Abstract
Brassica species include important oil crops and vegetables in the world. The R2R3-MYB gene participates in a variety of plant functions, including the activation or inhibition of anthocyanin biosynthesis. Although previous studies have reported its phylogenetic relationships, gene structures, and expression patterns in Arabidopsis, the number and sequence variation of this gene family in Brassica crops and its involvement in the natural quantitative variation in anthocyanin biosynthesis regulation are still largely unknown. In this study, by using whole genome sequences and comprehensive genome-wide comparative analysis among the six cultivated Brassica species, 2120 R2R3-MYB genes were identified in six Brassica species, in total These R2R3-MYB genes were phylogenetically clustered into 12 groups. The R2R3-MYB family between A and C subgenomes showed better collinearity than between B and C and between A and B. From comparing transcriptional changes of five Brassica species with the purple and green leaves for the detection of the R2R3-MYB genes associated with anthocyanin biosynthesis, 7 R2R3-MYB genes were co-differentially expressed. The promoter and structure analysis of these genes showed that some variations between non-coding region, but they were highly conserved at the protein level and spatial structure. Co-expression analysis of anthocyanin-related genes and R2R3-MYBs indicated that MYB90 was strongly co-expressed with TT8, and they were co-expressed with structural genes F3H, LDOX, ANS and UF3GT at the same time. These results further clarified the roles of the R2R3-MYBs for leaf coloration in Brasica species, which provided new insights into the functions of the R2R3-MYB gene family in Brasica species.
Similar content being viewed by others
Introduction
The roles of transcription factors (TFs) in the regulation of transcript levels of structural genes controls many crucial biological processes [2]. The TFs usually recognize target DNA in a sequence-specific manner and regulate the frequency of initiation of transcription upon binding to specific sites/motifs in the promoter of target genes. Generally, TFs can work as activators, repressors, or both to regulate the expression of target genes. MYB transcription factors have since been found in all plants and comprised one of the largest of transcription factors family [27]. MYB transcription factor contains a conserved DBD (DNA-binding domain), which is generally composed of 1–4 imperfect repeats, named R repeat [14, 27]. Each R repeat covers 50–55 amino acids in length that fold into three α-helices, when bound to target gene specific promoter sequences, the second and third α-helices were form a helix-turn-helix (HTH) structure [27, 31], and the third α-helix usually play a recognition role in binding to a short DNA sequence [43].
For decades, it has been identified and reported in many plants that the R2R3-MYB transcription factor is widely involved in plant development and metabolic regulation [13, 28, 43, 45,46,47]. Recently, the increasing availability of plant genome sequences have facilitated a better understanding of this large gene family [28, 45,46,47]. The first plant R3R3-MYB gene COLORED1 (C1) was isolated from maize (Zea mays) which encode a regulatory protein involved in anthocyanin biosynthesis [32]. Subsequently, the R2R3-MYB-mediated anthocyanin synthesis pathway was revealed in the model plant Arabidopsis, PRODUCTION OF ANTHOCYANIN PIGMENT 1/2 (PAP1/2, MYB75/90) are reported to be involved in the transcriptional regulation of anthocyanins in vegetative tissues, while TRANSPARENT TESTA 2 (TT2, MYB123) is involved in the biosynthesis and accumulation of anthocyanins in seed coats [19, 52]. Nevertheless, compared to Arabidopsis, relatively few members of the Brassica species R2R3-MYB gene family have been well identified and functional characterized. Moreover, the R2R3-MYBs characterized in Brassica species are limited in the transcriptional regulatory mechanism of anthocyanin metabolism pathway to date.
Anthocyanins are secondary metabolites of flavonoids, which can change from red, purple to blue, and are the main substances for plant coloring in nature. Anthocyanins play an important role in plant growth and development, especially against biotic and abiotic stress. For humans, previous studies have found that anthocyanins can scavenge free oxygen, improve vision, delay aging and used in cancer treatment. In recent years, R2R3-MYB transcription factors have been shown to be involved in phenylpropanoid metabolism [23, 41] and activate the structural genes in the anthocyanin biosynthetic pathway in many plants [1, 20, 51]. In Brassica species, the R2R3-MYB transcription factors play a key role in anthocyanin biosynthesis pathway. In B. oleracea, BoMYB2 with different types of mutations in the promoter region determines the different color types [53]. Similarly, the activated expression of the BrMYB2 gene in purple-leaf B. rapa and the BjPur gene in purple-leaf B. juncea is due to the large deletion of the first intron [21, 22]. In B. napus, BnaPAP2.A7 has been found to work as a key transcription factor regulating the synthesis of anthocyanins in the leaves [8]. However, both BoMYB2, BrMYB2, BjPur and BnaPAP2.A7 are homologous of Arabidopsis AtPAP2, while other R2R3-MYB transcription factors regulate the synthesis of anthocyanins in Brassica crops are rarely need to be explored.
