Abstract
Background
Blueberries (Vaccinium sp.) are native to North America and breeding efforts to improve blueberry fruit quality are focused on improving traits such as increased firmness, enhanced flavor and greater shelf-life. Such efforts require additional genomic resources, especially in southern highbush and rabbiteye blueberries.
Results
We generated the first full-length fruit transcriptome for the southern highbush and rabbiteye blueberry using the cultivars, Suziblue and Powderblue, respectively. The transcriptome was generated using the Pacific Biosciences single-molecule long-read isoform sequencing platform with cDNA pooled from seven stages during fruit development and postharvest storage. Raw reads were processed through the Isoseq pipeline and full-length transcripts were mapped to the ‘Draper’ genome with unmapped reads collapsed using Cogent. Finally, we identified 16,299 and 15,882 non-redundant transcripts in ‘Suziblue’ and ‘Powderblue’ respectively by combining the reads mapped to Northern Highbush blueberry ‘Draper’ genome and Cogent analysis. In both cultivars, > 80% of sequences were longer than 1,000 nt, with the median transcript length around 1,700 nt. Functionally annotated transcripts using Blast2GO were > 92% in both ‘Suziblue’ and ‘Powderblue’ with overall equal distribution of gene ontology (GO) terms in the two cultivars. Analyses of alternative splicing events indicated that around 40% non-redundant sequences exhibited more than one isoform. Additionally, long non-coding RNAs were predicted to represent 5.6% and 7% of the transcriptomes in ‘Suziblue’ and ‘Powderblue’, respectively. Fruit ripening is regulated by several hormone-related genes and transcription factors. Among transcripts associated with phytohormone metabolism/signaling, the highest number of transcripts were related to abscisic acid (ABA) and auxin metabolism followed by those for brassinosteroid, jasmonic acid and ethylene metabolism. Among transcription factor-associated transcripts, those belonging to ripening-related APETALA2/ethylene-responsive element-binding factor (AP2/ERF), NAC (NAM, ATAF1/2 and CUC2), leucine zipper (HB-zip), basic helix-loop-helix (bHLH), MYB (v-MYB, discovered in avian myeloblastosis virus genome) and MADS-Box gene families, were abundant.
Further we measured three fruit ripening quality traits and indicators [ABA, and anthocyanin concentration, and texture] during fruit development and ripening. ABA concentration increased during the initial stages of fruit ripening and then declined at the Ripe stage, whereas anthocyanin content increased during the final stages of fruit ripening in both cultivars. Fruit firmness declined during ripening in ‘Powderblue’. Genes associated with the above parameters were identified using the full-length transcriptome. Transcript abundance patterns of these genes were consistent with changes in the fruit ripening and quality-related characteristics.
Conclusions
A full-length, well-annotated fruit transcriptome was generated for two blueberry species commonly cultivated in the southeastern United States. The robustness of the transcriptome was verified by the identification and expression analyses of multiple fruit ripening and quality–regulating genes. The full-length transcriptome is a valuable addition to the blueberry genomic resources and will aid in further improving the annotation. It will also provide a useful resource for the investigation of molecular aspects of ripening and postharvest processes.
Similar content being viewed by others
Background
Blueberry (Vaccinium sp.) is in the Ericaceae family and native to North America. Blueberries are gaining popularity due to increased awareness of their health benefits, such as lowering the risk of cardiovascular diseases and damage due to aging [1, 2]. In 2020, its total utilized production was 637 million pounds, which included fresh market fruit at 350 million pounds and fruit for processing at 288 million pounds [3]. In the United States, cultivated blueberries ranked fifth after grape, apple, strawberry, and sweet cherry in the value of utilized production in the non-citrus fruit and nuts category totaling 904 million dollars [3]. There are several important cultivated blueberry species in the United States. They vary in their cold hardiness and chill hour requirements for flowering. Lowbush (Vaccinium angustifolium Ait.) and northern highbush (Vaccinium corymbosum L.) are grown mainly in the northern US, while rabbiteye (V. virgatum Ait.) and southern highbush (hybrids of V. corymbosum, V. virgatum, V. darrowii Camp.) are predominantly cultivated in the southern US [4, 5].
To facilitate blueberry breeding, it is important to generate and enhance genetic and genomic resources. Originally a draft blueberry genomic sequence was generated using a diploid V. corymbosum accession W8520 [6]. Subsequently RNA-Seq reads from fruit developmental stages were assembled onto this draft blueberry genome, which predicted around 60,000 gene models. Of these, 58% were functionally annotated by homologous protein search and around 24% were assigned GO terms [7]. Subsequently an improved version of the haploid-phased northern highbush blueberry genome was generated from a tetraploid northern highbush cultivar ‘Draper’, which predicted 32,140 protein coding genes per haplotype. The GO annotation for this genome was around 57%, which was a significant improvement from the previous draft genome. More recently, a reference genome for Vaccinium darrowii, an evergreen wild blueberry has been generated [8, 9]. However, there is still no available genome for southern highbush or rabbiteye blueberries, those usually grown in the southeastern parts of the US.
Transcriptome, RNA expressed from the genome, is an invaluable resource for genome assembly and annotation [10], linking genes to their function [11], and is an alternative resource when the complete genome is not available [12]. Pacific Biosciences (PacBio) single-molecule long-read isoform sequencing (Iso-seq) is a method for full-length transcript sequencing without the need for further assembly [13]. This approach can provide accurate information of transcript structure and alternative splicing. In fruit crops, this sequencing technology has been used to identify potential genes related to disease resistance in apple [14], proanthocyanidin accumulation in persimmon [15], carotenoid biosynthesis in avocado [16], and sugar-metabolism in Annona squamosa [17]. In this study, we developed the first full-length fruit transcriptome for two blueberry cultivars, a tetraploid southern highbush ‘Suziblue’ and hexaploid cultivar ‘Powderblue’. In addition, to evaluate the robustness of the fruit transcriptome, we measured three fruit ripening and quality-related traits (ABA and anthocyanin concentrations, and texture) and the trait-related gene expression by qRT-PCR based on the transcriptome sequences. This work will provide additional valuable genomic resources that can be exploited by the blueberry community.
Results
Transcriptome sequencing and annotation
Iso-Seq generated 29.48 Gb raw reads in ‘Suziblue’ and 25.82 Gb in ‘Powderblue’ (Table 1). Downstream analyses by the SMRT-Link Isoseq3 pipeline (Fig. 1), produced 541,220 circular consensus sequences (CCS) in ‘Suziblue’ and 482,718 CCS in ‘Powderblue’ (Table 1). This resulted in 31,846 high quality and 273 low quality full-length transcripts in ‘Suziblue’, and 31,091 high quality and 232 low quality full-length transcripts in ‘Powderblue’. Next, both high- and low-quality full-length transcripts were mapped to the ‘Draper’ genome to reduce redundancy of transcripts (Fig. 1). These analyses resulted in mapping of 89% of ‘Suziblue’ and 92% of ‘Powderblue’ full length transcripts to the ‘Draper’ genome. The remaining unmapped transcripts were collapsed by Cogent (Fig. 1). Non-redundant transcripts generated after mapping to the Draper genome and Cogent were combined (Table 1). Ultimately, 16,299 and 15,882 non-redundant transcripts were identified in ‘Suziblue’ and ‘Powderblue’ respectively (Table 1). The distribution of the lengths of non-redundant transcripts ranged from 75–7,650 nucleotides (nt) in ‘Suziblue’ and 60–7,945 nt in ‘Powderblue’ (Fig. 2). More than 80% of sequences have length greater than 1,000 nt, and the median length of the transcripts was around 1,700 nt in both cultivars (Fig. 2).
