Abstract
Background
The carA and carB genes code the small and large subunits of carbamoyl-phosphate synthase (CPS) that responsible for arginine and pyrimidine production. The purpose of this work was to study the gene organization and expression pattern of carAB operon, and the biological functions of carA and carB genes in Xanthomonas citri subsp. citri.
Methods
RT-PCR method was employed to identify the full length of carAB operon transcript in X. citri subsp. citri. The promoter of carAB operon was predicted and analyzed its activity by fusing a GUS reporter gene. The swimming motility was tested on 0.25 % agar NY plates with 1 % glucose. Biofilm was measured by cell adhesion to polyvinyl chloride 96-well plate.
Results
The results indicated that carAB operon was composed of five gene members carA-orf-carB-greA-rpfE. A single promoter was predicted from the nucleotide sequence upstream of carAB operon, and its sensitivity to glutamic acid, uracil and arginine was confirmed by fusing a GUS reporter gene. Deletion mutagenesis of carB gene resulted in reduced abilities in swimming on soft solid media and in forming biofilm on polystyrene microtiter plates.
Conclusions
From these results, we concluded that carAB operon was involved in multiple biological processes in X. citri subsp. citri.
Similar content being viewed by others
Background
Carbamoyl-phosphate synthase (CPS) catalyzes biocarbonate, ATP and glutamine to produce carbamoyl-phosphate, which serves as a precursor for synthesis of arginine and pyrimidine nucleotides in an alternative pathway [1]. In eukaryotes, CPS is responsible for removal of the excess and potentially neurotoxic ammonia via the urea cycle [2]. CPS deficiency results in an autosomal recessive disorder of urea cycle that leads to life threatening hyperammonemia [3]. Its primary localization in mitochondria makes it an ideal marker for mitochondrial damage and also during subacute phase of cecal ligation in the liver and in puncture sepsis [4, 5].
In most prokaryotes, CPS is composed of one minor and one major subunit. The smaller subunit CPSI is coded by the carA gene, and the larger subunit CPSII is coded by the carB gene [6–9]. CPSII mutant retains the ability to invade hosts but do not replicate in vivo [10]. CPSI and CPSII are proposed to combine with each other to form a tetrameric (αβ)4 protein, possessing an ammonia tunnel and a carbamate tunnel [11]. In prokaryotes, urea cycle does not occur and therefore the biological function of CPS is for arginine and the pyrimidine production.
The expression of carbamoyl phosphate synthetase is controlled by various metabolites along the pathways for pyrimidine and arginine synthesis. There exist two promoters in the promoter region of carAB operon in Eschericha coli [12, 13]. The upstream promoter P1 responds to pyrimidine and the downstream promoter P2 responds to arginine. The P1 is regulated by at least five transcription factors IHF, CarP, PyrH, PurR and RutR [14–16], while the P2 promoter is under the control of an arginine sensor ArgR [17]. Disruption of the repressors increases the carAB expression levels, which is positively correlated with pyridine production [18]. A recent work infers that carAB operon in E. coli is regulated through high intracellular levels of UTP that promote reiterative transcription to add extra U residues to the 3′ end of a nascent transcript during transcription initiation [19].
In plant bacterial pathogens, CPSI and CPSII are universal [8, 9, 20, 21]. However, their biological functions are not well understood. This research was carried out on X. citri subsp. citri, the causal agent of citrus canker disease. This is a severe bacterial disease affecting most commercial citrus cultivars grown in subtropical producing regions of the world. In the genome sequence of X. citri subsp. citri strain 306, XAC1861 and XAC1862 encode the small and large subunits CPSI and CPSII, respectively [9]. CPSII is thought to be involved in type II and type III secretion systems [9]. In this study, we reported the full-length transcript of carAB operon in X. citri subsp. citri. Our data showed that five genes carA, orf, carB, gerA and rpfE formed the carAB operon. The promoter was sensitive to the CPS-catalyzed intermediate and their final products namely glutamic acid, uracil and arginine. In addition, the loss of CPSII resulted in the phenotypic alterations in bacterial growth, swimming motility and biofilm formation. These supported the idea that carAB operon was involved in multiple biological processes in Xanthomonas citri subsp. citri.