In U’s Triangle Brassica species, B.rapa, B.oleracea and B.nigra genomes (designated A, B and C) share mesohexapolyploid ancestry and occur both singly and in each pairwise combination to define the Brassica species, which is the excellent model for studying the evolution of gene families. In this study, we performed genome-wide identification of MYB transcription factors of the six cultivated Brassica species. R2R3-MYB and R1R2R3-MYB subfamilies were identified and analyzed, including chromosomal distribution, synteny and evolutionary relationships. We also analyzed the spatial and temporal expression profiles as well as differential expression profiles of R2R3-MYB genes in five Brassica species (except B. nigra) with green and purple leaves, and then the gene structure, conserved domains and original promoters. These findings will provide a comprehensive understanding of novel R2R3-MYB genes involved in anthocyanin biosynthesis and assist further exploration of the functional characteristics of R2R3-MYB transcription factors in Brassica species.
Materials and methods
Identification of R2R3-MYB transcription factors in Brassica
In this study, the genome and protein sequences of the B. rapa (Chiifu), B. oleracea (TO1000), B. nigra (Ni100), B. napus (ZS11) were downloaded from the BRAD database ([9, 10]; http://brassicadb.cn), B. juncea (SY) genome sequence from NCBI PRJNA615316 (Kang et al., 2021; https://www.ncbi.nlm.nih.gov/), B. carinata (zd-1) genome sequence from GenBank JAAMPC000000000 (Song et al., 2021; https://www.ncbi.nlm.nih.gov/), and the corresponding R2R3-MYB genome and protein sequences from the Arabidopsis database (TAIR; http://www.Arabidopsis.org/).
The candidate R2R3-MYB members in the six Brassica species were identified by a local BLASTP search with 125 R2R3-MYB genes from Arabidopsis to identify candidates with E-value < 1e-10, then all putative R2R3-MYB genes were identified in the PFAM protein family database (https://pfam.xfam.org/) using the HMMER software version 3.0 [15] and using the hidden Markov model of the MYB-DNA-binding domain (PF00249) and Myb_DNA-bind_6 domain (PF13921) to search against the six Brassica species genome to identify candidates with E-value < 1e-10 [17]. All protein sequences were further investigated using different online tools, including the motif search (https://www.genome.jp/tools/motif/), SMART (http://smart.embl-heidelberg.de/), ScanProsite (https://prosite.expasy.org/scanprosite/), NCBI-Conserved Domains Database (CDD) web server (https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml), MEME Suite (https://meme-suite.org/meme/doc/meme-format.html) [28, 45].
Phylogenetic analysis
The R2R3-MYB protein sequences of the six Brassica species and Arabidopsis were used to generate phylogenetic trees via ClustalX [26] and MAFFT sofaware [24] multiple sequence alignments with the default parameters. A maximum likelihood (ML) phylogenetic tree was constructed using FastTree2 software (v2.1.11), in which JTT (Jones-Taylor-Thornton) model was the best substitution model [34]. Additionally, itol (https://itol.embl.de/) and Adobe Illustrator CS6 software (v16.0.0) were used to modify the evolutionary tree.
Chromosomal location and synteny analysis
The genome annotation data were collected and mapped on the chromosomes using the TBtools software (v0.67) to identify the physical chromosomal location of all R2R3-MYB genes in six Brassica crops and Arabidopsis [7]. The collinearity of intraspecific and interspecific genes was determined using the BLASTP (E-value: 1e-10, max_target_seqs:1) and Multiple Collinearity Scan toolkit (MCSscanX, gap_penalty: -1, E-value: 1e-10) [49], TBtools software (v0.67) was used to drop the collinearity genes on each chromosome [7].
Expression profiles analysis based on RNA-seq data
The raw data of RNA-seq were downloaded from NCBI SRA database with item number PRJNA298501 (B.rapa, B.oleracea, B.napus, B.juncea, B.carinata, green and purple leaves), PRJNA430791, PRJNA359160 (B.rapa, green and purple leaves), PRJNA474411 (B.oleracea, green and purple curds), PRJNA558197 (B.rapa, green and purple leaves), PRJNA312129 (B.rapa, green and purple leaves) and PRJNA560282 (B.oleracea, green and purple leaves). And 144 seed coats RNA-seq data of six Brassica species (B.rapa, B.oleracea, B.nigra, B.napus, B.juncea, B.carinata) were collected from Gene Expression Omnibus under accession no.GSE153257. Low-quality reads were removed from the raw reads using Cutadapt and Trimmomatic software to get clean reads [4, 29]. Clean reads were mapped to the corresponding reference genome using HISAT2 software [33]. Gene expression levels of each gene were calculated using StringTie and Ballgown software [33]. The read counts of each gene were calculated using the htseq-count function in htseq software [3]. The R package DEseq2 (v1.16.1) was used to identify the differentially expressed genes (DEGs) between leaves of different colors based on the following criteria: padj < 0.05 & log2FoldChange > 2 [8].
