Abstract
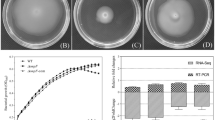
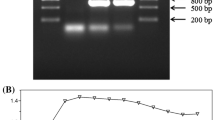
The genus Citrobacter is commonly found in environmental and industrial settings, some members of which have been used for bioremediation of heavy metals owing to the absorption ability of their biofilms. Although our previous studies have found that the outer membrane protein A (OmpA) contributes to the process of Citrobacter werkmanii biofilm formation, the underlying mechanisms remain elusive. Therefore, we deleted ompA from the genome of C. werkmanii and investigated its phenotypes in comparison to the wild type strain (WT) and the complementary strain using biochemical and molecular techniques including RNA-Seq. Our results demonstrated that the deletion of ompA led to an increase in biofilm formation on both polystyrene and glass surfaces due to upregulation of some biofilm formation related genes. Meanwhile, swimming ability, which is mediated by activation of flagellar assembly genes, was increased on semi-solid plates in the ∆ompA strain when compared with WT. Additionally, inactivation of ompA also caused increased 1,2-benzisothiazolin-3-one (BIT) resistance, differential responses to Ca2+ stress, curli protein expression and cellulose production. Finally, ∆ompA caused differential expression of a total of 1470 genes when compared with WT, of which 146 were upregulated and 1324 were downregulated. These genes were classified into different Gene Ontology (GO) and KEGG pathways. In summary, ompA in C. werkmanii contributes to a variety of biological functions and may act as a target site to modulate biofilm formation.
Key points
• ompA is a negative regulator for biofilm formation by C. werkmanii.
• ompA inhibits swimming motility of C. werkmanii.
• ompA deletion causes different expression profiles in C. werkmanii.







Similar content being viewed by others
Data availability
The Raw RNA-Seq data of both WT and ∆ompA strains have been deposited in the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA648788.
References
Barrios AFG, Zuo R, Ren D, Wood TK (2006) Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnol Bioeng 93(1):188–200
Bokranz W, Wang XD, Tschape H, Romling U (2005) Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J Med Microbiol 54(12):1171–1182
Bootman MD, Berridge MJ (1995) The elemental principles of calcium signaling. Cell 83(5):675–678
Bunpa S, Chaichana N, Teng JLL, Lee HH, Woo PCY, Sermwittayawong D, Sawangjaroen N, Sermwittayawong N (2019) Outer membrane protein A (OmpA) is a potential virulence factor of Vibrio alginolyticus strains isolated from diseased fish. J Fish Dis 43(2):275–284
Confer AW, Ayalew S (2013) The OmpA family of proteins: roles in bacterial pathogenesis and immunity. Vet Microbiol 163(3–4):207–222
Domka J, Lee J, Wood TK (2006) YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl Environ Microbiol 72(4):2449–2459
Feng S-X, Ma J-C, Yang J, Hu Z, Zhu L, Bi H-K, Sun Y-R, Wang H-H (2015) Ralstonia solanacearum fatty acid composition is determined by interaction of two 3-ketoacyl-acyl carrier protein reductases encoded on separate replicons. BMC Microbiol 15:223
Finlay JA, Allan VJM, Conner A, Callow ME, Basnakova G, Macaskie LE (1999) Phosphate release and heavy metal accumulation by biofilm-immobilized and chemically-coupled cells of a citrobacter sp. pre-grown in continuous culture. Biotechnol Bioeng 63(1):87–97
Gaddy JA, Tomaras AP, Actis LA (2009) The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun 77(8):3150–3160
Geesey GG, Wigglesworth-Cooksey B, Cooksey K (2000) Influence of calcium and other cations on surface adhesion of bacteria and diatoms: a review. Biofouling 15(1–3):195–205
Hegde S, Nilyanimit P, Kozlova E, Anderson ER, Narra HP, Sahni SK, Heinz E, Hughes GL (2019) CRISPR/Cas9-mediated gene deletion of the ompA gene in symbiotic Cedecea neteri impairs biofilm formation and reduces gut colonization of Aedes aegypti mosquitoes. PloS Neglect Trop Dis 13(12):e0007883