Flow chart for reconstruction of fruit specific and full-length transcriptomes in southern highbush blueberry ‘Suziblue’, and rabbiteye blueberry ‘Powderblue’ with Iso-Seq. CCS: circular consensus sequences, FLNC: full-length, non-concatemer reads, Cogent: Coding genome reconstruction tool, LncRNA: long non-coding RNA
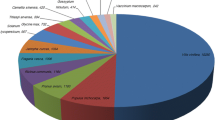
The non-redundant transcripts were subsequently annotated by Blast2GO. There were 15,172 and 12,734 transcripts that were functionally annotated by performing blastx against NCBI non-redundant protein database in ‘Suziblue’ and ‘Powderblue’ respectively (Table 2, Additional file 1: Table S1, S2). The functionally annotated transcripts were greater than 92% in both cultivars (Table 2). Additionally, 78% of these transcripts have at least one GO annotation, and more than 60% of transcripts have a GO term associated with cellular component, molecular function, and biological process (Table 2). Overall, the distribution of the GO terms in ‘Suziblue and ‘Powderblue’ are similar (Fig. 3). The top categories in cellular, molecular and biological processes were integral component of membrane, ATP binding, and oxidation reduction process respectively (Fig. 3).
Alternative splicing analysis
Overall, the number of isoforms' distributions were similar in ‘Suziblue’ and ‘Powderblue’ (Fig. 4A). Approximately 60% of non-redundant sequences had no additional isoforms, while 40% had more than one isoform, suggesting alternative splicing (Fig. 4A). About 20% of the non-redundant sequences had 2 isoforms, and about 3.5% had more than 6 isoforms (Fig. 4A). Further, intron-retention was the most common alternative splicing event, which was 46% in ‘Suziblue’ and 45% in ‘Powderblue’ (Fig. 4B). The other alternative splicing events in the descending order were alternative acceptor site, alternative donor site, and exon skipping (Fig. 4B).
Alternative splicing analysis of ‘Suziblue’ southern highbush and ‘Powderblue’ rabbiteye blueberry with Iso-Seq fruit-specific full-length transcripts. A Distribution of the number of isoforms for each non-redundant transcript is presented. The number above each bar indicates the number of non-redundant transcripts. B Identification of alternative splicing events by AStalavista. Contribution of various pathways to generation of alternatively spliced transcripts s presented
Long non-coding RNA analysis
There were 919 and 1,116 transcripts predicted as long non-coding transcripts with a length longer than 200 nt in ‘Suziblue’ and ‘Powderblue’ (Fig. 5, Additional file 1: Table S3). These constituted 5.6% and 7% the transcripts respectively in the two genotypes (Fig. 5).
Phytohormone-related and TF abundance in the transcriptomes
Overall the number and distribution among various phytohormone categories between the two cultivars were similar (Fig. 6A, B). The highest number of transcripts were related to ABA and auxin metabolism in both cultivars (Fig. 6A, B). The transcripts in these categories were assigned to response to hormones or hormone-activated/mediated signaling pathway (Additional file 1: Table S4, S5). These were followed by transcripts related to brassinosteroids and jasmonic acid metabolism and subsequently by transcripts associated with ethylene metabolism (Fig. 6A, B). Overall, the number of transcripts related to gibberellin, salicylic acid, cytokinin, and strigolactone were less abundant (Fig. 6A, B ).
Summary of the number of transcripts belonging to hormone categories (A, B) and transcription factor families in (C, D) Suziblue (A, C) and Powderblue (B, D). For genes related to phytohormones, the GO category for biological processes was searched using hormone-related key words. For transcription factors, sequences from tomato and strawberry were retrieved from the Sol Genomics Network and NCBI, and their homologs in blueberry transcriptome were identified using BLAST analysis (tblastx function)
The fruit ripening related TFs were similarly distributed in both cultivars (Fig. 6C, D). Transcription factors belonging to the APETALA2/ethylene-responsive element-binding factor (AP2/ERF) and NAC (NAM, ATAF1/2 and CUC2) family were the most abundant followed by homeodomain leucine zipper (HB-zip), basic helix-loop-helix (bHLH), MYB (v-MYB, discovered in avian myeloblastosis virus genome) and MADS-Box family (Fig. 6C, D). Transcription factors encoding the SQUAMOSA promoter binding protein (SBP)-Box and WD40 proteins were the least abundant (Fig. 6C, D).
ABA and anthocyanin content, and biosynthesis-related gene expression analyses
‘Suziblue’ and ‘Powderblue’ displayed an increase in ABA concentration from Immature Green (IMG) to Pink stage and then decreased during the Ripe stage (Fig. 7A). In ‘Suziblue’, ABA concentrations increased by 1.6-fold between IMG to Green, and Green to Pink stage and then decreased by 1.4-fold between Pink and the Ripe stage (Fig. 7A). In ‘Powderblue’, ABA concentrations increased 2.6-fold between IMG and Green stage, remained similar between Green and Pink stage and then declined 3.1-fold between Pink and Ripe stage (Fig. 7A). The transcriptome database generated in this study was used to identify candidates associated with ABA metabolism and their transcript abundance was measured. Among transcripts associated with ABA biosynthesis, abundance of 9-CIS-EPOXYCAROTENOID DIOXYGENASE1 (NCED1) was substantially lower than that of NCED2 (based on the Ct value; data not shown), suggesting that NCED2 was the predominantly expressed gene. NCED1 expression was higher between IMG and Pink stage in both cultivars (Fig. 7C). The pattern of NCED2 abundance in ‘Suziblue’ was similar across the fruit developmental stages; whereas in ‘Powderblue’ the transcript abundance increased by 3.3-fold between IMG and Green and then remained similar through advanced stages of ripening (Fig. 7D).