Results
The carAB operon was composed of carA, orf, carB, gerA and rpfE genes
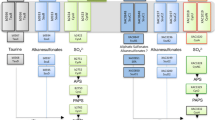
In the genome of X. citri subsp. citri strain 306, the CPS small subunit carA gene is annotated as ORF XAC1861, which is downstream of the dihydrodipicolinate reductase dapB gene (XAC1860). Even though the carB gene is annotated as XAC1862, there exists a 444-bp putative ORF between carA and carB genes. The nucleotide sequence of this orf overlaps one base at the 3′ end of carA gene. The greA, rpfE and recJ are consequently localized downstream of carB gene (Fig. 1a). Twelve specific primers were first applied to produce 1129 bp BA, 834 AP, 579 bp PB, 1254 bp BG, 957 GE and 963 bp EJ fragments by using gDNA as template (Fig. 1b; Additional file 1: Table S1). To detect the full length of the mRNA of carAB operon, the corresponding primer sets were applied to run RT-PCR from first strand of cDNA. Figure 1c showed that the primer combination D.F/A.R could not amplify the 1129 bp BA fragment covering partial dapB and carA genes. Meanwhile, the primer combination E.F/J.R did not produce the 963 bp EJ fragment containing partial rpfE and partial recJ genes. By contrast, the left AP, PB, BG and GE fragments were successfully amplified from the reverse transcript cDNA. For further confirmation, primer combinations A.F/B.R, B.F/E.R, B.F/J.R were used to amplify the desired 1423 bp AB, 1884 bp BE and 2582 bp BJ fragments from cDNA. We successfully amplified the AB and BE fragments, but not the 2582 bp BJ fragment (Fig. 1d). This indicated that carA, orf, carB, gerA and rpfE were localized within carAB operon.
Schematic representation of the carAB operon and the RT-PCR strategy. a The grey rectangles depicted the open reading frames of the operon and their lengths in base pairs. The arrows represent the sizes and approximate locations in the PCR analysis with primer sets. All forward and reverse primers were gene-specific. b PCR products by using gDNA as template. c and d PCR products from first strand cDNA. The DNA marker was DL5000
The transcription of carAB operon was suppressed by glutamic acid, uracil and arginine
We analyzed 0.5 kb DNA sequence upstream of carA gene in Neutral Network Promoter Prediction (http://fruitfly.org/seq_tools/promoter.html), and predicted one putative promoter at 102 bp from the translation start codon of carA gene (Fig. 2a). The promoter region was fused with GUS reporter gene and cloned into cosmid pUFR034 to monitor promoter activity. The recombinant construct pUGPcarAB was introduced into wild type X. citri subsp. citri strain Xac 29–1. Since glutamic acid, uracil and arginine are the biosynthetic products of CPS, 0.5 % of each one of them was added into minimal medium plates as substrate for promoter activity examination. On solid MMX plates, the GUS activities were clearly viewed from minimal media at 3 days after inoculation. All the three products had suppression effect on GUS activities (Fig. 2b). Further quantification experiments revealed that GUS activity was suppressed in the presence of uracil and glutamic acid by 37 and 48 % respectively. In the media with arginine, it was just 36 % of that in minimal media (Fig. 2c). These data suggested that the expression of carAB operon was suppressed by glutamic acid, uracil and arginine.