Analysis of the promoter characteristics, gene structure, conserved motif and co-differentially expression of R2R3-MYBs
Five groups green and purple leaves RNA-seq data were used to co-differentially expression R2R3-MYBs analyze, they are B.rapa (PRJNA359160), B.oleracea (PRJNA474411), B.napus (PRJNA298501), B.juncea (PRJNA430791), B.carinata (PRJNA298501). The co-differentially expression R2R3-MYB promoter regions of 2000 bp regions upstream of the translational start sites ATG were examined based on their positions in the genomes of six Brassica species and Arabidopsis using Samtools software (v 1.8), which was used to identify the cis-elements in the promoters according to the online PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The gene structures of all co-differentially expression R2R3-MYB genes were analyzed according to the GFF annotation file of the gene position information in the six Brassica plants and Arabidopsis database. TBtools software (v0.67) was used to locate the co-differentially expression R2R3-MYB to the different chromosomes of each species, including exon and intron numbers and lengths [7]. The MEME online tool (https://meme-suite.org/meme/) was used to investigate conserved domains, and the WEBLoGo online tool (https://weblogo.berkeley.edu/) and AlphaFold Protein Structure Database (https://www.alphafold.ebi.ac.uk/) were used to analyze the R2 and R3 motif conserved sequence and SWISS-MODEL online tool (https://swissmodel.expasy.org/) was used to draw spatial structure. The RNA-seq data were used to perform co-expression network analysis using R language (v3.6.1). In order to calculate the adjacent order function formed by the gene network and the difference coefficients of different nodes, the TOM similarity algorithm calculates the co-expression correlation matrix to express the gene correlation in the network. The correlation network diagram was drawn by extracting the non-weight coefficients (weight) of R2R3-MYB and anthocyanin-related genes in the matrix. STRING software (https://version-11-5.string-db.org/) was used to reveal a co-expression plot [28, 45].
Plant materials, RNA extraction, and qRT-PCR analysis
The B. rapa, B. oleracea, B. napus, B. juncea and B. carinata green and purple leaves of seven-leaf stage with three biological replicates were collected, all samples were collected and immediately frozen in liquid nitrogen for RNA extraction and qRT-PCR analysis. Six genes (BraA07g032100.3 C, Bo3g081880, Bo6g100940, BnaA07T0287000ZS, BnaC06T0329100ZS and BcaB05g24263) have been used the primer (RTPAPA7-3-F: GCATTGATAAGTATGGAGAAGG, RTPAPA7-744-R3: GACCACCTGTTTCCTAAAAGC) for qRT-PCR analysis, the primer (Bju-PAP2-qF: ATGGATGATCTGTCGTATAGG, Bju-PAP2-qR: CAGCTCTTGCAGGAACTA) was used to qRT-PCR analysis for BjuB05g10740S, and the primer (Bca-PAP2C3-qF: CCTGGACTCAATGACACTA, Bca-PAP2C3-qR: AACCTCCGCGTTTGACGT) was used to qRT-PCR analysis for BcaB03g15272. The Actin3 gene (Bnaactin3-qF: TCCATCCATCGTCCACAG, Bnaactin3-qR: GCATCATCACAAGCATCCTT) as an internal reference gene control. For detailed methods, please refer to Chen et al. [8]. All materials of this research were collected from Gannan Normal University research base.
Results
Identification and characterization of R2R3-MYB family genes
The 125 R2R3-MYB genes protein sequences of Arabidopsis were downloaded and used as seed sequences to search the protein sequences database of six Brassica plants and Arabidopsis to identify homologous R2R3-MYB genes in six Brassica plants. Firstly, the candidate R2R3-type MYB members in the six Brassica plants were identified by a local BLASTP search with Arabidopsis to identify candidates with E-value < 1e-10. Secondly, Hidden Markov model (HMM) profile of the MYB domain Myb-DNA-binding domain (PF00249) and Myb_DNA-bind_6 domain (PF13921) queried the hmmsearch program (HMMER3.0 package) against the protein database of six Brassica plants and Arabidopsis. Subsequently, all protein sequences were further investigated using hmmscan, SMART, ScanProsite, NCBI-CDD search and MEME Suite. A total of 2472 MYB genes were identified, these included 297 1R-MYB genes, 2120 R2R3-MYB genes, 33 R1R2R3-MYB genes, and 22 R1R2R3R4-MYB genes (Table 1, Supplementary Table S1). Among them, the R2R3-MYBs were the largest MYB subgroup containing 2120 genes, including 130 in Arabidopsis, 236 in B. rapa, 247 in B.oleracea, 248 in B. nigra, 425 in B. napus, 422 in B. juncea and 412 in B. carinata (Table 1).