Jagmann N, Henke SF, Philipp B (2015) Cells of Escherichia coli are protected against severe chemical stress by co-habiting cell aggregates formed by Pseudomonas aeruginosa. Appl Microbiol Biotechnol 99(19):8285–8294
Jeong BC, Hawes C, Bonthrone KM, Macaskie LE (1997) Localization of enzymically enhanced heavy metal accumulation by Citrobacter sp. and metal accumulation in vitro by liposomes containing entrapped enzyme. Microbiology 143(7):2497–2507
Khan SR, Gaines J, Roop RM, Farrand SK (2008) Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol 74(16):5053–5062
Kim SI, Yoon H (2019) Roles of YcfR in biofilm formation in Salmonella Typhimurium ATCC 14028. Mol Plant-Microbe Interact 32(6):708–716
Kim SW, Oh MH, Jun SH, Jeon H, Kim SI, Kim K, Lee YC, Lee JC (2016) Outer membrane protein A plays a role in pathogenesis of Acinetobacter nosocomialis. Virulence 7(4):413–426
Klebensberger J, Lautenschlager K, Bressler D, Wingender J, Philipp B (2007) Detergent-induced cell aggregation in subpopulations of Pseudomonas aeruginosa as a preadaptive survival strategy. Environ Microbiol 9(9):2247–2259
Kumar A, Ting YP (2013) Effect of sub-inhibitory antibacterial stress on bacterial surface properties and biofilm formation. Colloids Surf B Biointerfaces 111C:747–754
Li Y, Hao G, Galvani CD, Meng Y, De La Fuente L, Hoch H, Burr TJ (2007) Type I and type IV pili of Xylella fastidiosa affect twitching motility, biofilm formation and cell-cell aggregation. Microbiology 153(3):719–726
Li L, Zhou G, Shi Q, Chen Y, Chen Y, Ouyang Y, Hu W (2014) Identification and biofilm formation characterization of Citrobacter werkmanii isolated from industrial spoilage. Microbiology China 41(1):2–7
Ling H, Kang A, Tan MH, Qi X, Chang MW (2010) The absence of the luxS gene increases swimming motility and flagella synthesis in Escherichia coli K12. Biochem Biophys Res Commun 401(4):521–526
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25(4):402–408
Ma Q, Wood TK (2009) OmpA influences Escherichia coli biofilm formation by repressing cellulose production through the CpxRA two-component system. Environ Microbiol 11(10):2735–2746
Macaskie LE, Empson RM, Lin F, Tolley MR (1995) Enzymatically-mediated uranium accumulation and uranium recovery using a Citrobacter sp. immobilised as a biofilm within a plug-flow reactor. J Chem Technol Biotechnol 63(1):1–16
Maltz MA, Weiss BL, Neill M, Wu Y, Aksoy S (2012) OmpA-mediated biofilm formation is essential for the commensal bacterium Sodalis glossinidius to colonize the tsetse fly gut. Appl Environ Microbiol 78(21):7760–7768
Orme R, Douglas CWI, Rimmer S, Webb M (2006) Proteomic analysis of Escherichia coli biofilms reveals the overexpression of the outer membrane protein OmpA. Proteomics 6(15):4269–4277
Park JS, Lee WC, Yeo KJ, Ryu KS, Kumarasiri M, Hesek D, Lee M, Mobashery S, Song JH, Kim SI, Lee JC, Cheong C, Jeon YH, Kim HY (2012) Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane. FASEB J 26(1):219–228
Patrauchan M, Sarkisova S, Sauer K, Franklin M (2005) Calcium influences cellular and extracellular product formation during biofilm-associated growth of a marine Pseudoalteromonas sp. Microbiology 151(9):2885–2897
Rashid MH, Kornberg A (2000) Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 97(9):4885–4890
Reusch RN (2012) Insights into the structure and assembly of Escherichia coli outer membrane protein A. FEBS J 279(6):894–909
Ritter A, Com E, Bazire A, Goncalves MDS, Delage L, Pennec GL, Pineau C, Dreanno C, Compère C, Dufour A (2012) Proteomic studies highlight outer-membrane proteins related to biofilm development in the marine bacterium Pseudoalteromonas sp. D41. Proteomics 12(21):3180–3192
Rollauer SE, Sooreshjani MA, Noinaj N, Buchanan SK (2015) Outer membrane protein biogenesis in Gram-negative bacteria. Philos Trans R Soc B-Biol Sci 370(1679):20150023