Analysis of carAB operon promoter and its activity. a Nucleotide sequence of carAB operon promoter region. The promoter sequence was shaded in grey colour, and the −35 and −10 promoter elements were overlined. Dotted-line arrows indicated the forward and reverse primers for promoter cloning. The transcription initiation was shown by a black arrow. b The promoter-monitored GUS activities on MMX plates. The applied substrates uracil, glutamic acid and arginine were indicated by the first letter as U, G and A. U, uracil; G, glutamic acid; A, arginine; UA, uracil and arginine; UG, uracil and glutamic acid; GA, glutamic acid and arginine; UGA, uracil, glutamic acid and arginine. c Quantification of GUS activities in MMX liquid media. The experiment was repeated three times, and similar results were obtained. The asterisks in horizontal data column indicate significant differences at P = 0.01 by t test
carB gene was required for bacterial biofilm formation
In a previous study, we constructed carA and carB gene mutants and revealed that only carB played a role in bacterial pathogenicity [9]. To detect their roles in biofilm formation, deletion mutants ΔcarA and ΔcarB were cultured in NB media to examine whether the mutations had any effect on bacterial growth (Fig. 3a). Results showed that loss of CPSI slightly increased bacterial growth a phenotype which was also observed in bacteria growing in MMX medium [9]. In contrast, the loss of CPSII led to remarkably reduced growth speed. At every observation time point after inoculation, the cell density of ΔcarB was lower than that of the wild type and ΔcarA though its OD600nm value reached 2.0 at 42 h after inoculation and increased to 3.0 at 60 h. For biofilm formation tests, we cultured all the strains to stationary growth stage, and adjusted the OD600nm to 1.0. The ΔcarA mutants showed no distinct difference from wild type strain in biofim formation and the cells were tightly adhered to polystyrene microtiter plate (Fig. 3b). By contrast, the ΔcarB showed a 70 % decrease in biofilm formation. However, when the strain was complemented, its ability to form biofilm was restored to the level of the wild type strain (Fig. 3c). The addition of exogenous uracil and arginine partially restored biofim formation in ΔcarB mutant (Fig. 3b-c).
Bacterial growth and biofilm formation. a Growth curve of ΔcarA and ΔcarB mutants in NB media. b Biofilms of ΔcarA and ΔcarB mutants formed in a microtiter plate and stained with crystal violet. c Quantitative measurements of biofilm formation. The experiments were repeated three times. The asterisks in horizontal data column indicate significant differences at P = 0.01 by t test
carB gene was involved in cell swimming
The cell motility ability was studied on 0.25 % agar NYG plates supplemented with 1 % glucose. Two days after incubation at 28 °C, the diffusion area of the wild type strain and ΔcarA mutant were clearly viewed near the colonies and there was no difference in their swimming ability. By contrast, mutant ΔcarB showed a considerable reduction in its motility (Fig. 4). The diameters derived from ΔcarB mutant reduced by over 50 % when compared with wild type. The complemented mutant strains showed a restoration of their motility confirming that the loss of CPSII affected flagellar-dependent swimming (Fig. 4).
Discussion
In our previous study, the mutations in carA and carB gene resulted in diverse effects on bacterial pathogenicity [9]. The ΔcarA retained the ability to produce citrus canker on host plant and to induce hypersensitive response on nonhost plants, while the ΔcarB mutant abolished neither pathogenicity nor extracellular enzyme activity [9]. In this work, we confirmed that the mutation in carA gene resulted in slightly increased bacterial growth speed. The maximum OD600nm value at stationary stage in NB broth was higher than wild type and was consistent with the results of our previous work that investigated effect of culturing bacteria in minimal media MMX [9]. Loss of CPSII led to a remarkably reduction of nitrogen and carbon resource assimilation indicating carB gene was more important than carA in nucleotide and amino acid metabolism in X. citri subsp. citri. Additionally, the loss of CPSII led to a reduction of cell swimming ability by 50 % and biofilm formation was reduced by 70 %. This suggested that carB gene played the role in canker development during the early infection stage.