Phylogenetic analysis of R2R3-MYB genes in six Brassica species
To analyze the phylogenetic relationships and gene functions of the R2R3-MYB gene family members, a ML tree containing 2120 R2R3-MYB genes was constructed using mafft software for multiple sequence alignment of protein sequences, and using fasttree software a clustering tree was built. The 2120 R2R3-MYB genes could be divided into 12 subfamilies (T1-T12) and were drawn in different colors (Fig. 1). Compared with Arabidopsis 25 subfamilies, our T1 was the S21 subfamily, T2 contained the S22 and S23 subfamilies, T3 was the S25 subfamily, T5 contained the S6 and S15 subfamilies, T6 contained the S8, S18, S19 and S20 subfamilies, T7 contained the S13 and S16 subfamilies, T8 contained the S1, S2, S3, S5 and S12 subfamilies, T9 contained the S9, S10, S11 and S24 subfamilies, T10 was the S14 subfamily, T11 included the S4 and S7 subfamilies, T12 was the S17 subfamily (Supplementary Table S2). T4 subfamily contained one R2R3-MYB gene in Arabidopsis and 18 homologous in Brassica plants, which was separated into one subfamily (Supplementary Table S3). Previous research has shown that the group of R2R3-MYB genes in the same subclade mighty have a similar function [43, 52]. The R2R3-MYB gene functions in the T5, T8 and T11 subgroups were known to be involved in the phenylalanine metabolism pathway, including the regulation of anthocyanin and procyanidin synthesis [14]. For example, in T11 subgroup, AT1G22640 (AtMYB3), AT4G38620 (AtMYB4), AT2G16720 (AtMYB7), and AT4G34990 (AtMYB32) inhibited phenylalanine biosynthesis; AT2G47460 (AtMYB11), AT3G62610 (AtMYB12) and AT5G49330 (AtMYB111) had the ability to participate the PA biosynthesis. The T11 subfamily contained AT1G56650 (AtMYB75), AT1G66390 (AtMYB90), AT1G66370 (AtMYB113), and AT1G66380 (AtMYB114) that regulated anthocyanin biosynthesis in vegetative tissues [52].
Phylogenetic relationships of R2R3-MYB proteins between six Brassica crops and Arabidopsis. The lines represented genes from six Brassica crops and Arabidopsis, respectively. All 12 subfamilies of R2R3-MYBs were well separated in different clades and represented by different colors. The ML phylogenetic tree was generated using JTT algorithm with 1,000 bootstrap value via FastTree2
Chromosomal distribution and collinearity analysis of R2R3-MYBs in six Brassica species
All 2120 2R3-MYB genes were mapped onto the corresponding genomic chromosomes and were found to be located onto all chromosomes of each species, i.e., 130 genes in Arabidopsis to 5 chromosomes, 236 genes in B. rapa to 10 chromosomes, 247 genes in B. oleracea to 9 chromosomes, 248 genes in B. nigra to 8 chromosomes, 425 genes in B. napus to 19 chromosomes, 422 genes in B. juncea to 18 chromosomes and 412 genes in B. carinata to 17 chromosomes (Supplementary Fig. 1). The results showed that the R2R3-MYB gene was relatively evenly distributed in each subgenome of the six Brassica crops, and only relatively few in the B subgenome of B.carinata, with only 166 members (Table 2). Among the three subgenomes of A, B, and C, the A subgenome has the most distribution (Supplementary Fig. 1).
The comparison of interspecific synteny among six Brassica species and Arabidopsis were also analyzed to further explore the evolution of R2R3-MYB genes (Fig. 2). At the genome-wide level, three A subgenomes from B. rapa_(BraA), B. napus_(BnaA), B. juncea_(BjuA) had a good collinearity relationship, almost all collinearity blocks were distributed on the corresponding homologous chromosomes of the same number, showing that the A subgenome had the less degree of gene differentiation and better preservation of gene integrity during the evolution process (Fig. 2). In three C subgenomes from B. oleracea_(BolC), B. napus_(BnaC) and B. carinata_(BcaC), BolC and BnaC shared a good collinearity relationship, and all collinearity blocks were distributed in the corresponding homologous chromosomes of the same number. However, the chromosomal correspondence between the subgenome BnaC and BcaC was poor, which indicated that the C subgenome had a less degree of gene differentiation and better preservation of gene integrity during the evolution of B. napus, while the chromosomal level variation was higher in B. carinata. Similarly, in the three B subgenomes from B. nigra_(BniB), B. juncea_(BjuB) and B. carinata_(BcaB), BniB and BjuB gave a good collinearity relationship, and most of the collinearity blocks were distributed onto the corresponding same numbers on homologous chromosomes. However, the chromosomal correspondence between BniB and BcaB subgenome was poor, indicating that the B subgenome had a lower degree of gene differentiation and better preservation of gene integrity during the evolution of B. juncea, while the B. carinata had a larger chromosome level variation (Fig. 2).