Sarkisova S, Patrauchan M, Berglund D, Nivens D, Franklin M (2005) Calcium-induced virulence factors associated with the extracellular matrix of mucoid Pseudomonas aeruginosa biofilms. J Bacteriol 187(13):4327–4337
Shanks RMQ, Meehl MA, Brothers KM, Martinez RM, Donegan NP, Graber ML, Cheung AL, O'Toole GA (2008) Genetic evidence for an alternative citrate-dependent biofilm formation pathway in Staphylococcus aureus that is dependent on fibronectin binding proteins and the GraRS two-component regulatory system. Infect Immun 76(6):2469–2477
Sheng GP, Xu J, Li WH, Yu HQ (2013) Quantification of the interactions between Ca2+, Hg2+ and extracellular polymeric substances (EPS) of sludge. Chemosphere 93(7):1436–1441
Smani Y, Fabrega A, Roca I, Sanchez-Encinales V, Vila J, Pachon J (2014) Role of OmpA in the multidrug resistance phenotype of Acinetobacter baumannii. Antimicrob Agents Chemother 58(3):1806–1808
Sugawara E, Nikaido H (2012) OmpA is the principal nonspecific slow porin of Acinetobacter baumannii. J Bacteriol 194(15):4089–4096
Tang Q, Feng M (2007) DPS data processing system: experimental design, statistical analysis and data mining. Science Press, Beijing
Viale AM, Evans BA (2020) Microevolution in the major outer membrane protein OmpA of Acinetobacter baumannii. Microb Genomics 6(6):381
Wang Y (2002) The function of OmpA in Escherichia coli. Biochem Biophys Res Commun 292(2):396–401
Weber MM, French CL, Barnes MB, Siegele DA, McLean RJ (2010) A previously uncharacterized gene, yjfO (bsmA), influences Escherichia coli biofilm formation and stress response. Microbiology 156(1):139–147
Wood TK (2009) Insights on Escherichia coli biofilm formation and inhibition from whole-transcriptome profiling. Environ Microbiol 11(1):1–15
Xiong K, Chen ZJ, Xiang GM, Wang J, Rao XC, Hu FQ, Cong YG (2012) Deletion of yncD gene in Salmonella enterica ssp. enterica serovar Typhi leads to attenuation in mouse model. FEMS Microbiol Lett 328(1):70–77
Zhou G, Li L, Shi Q, Ouyang Y, Chen Y, Hu W (2013) Effects of nutritional and environmental conditions on planktonic growth and biofilm formation for Citrobacter werkmanii BF-6. J Microbiol Biotechnol 23(12):1673–1682
Zhou G, Li L, Shi Q, Ouyang Y, Chen Y, Hu W (2014) Efficacy of metal ions and isothiazolones in inhibiting Enterobacter cloacae BF-17 biofilm formation. Can J Microbiol 60(1):5–14
Zhou G, Shi Q, Huang X, Xie X (2016) Proteome responses of Citrobacter werkmanii BF-6 planktonic cells and biofilms to calcium chloride. J Proteome 133:134–143
Zhou G, Wang Y, Peng H, Huang X, Xie X, Shi Q (2018) Role of ttca of Citrobacter werkmanii in bacterial growth, biocides resistance, biofilm formation and swimming motility. Int J Mol Sci 19(9):2644
Acknowledgments
We would like to thank Prof. Hai-hong Wang of South China Agricultural University for the generous gift of E. coli S17-1. We are also grateful to Prof. Yan-guang Cong of Hospital of Traditional Chinese Medicine Affiliated to Southwest Medical University for providing plasmid pYG4 to us and also for his valuable experimental guidance.
Funding
This work was funded by the National Natural Science Foundation of China (No. 31770091), the GDAS’ Project of Science and Technology Development (Nos. 2019GDASYL-0104006 and 2017GDASCX-0102), and Natural Science Foundation of Guangdong Province (No. 2020A151501848), and Guangdong Science and Technology Program (No. 2017B030314045).
Author information
Authors and Affiliations
Contributions
GZ, XX, and QS conceived and designed research. GZ, YW, HP, SL, TS, and PS performed experiments. GZ, XX, QS, YW, and HP analyzed the data. GZ wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PDF 29 kb)
Rights and permissions
About this article
Cite this article
Zhou, G., Wang, Ys., Peng, H. et al. Roles of ompA of Citrobacter werkmanii in bacterial growth, biocide resistance, biofilm formation and swimming motility. Appl Microbiol Biotechnol 105, 2841–2854 (2021). https://doi.org/10.1007/s00253-020-11057-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-11057-1