Concentration of ABA (A) and anthocyanin (B) during fruit ripening in ‘Suziblue’ and ‘Powderblue’. Relative abundance of transcripts involved in ABA biosynthesis (C, D) and anthocyanin biosynthesis (E–H) during fruit ripening in the two cultivars. Values represent means and standard errors of at least three replicates. ANOVA was used to test for significance (α = 0.05) among stages within a cultivar and means separation was performed using Tukey’s HSD. Means followed by different letter are significantly different with upper and lower case for ‘Suziblue’ and ‘Powderblue’ respectively. NCED: 9-CIS-EPOXYCAROTENOID DIOXYGENASE, CHS: CHALCONE SYNTHASE, UFGT: ANTHOCYANIDIN 3-O-GLUCOSYLTRANSFERASE
ABA application in blueberry promotes anthocyanin biosynthesis and related gene expression [18, 19]. In this study, anthocyanins, accumulated during fruit ripening, especially from Pink to Ripe stage by 16.1-fold and 2.4-fold in ‘Suziblue’ and ‘Powderblue’, respectively (Fig. 7B). Further, multiple anthocyanin biosynthesis genes were identified using the transcriptome and these were further evaluated. CHALCONE SYNTHASE1 (CHS1) and CHS2 were 4.6- and 2.6-fold higher between Green and Pink stage in ‘Powderblue’ and remained similar between Pink and Ripe stage (Fig. 7E, F). In ‘Suziblue’, CHS1 and CHS2 exhibited a similar pattern as mentioned above with a 3.6- and 2.3-fold increase between Green and Pink stage (Fig. 7E, F). Compared to CHS, the expression of ANTHOCYANIDIN 3-O-GLUCOSYLTRANSFERASE (UFGT) was substantially lower (based on Ct value, data not shown). The expression of UFGT1 and UFGT2, increased between Pink and Ripe stage by 3.5- and 4-fold respectively, in ‘Powderblue’; such an increase was not observed in ‘Suziblue’ (Fig. 7G, H).
Fruit texture and transcript abundance of cell wall modification-related genes during fruit ripening
Fruit compression decreased by 2.4- and 1.4-fold, and puncture by 2- and 1.5-fold between IMG and Green, and Green and Pink stages, respectively (Fig. 8A, B). Multiple cell wall remodeling-related transcripts were identified and evaluated. XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE1 (XTH1) abundance decreased during ripening whereas that of XTH2 increased in both cultivars (Fig. 8C, D). Transcript abundance of XTH2 was highest between Green and Pink stage increasing by 5.8-fold and then declining at the Ripe stage in ‘Suziblue’, whereas in ‘Powderblue’, it increased by 6.4-fold between IMG and Pink stage and continued to remain high throughout ripening (Fig. 8D). The expression of 1,4-β-MANNOSIDASE1 (βMAN1) increased between the IMG and Pink stages by 3.8-fold and 1.9-fold in ‘Suziblue’ and ‘Powderblue’ respectively, and then declined at the Ripe stage (Fig. 8E). Overall, the transcript abundance of pectin-modifying POLYGALACTURONASE (PG) was low (data not shown) and its pattern of gene expression did not significantly change during fruit ripening (Fig. 8F). The transcript abundance of PECTINESTERASE1 (PE1) increased by 1.6-fold between IMG and Green in ‘Suziblue’ and then remained constant until the Ripe stage (Fig. 8G). In ‘Powderblue’, PE1 transcript abundance increased by 1.6-fold only between IMG and Pink stage (Fig. 8G). The expression of β-GALACTOSIDASE1 (βGAL1) increased steadily during ripening in ‘Suziblue’ with a 10.2-fold increase between IMG and the ripe stage, whereas in ‘Powderblue’ it increased by 1.8-fold between IMG and Green and then remained constant until the Ripe stage (Fig. 8H).
Fruit compression (A), puncture (B), and relative abundance of transcripts involved in cell wall modification (C-H) during fruit ripening in ‘Suziblue’ and ‘Powderblue’. Values represent means and standard errors of at least three replicates. ANOVA was used to test for significance (α = 0.05) among stages within a cultivar and mean separation was performed using Tukey’s HSD. Means followed by different letter are significantly different with upper and lower case for ‘Suziblue’ and ‘Powderblue’ respectively. XTHI: XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE, βMAN, 1,4-β-MANNOSIDASE, PG: POLYGALACTURONASE, PE, PECTINESTERASE, βGAL: β-GALACTOSIDASE
Discussion
The goal of this study was to generate a full-length blueberry fruit transcriptome that can serve as a resource for identifying important ripening, and postharvest-related genes. Further this transcriptome should provide a standalone reference to map short RNA seq-reads, specific to southern highbush and rabbiteye cultivars. The blueberry fruit transcriptome generated a total of 32,119 and 31,323 full-length transcripts in ‘Suziblue’ and ‘Powderblue’ respectively. The number of transcripts were similar to that observed in strawberry PacBio sequencing that generated 33,236 transcripts during fruit development [20]. Of the total unique transcripts, 16,299 and 15,882 non-redundant transcripts were generated in ‘Suziblue’ and ‘Powderblue’, respectively. The haplotype genome suggests 32,140 genes that encode proteins for a given haplotype with a projection of 128,559 total genes for a tetraploid blueberry [21]. ‘Suziblue’, a southern highbush blueberry is tetraploid, while ‘Powderblue’, a rabbiteye blueberry is hexaploid. Blueberry has been suggested to be an allopolyploid and dominance of one of the sub-genomes during various stages of development has been proposed, which has been the case for several allopolyploid species [21]. This may explain the similarity in the number of unique transcripts generated in the tetraploid and hexaploid cultivars used in this study.
The transcriptome assembled by IsoSeq is expected to be full-length and our analyses indicated greater than 80% of transcripts were more than 1,000 nt long. The length on an average ranged between 68 to 7,798 in both cultivars with median transcript length about 1,700 nt. In cotton, transcript length from the full-length transcriptome ranged from 200–10,000 nt [22]. Other studies reported an average length of 2,047 bp [23] and 2,177 bp [24] after full length sequencing. Thus, the results obtained in this study were well within the range obtained from other studies.
The blueberry draft genome generated using 454 and Illumina sequencing predicted about 24% of all the multi-exon genes to display transcript variants due to alternative splicing or promoter use [7]. In this study, 35.96% and 37.12% of non-redundant transcripts in the rabbiteye and the southern highbush cultivars, respectively, had at least one alternative splicing event. The detection of alternative splice events in multi-exon genes has been suggested to be more robust using SMRT sequencing (around 58%) compared with Illumina (around 35%) [20]. Thus, further exploration of alternative splicing events at a large-scale or with certain genes of interest is possible with the current dataset. In the current study, intron-retention was the most common alternative splicing event which was also the case in previous studies [22, 25,26,27,28,29]. However, the frequency of a certain splice event can change depending on the developmental stage. In strawberries intron retention was the most abundant alternative splicing event, however the proportion changed during fruit development with alternative acceptor sites being the major event after fertilization [20]. Splicing patterns have also been suggested to vary according to fruit developmental stages in blueberry [7].
Fruit ripening initiation and progression is coordinated by several hormones. Mainly levels of ABA and ethylene increase, and auxin declines during fruit ripening [30]. Therefore, it was not surprising that in the blueberry fruit transcriptome, among hormone-related categories, the highest number of transcripts were associated with ABA metabolism. A role for ABA in ripening initiation has been proposed in both climacteric fruits such as tomato and non-climacteric fruits such as strawberries and grapes [31,32,33]. In highbush blueberry, ABA application can promote anthocyanin biosynthesis and transcript abundance of anthocyanin biosynthesis related genes [18, 19]. In blueberries, ABA concentration increases during the onset of ripening after which the levels decline at the Ripe stage [34]. In the current study, ABA concentration was found to increase during ripening until the Pink stage and to decrease between the Pink and Ripe stages, in both cultivars. Overall, the expression patterns of the two transcripts that encode for ABA-biosynthesis genes, NCED1 and NCED2 was consistent with ABA production, similar to the results observed in other blueberry studies [18, 34]. Similarly, the expression patterns of two CHS and two UFGT transcripts coding for enzymes in the anthocyanin biosynthesis pathway reflected changes in anthocyanin concentration. Blueberries have been classified as exhibiting atypical climacteric physiology with a functional ethylene metabolism and signaling during ripening [35]. Further application of ethephon, an ethylene releasing compound can accelerate blueberry ripening [36]. Possibly like in other fruits there may be an interaction among various hormones during ripening in blueberries as well.