Interspecies synteny of R2R3-MYBs between the six species in U’s triangle and Arabidopsi. R2R3-MYBs colinearity of the genomes of the six species in U’s triangle and Arabidopsis, including three diploid species, B. rapa (A genome, BraA), B. nigra (B genome, BniB), and B. oleracea (C genome, BolC) and three tetraploid species, B. napus (AACC, BnaA, and BnaC subgenomes); B. juncea (AABB, BjuA and BjuB subgenomes); and B. carinata (BBCC, BcaB and BcaC subgenomes)
Analysis of the transcriptome and co-differentially expressed R2R3-MYBs
The different tissues with green, white or purple phenotype of B. rapa, B. oleracea, B. napus, B. juncea and B. carinata RNA-seq data were used to determine expression levels of R2R3-MYBs. Compared to the genome-wide analysis of the R2R3-MYB gene family, 41 R2R3-MYBs were found to be differentially expressed in transcriptome data. All these differentially expressed R2R3-MYBs (DE R2R3-MYBs) exhibited differential expressions between green and purple leaves. There were 9, 6, 11, 8, 7 DE R2R3-MYBs in B. rapa, B. oleracea, B. napus, B. juncea and B. carinata, respectively (Supplementary Table S4). Among them, 7 genes were differentially expressed in common, which were all homologous copies of Arabidopsis AT1G56650 (MYB75), AT1G66370 (MYB113), AT1G66380 (MYB114) and AT1G66390 (MYB90) genes (subfamily 6 of R2R3-MYBs). In five Brassica crops, they were BraA07g032100.3 C, Bo3g081880, Bo6g100940, BnaA07T0287000ZS, BjuB05g10740S, BcaB05g24263, BcaB03g15272 (Supplementary Table S4). In B. napus, the DE R2R3-MYBs BnaA07T0287000ZS and BnaC06T0329100ZS were all up-regulated in the purple leaf. Interestingly, of two differentially expressed genes in B. carinata, BcaB05g24263 was down-regulated in purple leaves, while BcaB03g15272 was up-regulated (Fig. 3).
Gene structural analysis and conserved motif identification of DE R2R3-MYBs
Gene structural analysis was helpful for the better understanding of its function and evolution. The intron numbers of the 7 DE R2R3-MYBs and Arabidopsis AT1G56650 (MYB75), AT1G66370 (MYB113), AT1G66380 (MYB114) and AT1G66390 (MYB90) were all three, except for four introns of Bo3g081880 (Supplementary Fig. 2). However, there were large variations in the length of exons and introns, especially for BraA07g032100.3 C with a large insert in the first intron.
The 2,000 base pairs (bp) upstream of R2R3-MYB gene sequences of the coding region were used to predict cis regulatory elements via the PlantCARE online tool (Supplementary Fig. 2). A total of 20 cis-regulatory elements of DE R2R3-MYBs were predicted, including 3 cellular development related cis-regulatory elements: cell cycle regulation, meristem expression, maximal elicitor-mediated activation, 5 hormone related cis-regulatory elements: abscisic acid responsiveness, auxin responsive, MeJA- responsiveness, gibberellin responsiveness, salicylic acid responsiveness. Similarly, 4 stress related cis-elements were also identified, including light responsiveness, defense and stress responsiveness, low-temperature responsiveness, dehydration, low-temp, salt stress. The MBS and MRE were specifically MYB binding sites involved in flavonoid biosynthetic, light responsiveness and drought-inducibility. AT-rich DNA binding protein, core promoter element around − 30, MYBHv1 binding, protein binding site and maximal elicitor-mediated activation. The numbers of cis-regulatory elements for each DE R2R3-MYBs varied, Bo6g100940 had only one and BcaB05g24263 had nine.
All DE R2R3-MYBs contained two highly conserved MYB binding domains. The motif logo of DE R2R3-MYBs had 51 amino acid residues in the R2 repeat and 45 amino acid residues in the R3 repeat, respectively (Supplementary Fig. 3). The HTH structure of these two domains as revealed by three-dimensional (3D) protein structural models showed that DE R2R3-MYBs matched the typical characteristics of the R2R3-MYB family (Supplementary Fig. 3).
Co-expression analysis of anthocyanin-related genes and R2R3-MYBs
The DEGs between the green, white or purple tissues of five Brassica species were used to predict candidate R2R3-MYBs related to anthocyanin biosynthesis (Supplementary Table S4). MYB90 (representing MYB75, MYB90, MYB113, MYB114 and eight DE R2R3-MYBs in Brassica plants) had a strong co-expression with TT8, and they were co-expressed with structural genes F3H, LDOX, ANS and UF3GT at the same time (Fig. 4). Simultaneously, the anthocyanin biosynthesis repressor CPC and MYBL2 were also co-expressed with TT8. These results showed that these eight DE R2R3-MYBs were the important regulation genes involved in anthocyanin biosynthesis processes.