In the model prokaryote Eschericha coli, dihydrodipicolinate reductase caiF is downstream of carB and has the same transcription orientation as carA and carB genes (Additional file 2: Figure S1). It is transcribed as a monocistronic mRNA under anaerobiosis independent of the presence of carnitine [22]. Solid evidence has been presented for the organization of carAB operon in Pseudomonas aeruginosa which was found to contain four gene members namely carA-orf-carB–greA [23]. The genetic function of greA gene has been well demonstrated [23]. In X. citri subsp. citri, the genetic organization of carAB operon is similar to that in P. aeruginosa (Additional file 2: Figure S1). However, it also has a putative protein coding ORF between carA and carB, even though both orfs showed no similarity (data not shown). In addition to the greA which is located downstream of carB gene, there are two genes rpfE and recJ having the same transcription orientation with carA and carB genes [20]. Through RT-PCR method, we demonstrated that X. citri subsp. citri has an extra fifth member in its carAB operon namely the rpfE gene. Thus, the carAB operon in X. citri subsp. citri is different from those found in P. aeruginosa and E. coli.
The transcriptional elongation factor GreA induces the endonucleolytic cleavage that occurs at the 3′ ends of arrested quarternary transcription complexes [23]. Mutation in greA gene results in loss of growth ability in minimal medium with arginine and nucleosides, but the mutant grows well in rich media [23]. The rpfE is required for full virulence, as well as swarming motility and production of cellulase and extracellular polysaccharide. In culture, rpfE mutant strain is unable to efficiently utilize sucrose or xylose as carbon sources [24]. Pyrimindine, the final product catalyzed by CPS, is involved in biofilm formation [25, 26]. Exogenous pyrimindine restores cellulose production in carB mutant, which is one of the extracellular adhesion and cell aggregation factors responsible for bacterial biofilm formation and maintenance [26]. Thus, the gene members in carAB operon are required for multiple cell life activities, including amino acid and nucleotide biosynthesis, cell motility, cellulose and extracellular polysaccharide production. Some of the processes are co-affected by the members in carAB operon, because CPSII and RpfE are both involved in cell motility and cellulose activity [9, 23].
There are two adjacent promoters for carAB operon in E. coli namely P1 and P2. The P1 promoter is located far from translation initiation site and is mainly regulated by pyrimidines while the P2 promoter which overlaps a pair of ARG boxes is regulated by arginine and the arginine repressor ArgR [13, 17]. A single promoter has been identified from carAB operon in P. aeruginosa, which is controlled by pyrimidines and arginine [7]. In this work, only one promoter was identified from carAB operon in X. citri subsp. citri. The promoter activity was suppressed by pyrimidines, arginine and glutamic acid, which was similar with the result obtained from P. aeruginosa. It appeared that the regulation mechanism of carAB operon in X. citri subsp. citri resembled that in P. aeruginosa, but differed from that in E. coli.
Conclusions
In this study we found out that X. citri subsp. citri carAB operon was made up of five genes carA, orf, carB, gerA and rpfE, and that its transcription was suppressed by glutamic acid, uracil and arginine. We also found out that the loss of the large subunit CPSII resulted to phenotypic alterations in bacterial growth, biofilm formation and swimming.
Methods
Bacterial strains, plasmids and culture conditions
Bacterial strains and plasmids used in this study are listed in Table 1. The X. citri spp. citri strains 29–1 (Xac 29–1) were cultivated in NB nutrient broth or NB with 1.5 % agar at 28 °C [9]. E. coli strains were cultured in Luria-Bertani medium (LB) at 37 °C. Three antibiotics were used at the following concentrations: ampicillin (Ap), 100 μg/ml; kanamycin (Km), 25 μg/ml and Gentamycin (Gm), 50 μg/ml.