During fruit ripening, the coordinated action of many cell wall modification enzymes facilitates the softening process. These enzymes are coded by large gene families, with some members displaying temporal regulation during fruit ripening [22, 37,38,39]. In this study we selected a subset of cell wall modification enzymes that have important roles during fruit development, including ripening. Consistent with progression of ripening and fruit softening, fruit firmness declined at the initiation of ripening and continued to decrease during its progression. Four transcripts associated with cell-wall modification, XTH2, βMAN1, PE1, and βGAL1, displayed an increase in abundance during various periods during ripening, suggesting their importance in facilitating cell wall remodeling during ripening. XTH enzymes display xylogucan transglucosylase and/or hydrolase activity, and are important for fruit softening, for example in persimmon [40, 41]. Of the two XTH transcripts characterized in this study only XTH2 expression increased during fruit ripening. The transcript abundance of XTH1 was higher in the IMG fruit suggesting its role in cell wall modification during early fruit development. Such a temporal regulation for XTH gene family members have been previously reported in tomato [42]. Another enzyme similar to XTH in function, βMAN, displayed high activity during fruit ripening in tomato [43, 44]. Similarly, the βMAN1 transcript selected in this study exhibited higher transcript abundance at the Pink stage. In addition, pectin modifying enzymes such as PG, PME and βGal play a positive role in fruit softening during ripening and postharvest storage [45,46,47,48]. In this study PME1 and βGal1 were induced during blueberry ripening, however the transcript abundance of PG1 was low in the southern highbush cultivar and almost undetectable in the rabbiteye cultivar. PG activity in highbush blueberry is high during initial ripening stages and then declines as ripening progresses and is correlated with pectin depolymerization [49]. While the current study does not support such a role in southern highbush and rabbiteye blueberry, it is possible that ripening specific PGs were not targeted for gene expression analysis here.
Upstream of ripening-related genes, transcription factors such as NAC, MADS-box, and CNR play a key role in ripening in tomato [50]. Multiple potential ripening-related transcription factors were identified from the full-length transcriptome. NAC transcription factors play a role in ripening and in leaf and fruit senescence [51,52,53,54]. One of the well-studied members of the NAC transcription factor family, NON-RIPENING (NOR), and another member NOR-like 1 positively regulates ripening upstream of ethylene [50,51,52, 55]. More recently, NAC transcription factors were shown to downregulate the transcript abundance of the abscisic acid biosynthesis gene, NCED in citrus, suggesting a potential negative role for some members within this family [56]. In tomato, RIN, a MADS-box transcription factor, and CNR, a member of the SBP-box gene family are important in ripening initiation [57,58,59,60]. In addition, the transcriptome also contained anthocyanin-related transcription factors. Transcriptional regulation of anthocyanin biosynthesis is coordinated via a protein complex consisting of MYB, bHLH, and WD40 [61]. This complex was also shown to regulate anthocyanin biosynthesis in blueberry fruit [62].
Collectively, the expression of ABA and anthocyanin biosynthesis related genes, and cell wall modification genes indicate that the current transcriptome is robust and offers full length gene sequences for investigating molecular aspects of ripening and postharvest processes.
Conclusions
This study describes the first full-length fruit transcriptome for southern highbush and rabbiteye type of blueberry. This transcriptome is a useful resource and can be used as a standalone reference for mapping short RNA seq-reads specific to southern highbush and rabbiteye blueberry cultivars. Further, this study demonstrates the robustness of the transcriptome with respect to identification of genes associated with fruit ripening and quality traits. Together, the full-length transcriptome developed here offers a valuable genomic resource that can help to facilitate breeding of fruit quality and other related traits in blueberry.
Methods
Sample collection
Fruit from southern highbush ‘Suziblue’ (Vaccinium sp.) and rabbiteye blueberry ‘Powderblue’ (V. virgatum) were collected at various stages of ripening and postharvest storage. Fruit used in this study were from cultivated species and appropriate permissions were obtained for fruit collection. All experimental procedures including use and collection of the fruits comply with the ethical standards and legislations. Ripening stages included Immature Green (IMG), Green, Pink, and Ripe. IMG stage was defined as a combination of S4 (7–9 mm) and S5 (9–13 mm) based on size [34]. Green stage was based on size and color (< 13 mm or < 30% pink skin). The remaining two stages were harvested based on surface coloration with Pink stage being predominantly pink (> 50%) and Ripe fruit being fully blue. Fruit from these stages were frozen in liquid N2 immediately after harvest in the field and stored at -80 °C. To collect fruit at various postharvest stages, ripe fruit were hand harvested and stored in 1-pint clamshells in a walk-in cooler at 4 °C and approximately 90% relative humidity. Fruit were taken at three stages during storage, left at room temperature (~ 25 °C) for 2 h, subsequently frozen in liquid N2 and stored at -80 °C until further use. The three postharvest (PH) stages of ‘Suziblue’ were 3 d after storage (PH3), 8 d after storage (PH8) and 13 d after storage (PH13), and for ‘Powderblue’ PH4, PH8, and PH14 (4, 8 and 14 d after storage). For ‘Suziblue’ all fruit were collected from the UGA Blueberry Research farm in Alapaha, GA (31.345288, -83.240317) in 2015, except for the IMG stage collected from the same farm in 2018. All ‘Powderblue’ fruit were collected from the Durham Horticulture Research farm in Watkinsville, GA (33.8872879, -83.4206120) in 2018. In each cultivar, for every stage multiple fruit (~ six) were pooled for RNA extraction.
RNA extraction and PacBio iso-seq
All samples were ground into a fine powder using a mortar and pestle with liquid N2 for RNA extraction. RNA extraction was performed by using a modified CTAB-based method [63]. RNA was analyzed on a Bioanalyzer (Agilent Technologies, 2100, CA) to ensure high quality of RNA (RIN > 8). For each cultivar, 2 µg of RNA from all developmental and postharvest stages described above were pooled. Libraries of the two samples were constructed following Pacific Biosciences’s standard protocol for Iso-Seq (Iso-Seq Template Preparation for Sequel Systems). Each library was sequenced on 1 SMRT cell on a PacBio Sequel Systems (Pacific Bioscience, CA).