Expression pattern of R2R3-MYB DEGs and anthocyanin related genes in five Brassica species
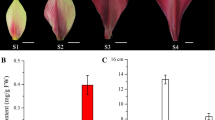
In order to confirm the transcriptional pattern revealed by the RNA-seq analysis, we performed qRT-PCR analysis for 7 DE R2R3-MYBs (Fig. 5). Six genes showed a similar relative expression level in green / white and purple tissues as revealed by RNA-seq analysis. The MYB90 was hardly expressed in the green / white tissues of five Brassica species, but significantly upregulated in the purple/red tissues, which was consistent with the transcriptome and phenotype, respectively. However, BcaB03g15272 is not expressed in purple and green leaves, which is different from the results of the transcriptome.
qRT-PCR analysis of AtPAP2 homologous relative expression in five Brassica species with green and purple leaves. For qRT-PCR analysis, both five Brassica species green and purple samples are with three biological replicates, the point represents the mean value of three technical replicates in a representative biological experiment, the error bars indicate s.d, student’s t-test, **P<0.01,*P<0.05
Discussion
The R2R3-MYB gene family in Brassica species
The R2R3-MYB is the largest subfamily of the MYB transcription factor family. To date, several R2R3-MYB families have been identified and analyzed. In the model plant (A) thaliana, 125 R2R3-MYB genes were found [43], and then 126 R2R3-MYB genes detected from the whole genome MYB transcription factor identification and divided into 26 subfamilies [14], which provides a reference for the identification and analysis of R2R3-MYB transcription factors in other plants. In recent years, numerous R2R3-MYB transcription factors have been identified and functionally analyzed, such as in Oryza sativa [11], Zea mays [13], (B) napus [20] and horticultural plants Petunia [9, 10], Strawberry [28] and Capsicum annuum [45]. Importantly, more and more biological pathways involved in R2R3-MYB have been revealed, such as the transcriptional regulation of anthocyanins [56], fiber development [45] and root hair development [42].
Six Brassica species of three diploids and three derived allotetrploids in U’s triangle are excellent models for studying gene families. With the completion of the sequencing of the B. carinata genome, the genomes of all six species have been published, providing a useful tool for genome-wide analysis of the R2R3-MYB gene family. To date, no full analysis of the R2R3-MYB gene family is reported in these six Brassica crops and most functions remain unclear. In this study, we identified 2120 R2R3-MYB genes from the genomes of Arabidopsis and six Brassica crops, including 130 in Arabidopsis, 236 in B. rapa, 247 in B. oleracea, 248 in B. nigra, 425 in B. napus, 422 in B. juncea and 412 in B. carinata (Table 1), indicating the expansion of this gene family along with genome duplication in Brassica crops. The present numbers of R2R3-MYBs in Arabidopsis, B. rapa and B. napus were different from those of previous studies, 126 in Arabidopsis [43], 256 in the B. rapa [48], and 249 in the B. napus [20]. The main reason was probably that with the development of three-generation sequencing, genome information became more complete.
Evolution of the R2R3-MYB gene family
Gene duplication is a major factor responsible for the expansion of gene families and the generation of new genes, which has also led to changes in the number and function of duplicated genes, and also provides an excellent example for studying the inheritance and evolution of gene family. The six Brassica crops of U’s triangle provides an excellent model for gene duplication and evolution research, because of the natural cross between the two diploids and the independent evolution of the parents and the allopolyploidy hybrids. To understand the evolutionary relationship of R2R3-MYB family genes in U’s Triangle six Brassica species, we constructed the phylogenetic tree with R2R3-MYB family genes from the U’s Triangle six Brassica species and Arabidopsis. All 2120 R2R3-MYB family genes from Arabidopsis and six Brassica crops were classified into 12 different classes with 130 R2R3-MYB family genes from Arabidopsis (Fig. 1), each cluster corresponds to one or more R2R3-MYB subfamily of Arabidopsis (Supplementary Table S3), which was in accordance with that the R2R3-MYB family was relatively conservative between different plants in evolution. Chromosomal distributions indicated that they were relatively evenly distributed on the A, B, and C subgenomes, but the abundance of the A subgenome was higher than that of the B and C subgenomes (Supplementary Fig. 1). Collinearity analysis of R2R3-MYBs in six Brassica species showed that the collinearity between A and C subgenomes was better than that between B and A or C subgenomes (Fig. 2), which was consistent that their genomic relationships [38]. Therefore, the results offered a useful framework for future research to understand the evolution of the R2R3-MYB gene family.
The expression level of R2R3-MYBs related to anthocyanin biosynthesis
Brassica species present color variations caused by the accumulation of anthocyanins in tissues or organs, such as leaves, stems, flowers, siliques and seeds, from B. campestris L. var. purpuraria L. H. Bailey, purple cabbage, purple cauliflower, purple B. juncea, purple rapeseed. In our study, we analyzed the transcriptional profiles of co-differentially expressed R2R3-MYBs for green and purple leaves in five Brassica crops (except B. nigra), and a totally of 41 R2R3-MYBs were differentially expressed (Table 2). Of these, eight co-DE R2R3-MYBs were found in five Brassica crops (Fig. 3), and were homologous genes of MYB75/MYB90/MYB113/MYB114 in Arabidopsis, and were the subfamily 6 of R2R3-MYB [14, 43]. The co-expression analysis of anthocyanin-related genes and R2R3-MYBs found that MYB90 have a strong interaction with TT8 to regulate the structural genes F3H, LDOX, ANS and UF3GT at the same time (Fig. 4). Interestingly, the anthocyanin biosynthesis repressor CPC and MYBL2 were also co-expressed with TT8. In this research, comprehensive transcriptome results, phenotypic data and qRT-PCR analysis showed that these six DE R2R3-MYBs are the important regulation genes involved in anthocyanin biosynthesis processes (Fig. 5).