RT-PCR
To detect the full length mRNA of carAB operon, specific primers (Additional file 1: Table S1) were designed according to genome sequence information. After wild type Xac 29–1 was cultured in liquid NB broth, RNAs were extracted from cells by the RNA prep pure Cell/Bacteria Kit (Tiangen Biotech, Beijing, China). The total RNAs were quantified by measuring the OD260/OD280 ratio and then analyzed by gel electrophoresis to find out whether they were intact. To ensure genomic DNA was not contaminated, the PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa-bio, Dalian, China) was used before reverse transcription. 2 μg total RNA was reverse transcribed to first strand cDNA by AMV reverse transcriptase (TaKaRa-bio, Dalian, China). The PCR thermal cycle consisted of an initial denaturation at 94 °C for 5 min, 32 cycles of DNA denaturation at 94 °C for 30 s, primer annealing at 52 °C for 40 s, and primer extension at 72 °C for 1 min, and followed by a final elongation step at 72 °C for 10 min. The expression of gyrA was used as a control to verify the quality of cDNA.
GUS activity assays
To construct the promoter β-glucuronidase (GUS) fusion, a 336 bp promoter region upstream of carA gene was PCR amplified from genome DNA by primer carAB.P.F and carAB.P.R (Additional file 1: Table S1). The gusA gene was amplified by primer GUSA.F and GUSA.R and ligased into pUFR034, together with the 336 bp promoter fragment (Additional file 1: Table S1). Recombinant pUGPcarAB was introduced into wild type Xac 29–1 to generate Xac 29-1/pUGPcarAB. The strain was cultured in NB until OD600nm reached 0.8. After centrifugation at 6000 rpm for 10 min at 4 °C, the cell pellets were re-suspended in NB broth to OD600nm = 1.0. About 1.5 μl of the cell suspension was added to MMX medium plates containing 50 μg/ml of X-gluc. To assess the effects on carAB transcription, 0.5 % of glutamic acid, uracil or arginine was applied to MMX medium plates. At 3 days after inoculation, colony color on plates was observed to determine GUS activities. In the parallel experiments, 3 ml liquid MMX media was used to culture the cells at 28 °C for 12 h induction. The cells were then collected for GUS activity analysis [27]. GUS activities were determined at 30 min intervals for 3 h by measuring absorbance at 415 nm (A 415) using p-nitrophenyl-D-glucuronide as the substrate. One unit was defined as 1 nmol of 4-methyl-umbelliferone produced per min per bacterium. Assays were repeated three times independently.
Swimming motility assay
The swimming motility was performed as described previously [28]. In brief, the cultured X. citri subsp. citri strains were suspended in sterile distilled water to a final concentration of OD600nm = 1.0. 1.5 μL of each cell sample was dropped to 0.25 % agar NY plates with 1 % glucose. The plates were stationary incubated at 28 °C. The motile ability was measured from the diameter of each colony 2 days post cultivation. The tests were repeated three times.
Determination of bacterial growth
The cultured cells were washed twice with sterilized water, and then re-suspended in sterilized water to OD600nm = 1.0. The re-suspended cells were subcultured (1:100) in NB broth media. The OD600nm values were tested after every 6 h post sub-culturing. All the experiments were repeated at least three times.
Biofilm assays
Biofilm was measured by cell adhesion to poly (vinyl chloride) 96-well plate (Falcon 353913, Becton Dickinson). All the cultured strains were re-suspended in NB to an OD600nm of 1.0. To assess the effects of uracil and arginine on biofilm formation, 0.5 % of each chemical was applied to NB medium. Typically, 100 μl cell suspensions were dropped into one well of a microtiter plate. The plates were sealed with plastic wrap and incubated without shaking for 72 h at 28 °C. Bacterial adhesion was measured after repetitive washing of the plates and staining with 1 % crystal violet for 15 min at room temperature. Excess stain was removed by gently washing with distilled water, and the crystal violet stain was solubilized by the addition of 150 μl of 95 % ethanol to each well. Crystal violet was then quantified with a microplate reader at A570 absorption wavelength. All the experiments were repeated at least four times and the average for each strain was checked by T-test.
Abbreviations
- CPS:
-
Carbamoyl-phosphate synthase
- CPSI:
-
Carbamoyl-phosphate synthase small subunit
- CPSII:
-
Carbamoyl-phosphate synthase large subunit
- GUS:
-
β-Glucuronidase
- ORF:
-
Open reading frame
- RT-PCR:
-
Reverse transcription polymerase chain reaction
References
Raushel FM, Thoden JB, Holden HM. The aminotransferase family of enzymes: molecular machines for the production and delivery of ammonia. Biochemistry. 1999;38:7891–9.