Transcriptome reconstruction and annotation
The full-length transcripts were characterized by the isoseq3 tool in SMRT Link v8.0, a software designed by PacBio to analyze Single Molecule, Real-Time (SMRT) sequencing data, with the following steps suggested by the SMRT Tools Reference Guide: 1) circular consensus sequences (CCS) were generated from the raw data sub-reads. 2) the IsoSeq v2 primer sets at both ends of the sequences were removed. 3) noises including poly A tails and concatemers were removed generating full-length, non-concatemer (FLNC) reads. 4) full-length unpolished transcripts were generated by clustered consensus sequences. 5) full-length unpolished transcripts were polished by using raw subreads, including non-full-length transcripts, to generate full-length transcripts (Fig. 1). Next to remove the redundancy, the polished full-length transcripts were mapped to the northern highbush blueberry ‘Draper’ (V. corymbosum) genome [21] and then collapsed transcripts by genomic mapping. The transcripts unmapped to the genome were collapsed by Cogent (https://github.com/Magdoll/Cogent/). Finally, non-redundant transcripts were generated after combining the transcripts collapsed after mapping to the genome and those from Cogent analyses (Fig. 1).
The non-redundant transcripts were further annotated by Blast2GO [64]. The annotation process included 1) performing Blastx against NCBI non-redundant protein database with flowering plants as taxonomy filter, 2) retrieving domain/motif information by InterProScan, and 3) assigning gene ontology (GO) terms and enzyme codes. Except for the parameters mentioned above, all steps were performed with the default settings.
Alternative splicing identification and long non-coding RNA analysis
The generic feature format output generated by Isoseq3 and Cogent were used to classify alternative splicing events by alternative splicing transcriptional landscape visualization tool (AStalavista) [65]. To identify long non-coding RNA, the non-redundant transcriptomes for each cultivar were analyzed by Coding-Non-Coding Identifying tool (CNIT) [66] with the setting of the plant mode (Fig. 1).
Identification of phytohormone-related genes and ripening-related transcription factors
For phytohormone-related genes, the GO category for biological processes was searched using key words for hormones which included, ABA, auxin, brassinosteroid, jasmonic acid, ethylene, gibberellin, salicyclic acid, cytokinin, and strigolactone (Additional file 1: Table S4, S5). For mining of transcription factors, sequences were retrieved from the Sol Genomics Network (https://solgenomics.net/) for tomato and from NCBI (https://www.ncbi.nlm.nih.gov/) for strawberries. The transcription factors belonged to various gene families with known roles in ripening. The tomato MADS-Box genes included, RIPENING INHIBITOR (RIN: Solyc05g012020), TOMATO AGAMOUS-LIKE 1 (TAGL1: Solyc07g055920), MADS1 (Solyc03g114840), FRUITFUL1 (FUL1: Solyc06g069430), and FRUITFUL2 (FUL2: Solyc03g114830), and from strawberry, MADS-RIN (AF484683.1). Also, transcription factors belonging to other families such as, NAC (NAC4: Solyc11g017470, NOR: Solyc10g006880), AP2/ERF (AP2: Solyc03g044300; ERF: Solyc01g065980), SBP-Box (SPL-CNR: Solyc02g077920), and HB-zip (HB1: Solyc02g086930) were identified from tomato. Transcription factors related to anthocyanin biosynthesis included the MYB (JQ989281), bHLH domain protein (JQ989284), and WD40 protein (JQ989287) from the strawberry. These transcription factors were used to identify blueberry homologs in the ‘Powderblue’ and ‘Suziblue’ transcriptomes using BLAST analysis (tblastx function). For all the blueberry homologs identified, reciprocal blast analysis was performed to confirm their gene identity.
ABA quantification
For ABA quantification, 0.6 g of fine powdered frozen fruit sample was shipped in dry ice to the University of North Texas (UNT) BioAnalytical Facility. The protocol used by UNT was based on [67]. Briefly, samples were lyophilized, and 10 mg of sample extracted with 400 µL of 10% methanol containing 1% acetic acid with an internal standard, 13C-benzoic acid. Subsequently samples were disrupted using a Tissuelyzer bead mill, and then incubated in ice for 30 min. Samples were then centrifuged at 17,000 × g for 10 min at 4 °C and the supernatant collected. The pellet was re-extracted in methanol and acetic acid and the two supernatants pooled. The sample was filtered using ultra-centrifugal filter units (Amicon, Florida, USA). Finally, ABA was quantified by liquid chromatography with tandem mass spectrometry (LC–MS/MS).
Anthocyanin measurements
Fruit tissue was finely ground in liquid N2 using a mortar and pestle. 100 mg of ground sample was extracted using 2 mL of 100% methanol acidified with 0.1 mL HCl [68]. Samples were mixed briefly using a vortex and sonicated in dark for 10 min using a Bransonic 220 sonicator (Branson Ultrasonics Corp., Danbury, CT). Next, tubes were mixed in the dark for 1 h at 300 ppm using a Environ-shaker (Lab-Line Instruments Inc., Melrose Park, IL) and centrifuged at 2000 × g for 25 min at 20 °C. The supernatant was transferred into a new tube. Total anthocyanins were measured using the differential pH method. 40 µL of the supernatant along with 160 µL of 0.025 M potassium chloride (pH 1.0) and 160 µL of 0.4 M sodium acetate (pH 4.5) was transferred into a 96 well plate (Becton Diskinson, Franklin Lakes, NJ). A blank was prepared using 40 µL of extraction buffer (methanol acidified with HCl) along with the two buffers described above. The samples were mixed with the buffers using a multichannel pipette and the absorbance measured at 520 and 700 nm using a Biotek microplate reader (BioTek, Winooski, VT). Anthocyanin concentration was calculated as cyanidin-3-glucosides equivalents using the equation in [69, 70].
Fruit firmness measurements
Fruit compression and puncture in ‘Powderblue’ were measured as described in [36]. Briefly, 12 fruit per replicate (total four replicates) were used for compression and puncture analyses using a fruit texture analyzer (GS-15, Güss Manufacturing, Strand, South Africa). For compression analyses, a probe with a 15-mm diameter end plate was used to calculate the force required to compress the fruit by 1 mm. For puncture analyses, a 1.5-mm probe was used to measure the force required to puncture the fruit skin while traveling a distance of 3 mm.
