjaA genes, and TspE4.C2 fragment) described previously [47].
Statistical analysis
The statistical significance of the prevalence of bacteriocin determinants, E. coli phylogenetic groups, and virulence factors was performed by applying standard methods derived from the binomial distribution, including the two-tailed test. STATISTICA version 8.0 (StatSoft, Tulsa, OK, USA) was used for statistical calculations. The Bonferroni correction was used in analyses involving multiple comparisons.
Abbreviations
- E. coli :
-
Escherichia coli
- ExPEC:
-
Extraintestinal pathogenic Escherichia coli
- PCR:
-
Polymerase chain reaction
- SSTIs:
-
Skin and/or soft tissue infections
- MLST:
-
Multilocus sequence typing
- ESBL:
-
Extended Spectrum ß-Lactamase
- TY agar:
-
Tryptone yeast agar
References
Orskov I, Orskov F. Escherichia coli in extra-intestinal infections. J Hyg (Lond). 1985;95:551–75.
Eisenstein BI, Jones GW. The spectrum of infections and pathogenic mechanisms of Escherichia coli. Adv Intern Med. 1988;33:231–52.
Rasko DA, Rosovitz MJ, Myers GS, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, Henderson IR, Sperandio V, Ravel J. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol. 2008;190:6881–93.
Johnson JR, Russo TA. Extraintestinal pathogenic Escherichia coli: “the other bad E. coli”. J Lab Clin Med. 2002;139:155–62.
Leffler H, Svanborg-Edén C. Glycolipid receptors for uropathogenic Escherichia coli on human erythrocytes and uroepithelial cells. Infect Immun. 1981;34:920–9.
Mulvey MA. Adhesion and entry of uropathogenic Escherichia coli. Cell Microbiol. 2002;4:257–71.
Johnson JR. Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev. 1991;4:80–128.
Šmarda J, Šmajs D. Colicins - exocellular lethal proteins of Escherichia coli. Folia Microbiol (Praha). 1998;43:563–82.
Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubès R, Postle K, Riley M, Slatin S, Cavard D. Colicin biology. Microbiol Mol Biol Rev MMBR. 2007;71:158–229.
Azpiroz MF, Poey ME, Laviña M. Microcins and urovirulence in Escherichia coli. Microb Pathog. 2009;47:274–80.
Šmajs D, Micenková L, Šmarda J, Vrba M, Ševčíková A, Vališová Z, Woznicová V. Bacteriocin synthesis in uropathogenic and commensal Escherichia coli: colicin E1 is a potential virulence factor. BMC Microbiol. 2010;10:288.
Budič M, Rijavec M, Petkovšek Z, Zgur-Bertok D. Escherichia coli bacteriocins: antimicrobial efficacy and prevalence among isolates from patients with bacteraemia. PLoS One. 2011;6:e28769.
Šmajs D, Weinstock GM. Genetic organization of plasmid ColJs, encoding colicin Js activity, immunity, and release genes. J Bacteriol. 2001;183:3949–57.
Šmajs D, Weinstock GM. The iron- and temperature-regulated cjrBC genes of Shigella and enteroinvasive Escherichia coli strains code for colicin Js uptake. J Bacteriol. 2001;183:3958–66.
Riley MA, Gordon DM. The ecological role of bacteriocins in bacterial competition. Trends Microbiol. 1999;7:129–33.
Cornut G, Fortin C, Soulières D. Antineoplastic properties of bacteriocins: revisiting potential active agents. Am J Clin Oncol. 2008;31:399–404.
Micenková L, Štaudová B, Bosák J, Mikalová L, Littnerová S, Vrba M, Ševčíková A, Woznicová V, Šmajs D. Bacteriocin-encoding genes and ExPEC virulence determinants are associated in human fecal Escherichia coli strains. BMC Microbiol. 2014;14:109.
Davies DL, Falkiner FR, Hardy KG. Colicin V production by clinical isolates of Escherichia coli. Infect Immun. 1981;31:574–9.