Previous studies have indicated that R2R3-MYB genes play key roles in the anthocyanin biosynthesis in various plants, such as TT2, MYB75, MYB90, MYB113 and MYB114 in Arabidopsis ([52]), Ruby1 and Ruby2 in citrus (Butelli et al., 2012), AcMYB123 in kiwifruit [46, 47], PpMYB10.1 in peaches [35], MdMYB10 and MdMYB110a in Apple [6, 16], and VvMYBA1 and VvMYBA2 in grapes [44]. In Brassica, previous studies have shown that the sequence variation of the R2R3-MYB transcription factor in the promoter region or the gene intron region to drive its active expression is the main reason for the cumulative variation of Brassica anthocyanins. Different types of transposon insertions or sequence variations in the BoMYB2 promoter in purple B. oleracea drove its expression to increase significantly, resulting in different color phenotypes in different tissues or organs of different subspecies [53]. The activated expression of the BrMYB2 gene in purple leaf Chinese cabbage and the BjPur gene in purple leaf B. juncea was due to the deletion of the first large intron in the intergenic region [21, 22]. In B. napus, BnaPAP2.A7 was previously found to be a key transcription factor regulating the synthesis of anthocyanins in the leaves [8]. Interestingly, BoMYB2, BrMYB2, BjPur, and BnaPAP2.A7 were homologous of Bo6g100940, BraA07g032100.3 C, BjuB05g10740S and BnaA07T0287000ZS, which was highly consistent with our results. In addition, our results indicated that MYB90 was the key transcription factor that combined with TT8 transcription factor and TTG1 protein to form a MBW transcription regulatory complex to regulate the synthesis of anthocyanins in Brassica crops, when MYB90 was not expressed or MYBL2 inhibited MYB90 expression by competing with TT8 and TTG1, Brassica crops failed to complete the synthesis and accumulation of anthocyanins (Fig. 6).
Proposed ‘activator-and-repressor’ loop model for the regulation of anthocyanin accumulation in Brassica species. PAP2 acts as the activator by directly and positively regulating the expression of anthocyanin biosynthetic genes with TTG1 and TT8, whereas MYBL2 acts as the repressor by inhibiting the activity of the PAP2-TT8-TTG1 complex. The arrow and error symbol lines indicate activation or repression, respectively
In addition, we collected 144 seed coats RNA-seq data at different developmental stages of U’s Triangle six Brassica species [18]. Interestingly, co-DE R2R3-MYBs were found that 25 co-DE R2R3-MYBs in the groups UO and Torpedo of six Brassica species, but there was no co-differentially expressed R2R3-MYB gene in seed coats and leaves. The analysis of differentially expressed genes related to anthocyanin biosynthesis pathway found that there were many co-differentially expressed structural genes and transcription factors in all six Brassica species, such as DFR, LDOX, TT2, MYB5, EGL3 and so on, however, PAP1/PAP2/MYB113/MYB114 were not differentially expressed in these materials. Comparing the leaf and seed coat transcriptome analysis data of Brassica species, we found that the purple phenotype material anthocyanin synthesis pathway structural genes were significantly up-regulated in both leaf and seed coat, while the regulated MYB transcription factors were different. The differentially expressed R2R3-MYB transcription factor was PAP2 in leaves, but mainly TT2 and MYB5 in seed coats. Previously, BrTT8 and BrMYB5 may be involved in the regulation of anthocyanins in B.rapa seed coat [37],
BjuTT8 may be regulates the seed coat color variation of B.rapa, B.napus, and B.juncea [40], BnaC.TT2.a [57] and BnTT8 [55] regulates the rapeseed coat color variation. In conclusion, among Brassica species, the regulatory genes of anthocyanins in leaves and seed coats may be different, and the regulatory mechanisms need to be further explored.
Overall, the present study provided new insights into the roles of novel R2R3-MYB transcription factors in anthocyanin synthesis of Brassica species and into anthocyanin transcriptional regulation in plants.
Availability of data and materials
All materials and related data in this study are available upon request. If you need these materials and related data, you can contact Daozong Chen(chendaozong61@163.com).
References
Albert N, Davies K, Lewis D, et al. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell. 2014;26:962–80.
Allan A, Hellens R, Laing W. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008;13:99–102.
Anders S, Pyl P, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9.