Gene expression analysis
Four fruit developmental stages, IMG, Green, Pink, and Ripe stages were used for gene expression analyses. Fruit samples from the cultivar, Suziblue, were the same as that used for IsoSeq. For the cultivar, Powderblue, samples harvested in 2020 as previously described from the Durham Horticulture Research farm in Watkinsville, GA. The RNA extraction method was the same as described above. The synthesis of cDNA was conducted using 1 µg of total RNA. After removing any potential DNA from the RNA sample by DNase treatment, cDNA was synthesized by reverse transcription following the manufactures’ protocol (Promega, Madison, WI, USA) and the final volume was diluted to 100 µl. After that, real-time quantitative reverse transcription PCR (qRT-PCR) analysis was performed by using 1 μL of cDNA with PowerUp SYBR Green Master Mix reagent (Applied Biosystems, Foster City, CA, USA) and with Stratagene Mx3005P qRT-PCR instrument (Agilent Technologies, USA) to measure the expression of the following genes: NCED, CHS, UFGT, XTH, PG, PE, and βGAL (Additional file 2). Three reference genes, UBIQUITIN-CONJUGATING ENZYME (UBC28), RNA HELICASE-LIKE (RH8), and CLATHRIN ADAPTER COMPLEXES MEDIUM SUBUNIT FAMILY PROTEIN (CACSa) [63], were used to normalize the expression of the target gene. The concentration of forward and reverse primers was 0.2 μM for all genes except PG (0.15 μM). The reaction conditions were set at: 50 °C for 2 min, 95 °C for 5 min, followed by 95 °C for 15 s, and 60 °C for 1 min, repeated for 40 cycles for all genes, except NCED. For NCED, the annealing and extension temperatures were set at 64 °C. Melt curve analysis was performed at 95 °C for 1 min, 55 °C for 30 s, 95 °C for 30 s to check for primer specificity. Efficiency of the qRT-PCR reactions were determined using the LinRegPCR software [71]. The relative quantity (RQ) values were determined using the Ct (threshold cycle) value and were corrected for primer efficiency [72]. RQ values of a given sample were normalized using the geometric mean of the RQ values of reference genes for that sample to determine normalized RQ values (NRQ) [72]. The Pink Stage was used as the reference stage for NCED1 and PG and all data are expressed as fold-change in relation to gene expression at this stage. For all other genes, the IMG stage of ‘Suziblue’ was set as the reference stage, with fold-change for gene expression calculated using this stage. Standard error was calculated as described in [72]. Statistical analyses were performed on log2 transformed NRQ values. Statistical analysis was performed using one-way analysis of variance (ANOVA) to compare across stages within a given genotype. If significant (P ≤ 0.05), mean separation was performed using Tukey’s honestly significant difference (HSD) test in JMP Pro 15 (SAS Institute, Cary, NC, USA). All figures were made using SigmaPlot (ver. 14.0; SYSTAT, Palo Alto, CA, USA) and figures were compiled using Inkscape (ver. 1.0; Boston, MA, USA).
Availability of data and materials
Datasets used in the current study that support the conclusion are included within the article and as additional files. The raw reads are available in the NCBI Sequence Read Archive (SRA) BioProject database (BioProject ID PRJNA814709; http://www.ncbi.nlm.nih.gov/bioproject/814709). The link provided will be active upon acceptance of this manuscript. Other data sets generated in this study will be available from the corresponding author upon request.
Abbreviations
- ABA:
-
Abscisic acid
- AP2/ERF:
-
APETALA2/ethylene-responsive element-binding factor
- bHLH:
-
Basic helix-loop-helix
- CACSa :
-
CLATHRIN ADAPTER COMPLEXES MEDIUM SUBUNIT FAMILY PROTEIN
- CHS :
-
CHALCONE SYNTHASE
- CCS:
-
Circular consensus sequences
- CNIT:
-
Coding-non-coding identifying tool
- FUL1 :
-
FRUITFUL1
- FUL2 :
-
FRUITFUL2
- FLNC:
-
Full-length, non-concatemer
- GO:
-
Gene ontology
- HB-zip:
-
Homeodomain leucine zipper
- IMG:
-
Immature green
- Iso-Seq:
-
Isoform sequencing
- LC–MS/MS:
-
Liquid chromatography with tandem mass spectrometry
- NAC :
-
NAM, ATAF1/2 And CUC2
- NCED :
-
9-CIS-EPOXYCAROTENOID DIOXYGENASE
- NOR :
-
NON-RIPENING
- nt:
-
Nucleotides
- PacBio:
-
Pacific biosciences
- PE :
-
PECTINESTERASE
- PG :
-
POLYGALACTURONASE
- PH:
-
Postharvest
- qRT-PCR:
-
Real-time quantitative reverse transcription PCR
- RH8 :
-
RNA HELICASE-LIKE
- RIN :
-
RIPENING INHIBITOR
- SMRT:
-
Single molecule, real-time
- SBP:
-
SQUAMOSA promoter binding protein
- TAGL1 :
-
TOMATO AGAMOUS-LIKE 1
- UBC28 :
-
UBIQUITIN-CONJUGATING ENZYME
- UFGT :
-
ANTHOCYANIDIN 3-O-GLUCOSYLTRANSFERASE
- XTH :
-
XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE
- βGAL :
-
β-GALACTOSIDASE
- βMAN :
-
1,4-β-MANNOSIDASE
References
Basu A, Rhone M, Lyons TJ. Berries: emerging impact on cardiovascular health. Nutr Rev.2010;68(3):168-77.
Neto CC. Cranberry and blueberry: evidence for protective effects against cancer and vascular diseases. Mol Nutr Food Res.2007;51(6):652-64.
USDA-NASS. Noncitrus Fruits and Nuts 2020 Summary. 2021. https://downloads.usda.library.cornell.edu/usda-esmis/files/zs25x846c/sf269213r/6t054c23t/ncit0521.pdf
Lang GA. Southern highbush blueberries: Physiological and cultural factors important for optimal cropping of these complex hybrids. Acta Hort. 1993;72-80.
Rowland LJ, Ogden EL, Bassil N, Buck EJ, McCallum S, Graham J, Brown A, Wiedow C, Campbell AM, Haynes KG. Construction of a genetic linkage map of an interspecific diploid blueberry population and identification of QTL for chilling requirement and cold hardiness. Mol Breeding.2014;34(4):2033-48.
Bian Y, Ballington J, Raja A, Brouwer C, Reid R, Burke M, Wang X, Rowland LJ, Bassil N, Brown A. Patterns of simple sequence repeats in cultivated blueberries (Vaccinium section Cyanococcus spp.) and their use in revealing genetic diversity and population structure. Molecular Breeding. 2014;34(2):675–89.
Gupta V, Estrada AD, Blakley I, Reid R, Patel K, Meyer MD, Andersen SU, Brown AF, Lila MA, Loraine AE. RNA-Seq analysis and annotation of a draft blueberry genome assembly identifies candidate genes involved in fruit ripening, biosynthesis of bioactive compounds, and stage-specific alternative splicing. Gigascience.2015;4(1):5.
Cui F, Ye X, Li X, Yang Y, Hu Z, Overmyer K, Brosché M, Yu H, Salojärvi J. Chromosome-level genome assembly of the diploid blueberry Vaccinium darrowii provides insights into its subtropical adaptation and cuticle synthesis. Plant Commun. 2022;3(4):100307.
Yu J, Hulse-Kemp AM, Babiker E, Staton M. High-quality reference genome and annotation aids understanding of berry development for evergreen blueberry (Vaccinium darrowii). Hortic Res.2021;8:228.
Zhao H, Gao Z, Wang L, Wang J, Wang S, Fei B, Chen C, Shi C, Liu X, Zhang H. Chromosome-level reference genome and alternative splicing atlas of moso bamboo (Phyllostachys edulis). Gigascience.2018;7(10):giy115.
Adams J. Transcriptome: connecting the genome to gene function. Nat Educ.2008;1(1):195.
Chen X, Liu X, Zhu S, Tang S, Mei S, Chen J, Li S, Liu M, Gu Y, Dai Q. Transcriptome-referenced association study of clove shape traits in garlic. DNA Res.2018; 25(6):587-96.