Ahuja V, Powers-Lee SG. Human carbamoyl-phosphate synthetase: insight into N-acetylglutamate interaction and the functional effects of a common single nucleotide polymorphism. J Inherit Metab Dis. 2008;31:481–91.
Ono H, Suto T, Kinoshita Y, Sakano T, Furue T, Ohta T. A case of carbamoyl phosphate synthetase 1 deficiency presenting symptoms at one month of age. Brain Dev. 2009;31:779–81.
Weerasinghe SV, Jang YJ, Fontana RJ, Omary MB. Carbamoyl phosphate synthetase-1 is a rapid turnover biomarker in mouse and human acute liver injury. Am J Physiol Gastrointest Liver Physiol. 2014;307:G355–64.
Summar ML. Molecular genetic research into carbamoyl-phosphate synthase I: molecular defects and linkage markers. J Inherit Metab Dis. 1998;21:30–9.
Nyunoya H, Lusty CJ. The carB gene of Escherichia coli: a duplicated gene coding for the large subunit of carbamoyl-phosphate synthetase. Proc Natl Acad Sci U S A. 1983;80:4629–33.
Kwon DH, Lu CD, Walthall DA, Brown TM, Houghton JE, Abdelal AT. Structure and regulation of the carAB operon in Pseudomonas aeruginosa and Pseudomonas stutzeri: no untranslated region exists. J Bacteriol. 1994;176:2532–42.
Salzberg SL, Sommer DD, Schatz MC, Phillippy AM, Rabinowicz PD, Tsuge S, et al. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics. 2008;9:204–21.
Guo J, Song X, Zou LF, Zou HS, Chen GY. The small and large subunits of carbamoyl-phosphate synthase exhibit diverse contributions to pathogenicity in Xanthomonas citri subsp. citri. J Integr Agr. 2015;14:1338–47.
Dzierszinshi FS, Hunter CA. Advances in the use of genetically engineered parasites to study immunity to Toxoplasma gondii. Parasite Immunol. 2008;30:235–44.
Thoden JB, Raushel FM, Benning MM, Tayment I, Holden HM. The structure of carbamoyl phosphate synthetase determined to 2.1 A resolution. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 1):8–24.
Bouvier J, Patte JC, Stragier P. Multiple regulatory signals in the control region of the Escherichia coli carAB operon. Proc Natl Acad Sci U S A. 1984;81:4139–43.
Piette J, Nyunoya H, Lusty CJ, Cunin R, Weyens G, Crabeel M. DNA sequence of the carA gene and the control region of carAB: tandem promoters, respectively controlled by arginine and the pyrimidines, regulate the synthesis of carbamoyl-phosphate synthetase in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1984;81:4134–8.
Charlier D, Roovers M, Gigot D, Huysveld N, Piérard A, Glansdorff N. Integration host factor (IHF) modulates the expression of the pyrimidine-specific promoter of the carAB operons of Escherichia coli K-12 and Salmonella typhimurium LT2. Mol Gen Genet. 1993;237:273–86.
Charlier D, Hassanzadeh G, Kholti A, Gigot D, Piérard A, Glansdorff N. carP, involved in pyrimidine regulation of the Escherichia coli carbamoylphosphate synthetase operon, encodes a sequence-specific DNA-binding protein identical to XerB and PepA, also required for resolution of ColEI multimers. J Mol Biol. 1995;250:392–406.
Kholti A, Charlier D, Gigot D, Huysveld N, Roovers M, Glansdorff N. pyrH-encoded UMP-kinase directly participates in pyrimidine-specific modulation of promoter activity in Escherichia coli. J Mol Biol. 1998;280:571–82.