Kohoutová D, Šmajs D, Moravková P, Cyrany J, Moravková M, Forstlová M, Čihák M, Rejchrt S, Bureš J. Escherichia coli strains of phylogenetic group B2 and D and bacteriocin production are associated with advanced colorectal neoplasia. BMC Infect Dis. 2014;14:733.
Micenková L, Bosák J, Štaudová B, Kohoutová D, Čejková D, Woznicová V, Vrba M, Ševčíková A, Bureš J, Šmajs D. Microcin determinants are associated with B2 phylogroup of human fecal Escherichia coli isolates. Microbiol Open. 2016. doi:10.1002/mbo3.345.
Mokady D, Gophna U, Ron EZ. Virulence factors of septicemic Escherichia coli strains. Int J Med Microbiol. 2005;295:455–62.
Petkovsek Z, Elersic K, Gubina M, Zgur-Bertok D, Starcic Erjavec M. Virulence potential of Escherichia coli isolates from skin and soft tissue infections. J Clin Microbiol. 2009;47:1811–7.
Köhler CD, Dobrindt U. What defines extraintestinal pathogenic Escherichia coli? Int J Med Microbiol. 2011;301:642–7.
Fakruddin M, Mazumdar RM, Chowdhury A, Mannan KS. A preliminary study on virulence factors & antimicrobial resistance in extra-intestinal pathogenic Escherichia coli (ExPEC) in Bangladesh. Indian J Med Res. 2013;137:988–90.
Koga VL, Tomazetto G, Cyoia PS, Neves MS, Vidotto MC, Nakazato G, Kobayashi RK. Molecular screening of virulence genes in extraintestinal pathogenic Escherichia coli isolated from human blood culture in Brazil. Biomed Res Int. 2014;2014:465054. doi:10.1155/2014/465054 [Epub 2014 Apr 15].
Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun. 1999;67:546–53.
Santos AC, Zidko AC, Pignatari AC, Silva RM. Assessing the diversity of the virulence potential of Escherichia coli isolated from bacteremia in São Paulo, Brazil. Braz J Med Biol Res. 2013;46:968–73.
Mirzarazi M, Rezatofighi SE, Pourmahdi M, Mohajeri MR. Occurrence of genes encoding enterotoxins in uropathogenic Escherichia coli isolates. Braz J Microbiol. 2015;46:155–9.
Chakraborty A, Saralaya V, Adhikari P, Shenoy S, Baliga S, Hegde A. Characterization of Escherichia coli Phylogenetic Groups Associated with Extraintestinal Infections in South Indian Population. Indian J Pathol Microbiol. 2014;57:255–8.
Waters VL, Crosa JH. Colicin V virulence plasmids. Microbiol Rev. 1991;55:437–50.
Duquesne S, Destoumieux-Garzón D, Peduzzi J, Rebuffat S. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat Prod Rep. 2007;24:708–34.
Jeziorowski A, Gordon DM. Evolution of microcin V and colicin Ia plasmids in Escherichia coli. J Bacteriol. 2007;189:7045–52.
Šmajs D, Strouhal M, Matějková P, Čejková D, Cursino L, Chartone-Souza E, Šmarda J, Nascimento AM. Complete sequence of low-copy-number plasmid MccC7-H22 of probiotic Escherichia coli H22 and the prevalence of mcc genes among human E. coli. Plasmid. 2008;59:1–10.
Gordon DM, O’Brien CL. Bacteriocin diversity and the frequency of multiple bacteriocin production in Escherichia coli. Microbiology. 2006;152:3239–44.
Nielsen KL, Dynesen P, Larsen P, Frimodt-Møller N. Faecal Escherichia coli from patients with E. coli urinary tract infection and healthy controls who have never had a urinary tract infection. J Med Microbiol. 2014;63:582–9.
Moreno E, Andreu A, Pigrau C, Kuskowski MA, Johnson JR, Prats G. Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. J Clin Microbiol. 2008;46:2529–34.
Micenková L, Šišková P, Bosák J, Jamborová I, Černohorská L, Šmajs D. Characterization of human uropathogenic ESBL-producing Escherichia coli in the Czech Republic: spread of CTX-M-27-producing strains in a university hospital. Microb Drug Resist. 2014;20:610–7.