Gonzalez-Garay ML. Introduction to isoform sequencing using Pacific Biosciences technology (Iso-Seq). In: Transcriptomics and Gene Regulation. Translational Bioinformatics, Springer; 2016;9:141–160.
Liu X, Li X, Wen X, Zhang Y, Ding Y, Zhang Y, Gao B, Zhang D. PacBio full-length transcriptome of wild apple (Malus sieversii) provides insights into canker disease dynamic response. BMC Genomics.2021;22:52.
Zheng Q, Chen W, Luo M, Xu L, Zhang Q, Luo Z. Comparative transcriptome analysis reveals regulatory network and regulators associated with proanthocyanidin accumulation in persimmon. BMC Plant Biol.2021;21:356.
Ge Y, Cheng Z, Si X, Ma W, Tan L, Zang X, Wu B, Xu Z, Wang N, Zhou Z. Transcriptome profiling provides insight into the genes in carotenoid biosynthesis during the mesocarp and seed developmental stages of avocado (Persea americana). Int J Mol Sci.2019;20(17):4117.
Fang R, Weixiong H, Jinyan Y, Xing L, Ji Z, Shuangyun Z, Biao D, Wenzhong T, Zhenyu A. Characterization of full-length transcriptome and mechanisms of sugar accumulation in Annona squamosa fruit. Biocell.2020;44(4):737.
Chung SW, Yu DJ, Oh HD, Ahn JH, Huh JH, Lee HJ. Transcriptional regulation of abscisic acid biosynthesis and signal transduction, and anthocyanin biosynthesis in ‘Bluecrop’highbush blueberry fruit during ripening. PLoS ONE.2019;14(7):e0220015.
Oh HD, Yu DJ, Chung SW, Chea S, Lee HJ. Abscisic acid stimulates anthocyanin accumulation in ‘Jersey’highbush blueberry fruits during ripening. Food Chem.2018;244:403-07.
Li Y, Dai C, Hu C, Liu Z, Kang C. Global identification of alternative splicing via comparative analysis of SMRT‐and Illumina‐based RNA‐seq in strawberry. Plant J.2017;90(1):164-76.
Colle M, Leisner CP, Wai CM, Ou S, Bird KA, Wang J, Wisecaver JH, Yocca AE, Alger EI, Tang H. Haplotype-phased genome and evolution of phytonutrient pathways of tetraploid blueberry. Gigascience.2019;8(3):giz012.
Feng S, Xu M, Liu F, Cui C, Zhou B. Reconstruction of the full-length transcriptome atlas using PacBio Iso-Seq provides insight into the alternative splicing in Gossypium australe. BMC Plant Biol. 2019;19:365.
Kuang X, Sun S, Wei J, Li Y, Sun C. Iso-Seq analysis of the Taxus cuspidata transcriptome reveals the complexity of Taxol biosynthesis. BMC Plant Biol. 2019;19:210.
Wu Q, Zang F, Xie X, Ma Y, Zheng Y, Zang D. Full-length transcriptome sequencing analysis and development of EST-SSR markers for the endangered species Populus wulianensis. Sci Rep.2020;10:16249.
Shen Y, Zhou Z, Wang Z, Li W, Fang C, Wu M, Ma Y, Liu T, Kong L-A, Peng D-L. Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell.2014;26(3):996-1008.
Thatcher SR, Zhou W, Leonard A, Wang B-B, Beatty M, Zastrow-Hayes G, Zhao X, Baumgarten A, Li B. Genome-wide analysis of alternative splicing in Zea mays: landscape and genetic regulation. Plant Cell.2014;26(9):3472-87.
Wang B-B, Brendel V. Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci.2006;103(18):7175-80.
Zhu G, Li W, Zhang F, Guo W. RNA-seq analysis reveals alternative splicing under salt stress in cotton, Gossypium davidsonii. BMC Genomics.2018;19:73.
Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, Konagaya A, Shinozaki K. Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Res. 2004;32(17):5096-103.
Fenn MA, Giovannoni JJ. Phytohormones in fruit development and maturation. PlantJ. 2021;105(2):446-58.
Cantín CM, Fidelibus MW, Crisosto CH. Application of abscisic acid (ABA) at veraison advanced red color development and maintained postharvest quality of ‘Crimson Seedless’ grapes. Postharvest Biol Technol. 2007;46(3):237-41.
Jia H-F, Chai Y-M, Li C-L, Lu D, Luo J-J, Qin L, Shen Y-Y. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 2011;157(1):188-99.
Zhang M, Yuan B, Leng P. The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J Exp Bot. 2009;60(6):1579-88.
Zifkin M, Jin A, Ozga JA, Zaharia LI, Schernthaner JP, Gesell A, Abrams SR, Kennedy JA, Constabel CP. Gene expression and metabolite profiling of developing highbush blueberry fruit indicates transcriptional regulation of flavonoid metabolism and activation of abscisic acid metabolism. Plant Physiol. 2012;158(1):200-24.
Wang Y-W, Acharya TP, Malladi A, Tsai H-J, NeSmith DS, Doyle JW, Nambeesan SU. Atypical climacteric and functional ethylene metabolism and signaling during fruit ripening in blueberry (Vaccinium sp.). Front Plant Sci. 2022;13:932642.
Wang Y-W, Malladi A, Doyle JW, Scherm H, Nambeesan SU. The effect of ethephon, abscisic acid, and methyl jasmonate on fruit ripening in rabbiteye blueberry (Vaccinium virgatum). Horticulturae. 2018;4(3):24.
Brummell DA. Cell wall disassembly in ripening fruit. Funct Plant Biol. 2006;33(2):103-19.
Brummell DA, Harpster MH. Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant Mol Biol. 2001;47(1):311-40.
Posé S, Paniagua C, Matas AJ, Gunning AP, Morris VJ, Quesada MA, Mercado JA. A nanostructural view of the cell wall disassembly process during fruit ripening and postharvest storage by atomic force microscopy. Trends Food Sci Technol. 2019;87:47-58.
Han Y, Ban Q, Li H, Hou Y, Jin M, Han S, Rao J. DkXTH8, a novel xyloglucan endotransglucosylase/hydrolase in persimmon, alters cell wall structure and promotes leaf senescence and fruit postharvest softening. Sci Rep. 2016;6:39155.
Saladié M, Rose JK, Cosgrove DJ, Catalá C. Characterization of a new xyloglucan endotransglucosylase/hydrolase (XTH) from ripening tomato fruit and implications for the diverse modes of enzymic action. Plant J. 2006;47(2):282-95.
Ohba T, Takahashi S, Asada K. Alteration of fruit characteristics in transgenic tomatoes with modified expression of a xyloglucan endotransglucosylase/hydrolase gene. Plant Biotechnol. 2011;28:25-32.
Bewley JD, Banik M, Bourgault R, Feurtado JA, Toorop P, Hilhorst HW. Endo‐β‐mannanase activity increases in the skin and outer pericarp of tomato fruits during ripening. J Exp Bot. 2000;51(344):529-38.
Schröder R, Atkinson RG, Redgwell RJ. Re-interpreting the role of endo-β-mannanases as mannan endotransglycosylase/hydrolases in the plant cell wall. Ann Bot. 2009;104(2):197-204.