Devroede N, Thia-Toong TL, Gigot D, Maes D, Charlier D. Purine and pyrimidine-specific repression of the Escherichia coli carAB operon are functionally and structurally coupled. J Mol Biol. 2004;336:25–42.
Shimada T, Hirao K, Kori A, Yamamoto K, Ishihama A. RutR is the uracil/thymine-sensing master regulator of a set of genes for synthesis and degradation of pyrimidines. Mol Microbiol. 2007;66:744–57.
Han X, Turnbough Jr CL. Transcription start site sequence and spacing between the −10 region and the start site affect reiterative transcription-mediated regulation of gene expression in Escherichia coli. J Bacteriol. 2014;196:2912–20.
da Silva AC, Ferro JA, Reinach FC, Farah CS, Furlan LR, Quaggio RB, et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature. 2002;417:59–63.
Salanoubat M, Genin S, Artiguenave F, Gouzy J, Mangenot S, Arlet M, et al. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature. 2002;415:497–502.
Eichler K, Buchet A, Lemke R, Kleber HP, Mandrand-Berthelot MA. Identification and characterization of the caiF gene encoding a potential transcriptional activator of carnitine metabolism in Escherichia coli. J Bacteriol. 1996;178:1248–57.
Lu CD, Kwon DH, Abdelal AT. Identification of greA encoding a transcriptional elongation factor as a member of the carA-orf-carB-greA operon in Pseudomonas aeruginosa PAO1. J Bacteriol. 1997;197:3043–46.
Dow JM, Feng JX, Barber CE, Tang JL, Daniels MJ. Novel genes involved in the regulation of pathogenicity factor production within the rpf gene cluster of Xanthomonas campestris. Microbiology. 2000;146:885–91.
Ueda A, Attila C, Whiteley M, Wood TK. Uracil influences quorum sensing and biofilm formation in Pseudomonas aeruginosa and fluorouracil is an antagonist. Microb Biotechnol. 2009;2:62–74.
Garavaglia M, Rossi E, Landini P. The pyrimidine nucleotide biosynthetic pathway modulates production of biofilm determinants in Escherichia coli. PLoS One. 2012;7:e31252.
Li YR, Che YZ, Zou HS, Cui YP, Guo W, Zou LF, et al. Hpa2 required by HrpF to translocate Xanthomonas oryzae transcriptional activator-like effectors into rice for pathogenicity. Appl Environ Microbiol. 2011;77:3809–18.
Malamud F, Torres PS, Roeschlin R, Rigabo LA, Enrique R, Bonomi HR, et al. The Xanthomonas axonopodis pv. citri flagellum is required for mature biofilm and canker development. Microbiology. 2011;157(Pt 3):819–29.
DeFeyter R, Kado CI, Gabriel DW. Small, stable shuttle vectors for use in Xanthomonas. Gene. 1990;88:65–72.
Acknowledgement
This work was supported by the National Natural Science Foundation of China (31171832 to Zou).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TZ and WR carried out the molecular genetic studies and drafted the manuscript. XS participated in the promoter sequence and activity analysis. JG and XF performed the statistical analysis. HZ conceived of the study, and participated in its design and coordination and aided in the preparation of the manuscript draft. GK participated in the editing of the manuscript. All authors read and approved the final manuscript.
Tao Zhuo and Wei Rou contributed equally to this work.
Additional files
Additional file 1: Table S1.
Primers used in this study. (DOC 38 kb)
Additional file 2: Figure S1.
Alignment of carAB operons from Xanthomonas citri subsp. citri, Pseudomonas aeruginosa and Escherichia coli. The genetic information from each strain was based their genome information in GenBank (NC_003919.1, NC_002516.2 and NC_002655.2). (DOC 41 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhuo, T., Rou, W., Song, X. et al. Molecular study on the carAB operon reveals that carB gene is required for swimming and biofilm formation in Xanthomonas citri subsp. citri . BMC Microbiol 15, 225 (2015). https://doi.org/10.1186/s12866-015-0555-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-015-0555-9