Patzer SI, Baquero MR, Bravo D, Moreno F, Hantke K. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology (Reading, Engl). 2003;149:2557–70.
Martínez JL, Herrero M, de Lorenzo V. The organization of intercistronic regions of the aerobactin operon of pColV-K30 may account for the differential expression of the iucABCD iutA genes. J Mol Biol. 1994;238:288–93.
Schmidt H, Knop C, Franke S, Aleksic S, Heesemann J, Karch H. Development of PCR for screening of enteroaggregative Escherichia coli. J Clin Microbiol. 1995;33:701–5.
Yamamoto S, Terai A, Yuri K, Kurazono H, Takeda Y, Yoshida O. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol. 1995;12:85–90.
Kuhnert P, Hacker J, Mühldorfer I, Burnens AP, Nicolet J, Frey J. Detection system for Escherichia coli-specific virulence genes: absence of virulence determinants in B and C strains. Appl Environ Microbiol. 1997;63:703–9.
Paton AW, Paton JC. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol. 1998;36:598–602.
Paciorek J. Virulence properties of Escherichia coli faecal strains isolated in Poland from healthy children and strains belonging to serogroups O18, O26, O44, O86, O126 and O127 isolated from children with diarrhoea. J Med Microbiol. 2002;51:548–56.
López-Saucedo C, Cerna JF, Villegas-Sepulveda N, Thompson R, Velazquez FR, Torres J, Tarr PI, Estrada-García T. Single multiplex polymerase chain reaction to detect diverse loci associated with diarrheagenic Escherichia coli. Emerging Infect Dis. 2003;9:127–31.
Bírošová E, Siegfried L, Kmeťová M, Makara A, Ostró A, Gresová A, Urdzík P, Liptáková A, Molokácová M, Bártl R, Valanský L. Detection of virulence factors in alpha-haemolytic Escherichia coli strains isolated from various clinical materials. Clin Microbiol Infect. 2004;10:569–73.
Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–8.
Acknowledgments
We would like to thank Thomas Secrest (Secrest Editing, Ltd.) for the English editing of the manuscript.
Funding
This work was supported by the Grant Agency of the Czech Republic (16-21649S) and by a grant from the Ministry of Health of the Czech Republic (NT13413-4/2012) to DS.
Availability of data and material
All data generated or analyzed during this study are included in Additional files 1: Table S1, Additional files 2: Table S2, Additional files 3: Table S3, Additional files 4: Table S4, and Additional files 5: Table S5.
Authors’ contributions
DS designed the study and together with LM and JB wrote the manuscript. LM and JB performed bacteriocin and virulence testing of E. coli strains. DS and LM analyzed the data. LM, JB, MV, and AS contributed to isolation and characterization of the bacterial strains and gathered data. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
All human data used in the study were anonymized and the study was approved by the Ethics Committee of the Faculty of Medicine, Masaryk University, Czech Republic. All clinical samples were collected after patients gave written informed consent for participation in the study and for their samples to be used in research.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1:
Table S1. Complete list of ExPEC strains (n = 407) characterized in this study. (XLSX 90 kb)
Additional file 2:
Table S2. Distribution of E. coli phylogroups, virulence factors, bacteriocin producers, and the prevalence of bacteriocin determinants in ExPEC strains and fecal E. coli strains. (XLSX 13 kb)
Additional file 3:
Table S3. Detection of individual bacteriocin determinants in ExPEC and fecal bacteriocin producers within less virulent phylogenetic groups A + B1 compared to more virulent phylogroups B2 + D. (XLSX 11 kb)
Additional file 4:
Table S4. Differences in the distribution of virulence factors, E. coli phylogroups, bacteriocin production, and bacteriocin determinants between subgroups of ExPEC strains and fecal strains. (XLSX 14 kb)
Additional file 5:
Table S5. List of primers and the length of PCR products. (DOCX 19 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Micenková, L., Bosák, J., Vrba, M. et al. Human extraintestinal pathogenic Escherichia coli strains differ in prevalence of virulence factors, phylogroups, and bacteriocin determinants. BMC Microbiol 16, 218 (2016). https://doi.org/10.1186/s12866-016-0835-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-016-0835-z