Castillejo C, de la Fuente JI, Iannetta P, Botella MÁ, Valpuesta V. Pectin esterase gene family in strawberry fruit: study of FaPE1, a ripening‐specific isoform. J Exp Bot. 2004;55(398):909-18.
Paniagua C, Blanco-Portales R, Barceló-Muñoz M, García-Gago JA, Waldron KW, Quesada MA, Muñoz-Blanco J, Mercado JA. Antisense down-regulation of the strawberry β-galactosidase gene FaβGal4 increases cell wall galactose levels and reduces fruit softening. J Exp Bot. 2016;67(3):619-31.
Smith DL, Abbott JA, Gross KC. Down-regulation of tomato β-galactosidase 4 results in decreased fruit softening. Plant Physiol. 2002;129(4):1755-62.
Tieman DM, Handa AK. Reduction in pectin methylesterase activity modifies tissue integrity and cation levels in ripening tomato (Lycopersicon esculentum Mill.) fruits. Plant Physiol. 1994;106(2):429-36.
Chea S, Yu DJ, Park J, Oh HD, Chung SW, Lee HJ. Fruit softening correlates with enzymatic and compositional changes in fruit cell wall during ripening in ‘Bluecrop’highbush blueberries. Scientia Hort. 2019;245:163-70.
Giovannoni JJ. Genetic regulation of fruit development and ripening. Plant Cell. 2004;16(suppl 1):S170-80.
Barry CS, Giovannoni JJ. Ethylene and fruit ripening. J Plant Growth Regul. 2007;26(2):143.
Casals J, Pascual L, Cañizares J, Cebolla-Cornejo J, Casañas F, Nuez F. Genetic basis of long shelf life and variability into Penjar tomato. Genet Resour Crop Evol. 2012;59(2):219-29.
Kumar R, Tamboli V, Sharma R, Sreelakshmi Y. NAC-NOR mutations in tomato Penjar accessions attenuate multiple metabolic processes and prolong the fruit shelf life. Food Chem. 2018;259:234-44.
Ma X, Balazadeh S, Mueller-Roeber B. Tomato fruit ripening factor NOR controls leaf senescence. J Exp Bot. 2019;70(10):2727-40.
Gao Y, Wei W, Zhao X, Tan X, Fan Z, Zhang Y, Jing Y, Meng L, Zhu B, Zhu H. A NAC transcription factor, NOR-like1, is a new positive regulator of tomato fruit ripening. Hortic Res. 2018;5:75.
Zhu F, Luo T, Liu C, Wang Y, Zheng L, Xiao X, Zhang M, Yang H, Yang W, Xu R. A NAC transcription factor and its interaction protein hinder abscisic acid biosynthesis by synergistically repressing NCED5 in Citrus reticulata. J Exp Bot. 2020;71(12):3613-25.
Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D. A tomato HD‐Zip homeobox protein, LeHB‐1, plays an important role in floral organogenesis and ripening. Plant J. 2008;55(2):301-10.
Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38(8):948-52.
Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ. The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol. 2011;157(3):1568-1579.
Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. A MADS-box gene necessary for fruit ripening at the tomato Ripening-Inhibitor (rin) locus. Science 2002;296(5566):343-46.
Schaart JG, Dubos C, Romero De La Fuente I, van Houwelingen AM, de Vos RC, Jonker HH, Xu W, Routaboul JM, Lepiniec L, Bovy AG. Identification and characterization of MYB‐b HLH‐WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria× ananassa) fruits. New Phytol. 2013;197(2):454-67.
Zhao M, Li J, Zhu L, Chang P, Li L, Zhang L. Identification and characterization of MYB-bHLH-WD40 regulatory complex members controlling anthocyanidin biosynthesis in blueberry fruits development. Genes. 2019;10(7):496.
Vashisth T, Johnson LK, Malladi A. An efficient RNA isolation procedure and identification of reference genes for normalization of gene expression in blueberry. Plant Cell Rep. 2011;30(12):2167-76.
Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36(10):3420-35.
Foissac S, Sammeth M. ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res. 2007;35(suppl_2):W297-W299.
Guo J-C, Fang S-S, Wu Y, Zhang J-H, Chen Y, Liu J, Wu B, Wu J-R, Li E-M, Xu L-Y. CNIT: a fast and accurate web tool for identifying protein-coding and long non-coding transcripts based on intrinsic sequence composition. Nucleic Acids Res. 2019;47(W1):W516-W522.
Forcat S, Bennett MH, Mansfield JW, Grant MR. A rapid and robust method for simultaneously measuring changes in the phytohormones ABA, JA and SA in plants following biotic and abiotic stress. Plant Methods. 2008;4(1):1-8.
Karppinen K, Tegelberg P, Häggman H, Jaakola L. Abscisic acid regulates anthocyanin biosynthesis and gene expression associated with cell wall modification in ripening bilberry (Vaccinium myrtillus L.) fruits. Front Plant Sci. 2018;9:1259.
Lee J, Durst RW, Wrolstad RE. Determination of total monomeric anthocyanin pigment content of fruit juices, beverages, natural colorants, and wines by the pH differential method: collaborative study. J AOAC Int. 2005;88(5):1269-78.
Lee J, Rennaker C, Wrolstad RE. Correlation of two anthocyanin quantification methods: HPLC and spectrophotometric methods. Food Chem. 2008;110(3):782-6.
Ruijter J, Ramakers C, Hoogaars W, Karlen Y, Bakker O, Van den Hoff M, Moorman A. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37(6):e45-e45.
Rieu I, Powers SJ. Real-time quantitative RT-PCR: design, calculations, and statistics. Plant Cell. 2009;21(4):1031-1033.
Acknowledgements
We thank the Georgia Genomics facility for performing the sequencing and Magdy Alabady for providing consultation on data analysis. We thank Tej P. Acharya for helping with identification of blueberry transcription factors and fruit texture analysis. We would like to thank Rion Mooneyham for design of cell wall modification primers. We thank Anish Malladi for critically reviewing the manuscript.
Funding
This publication was supported by funds received from the College of Agriculture and Environmental Sciences at University of Georgia.
Author information
Authors and Affiliations
Contributions
Y-WW and SUN conceived the study and designed the experiments. Y-WW and SUN was involved in fruit collection, processing of samples for sequencing, and gene expression. Y-WW and SUN contributed to data analysis in bioinformatics and preparation of the final manuscript. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Table S1. The annotation of fruit-specific transcriptome in Suziblue. Table S2. The annotation of fruit-specific transcriptome in Powderblue. Table S3. List of long non-coding RNA in fruit-specific transcriptomes of Suziblue and Powderblue. Table S4. Summary of hormone-related transcripts in Suziblue fruit-specific transcriptome. Table S5. Summary of hormone-related transcripts in Powderblue fruit-specific transcriptome.
Additional file 2.
List of primers used for QRT-PCR analysis.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, YW., Nambeesan, S.U. Full-length fruit transcriptomes of southern highbush (Vaccinium sp.) and rabbiteye (V. virgatum Ait.) blueberry. BMC Genomics 23, 733 (2022). https://doi.org/10.1186/s12864-022-08935-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-022-08935-5