Bolger A, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Butelli E, Licciardello C, Zhang Y et al. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell. 2012;24(3):1242–55.
Chagné D, Lin-Wang K, Espley RV, et al. An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 2013;161:225–39.
Chen C, Chen H, Zhang Y, et al. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant. 2020;13:1194–202.
Chen D, Liu Y, Yin S, et al. Alternatively Spliced BnaPAP2.A7 Isoforms Play Opposing Roles in Anthocyanin Biosynthesis of Brassica napus L. Front Plant Sci. 2020;11:983.
Chen G, He W, Guo X, et al. Genome-wide identification, classification and expression analysis of the MYB transcription factor family in Petunia. Int J Mol Sci. 2021;22:4838.
Chen H, Wang T, He X, et al. BRAD V3.0: an upgraded Brassicaceae database. Nucleic Acids Res. 2021;10:1057.
Chen Y, Yang X, He K, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol. 2006;60:107–24.
Devic M, Guilleminot J, Debeaujon I, et al. The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development. Plant J. 1999;19:387–98.
Du H, Feng B, Yang S, et al. The R2R3-MYB transcription factor gene family in maize. PLoS One. 2012;7:e37463.
Dubos C, Stracke R, Grotewold E, et al. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–81.
Eddy S. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195.
Espley R, Hellens R, Putterill J, et al. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007;49:414–27.
Finn R, Bateman A, Clements J, et al. Pfam: the protein families database. Nucleic Acids Res. 2013;42:D222–30.
Gao P, Quilichini TD, Yang H, et al. Evolutionary divergence in embryo and seed coat development of U’s Triangle Brassica species illustrated by a spatiotemporal transcriptome atlas. New Phytol. 2022;233:30–51.
Gonzalez A, Zhao M, Leavitt J, et al. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–27.
Hajiebrahimi A, Owji H, Hemmati S. Genome-wide identification, functional prediction, and evolutionary analysis of the R2R3-MYB superfamily in Brassica napus. Genome. 2017;60:797–814.
He Q, Wu J, Xue Y, et al. The novel gene BrMYB2, located on chromosome A07, with a short intron 1 controls the purple-head trait of Chinese cabbage (Brassica rapa L.). Hort Res. 2020;7:97.
Heng S, Cheng Q, Zhang T, et al. Fine mapping of the BjPur gene for purple leaf color in Brassica juncea. Theor Appl Genet. 2020;133:2989–3000.
Hichri I, Barrieu F, Bogs J, et al. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot. 2011;62:2465–83.
Katoh K, Standley D. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80.
Kang L, Qian L, Zheng M, Chen L et al. Genomic insights into the origin, domestication and diversification of Brassica juncea. Nat Genet. 2021;53(9):1392–402.
Larkin M, Blackshields G, Brown N, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–8.
Lipsick J. One billion years of Myb. Oncogene. 1996;13:223–35.
Liu J, Wang J, Wang M, et al. Genome-wide analysis of the R2R3-MYB gene family in Fragaria × Ananassa and its function identification during anthocyanins biosynthesis in pink-flowered Strawberry. Front Plant Sci. 2021;12:702160.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–2.
Mushtaq MA, Pan Q, Chen D, et al. Comparative Leaves Transcriptome Analysis Emphasizing on Accumulation of Anthocyanins in Brassica: Molecular Regulation and Potential Interaction with Photosynthesis. Front Plant Sci. 2016;18:311.
Ogata K, Hojo H, Aimoto S, et al. Solution structure of a DNA-binding unit of Myb: a helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc Natl Acad Sci. 1992;89:6428–32.
Paz-Ares J, Ghosal D, Wienand U, et al. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987;6:3553–8.
Pertea M, Kim D, Pertea GM, et al. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–67.
Price M, Dehal P, Arkin A. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490.
Rahim M, Busatto N, Trainotti L. Regulation of anthocyanin biosynthesis in peach fruits. Planta. 2014;240:913–29.
Ramsay N, Glover B. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10:63–70.
Ren Y, Zhang N, Li R, et al. Comparative transcriptome and flavonoids components analysis reveal the structural genes responsible for the yellow seed coat color of Brassica rapa L. Peer J. 2021;9:e10770.
Schranz M, Lysak M, Mitchell-Olds T. The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 2006;11:535–42.
Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Shen S, Tang Y, Zhang C, et al. Metabolite profiling and transcriptome analysis provide insight into seed coat color in Brassica juncea. Int J Mol Sci. 2021;22:7215.
Shi M, Xie D. Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana. Recent Pat Biotechnol. 2014;8:47–60.
Slabaugh E, Held M, Brandizzi F. Control of root hair development in Arabidopsis thaliana by an endoplasmic reticulum anchored member of the R2R3-MYB transcription factor family. Plant J. 2011;67:395–405.
Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol. 2001;4:447–56.
Walker A, Lee E, Bogs J, et al. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 2007;49:772–85.
Wang J, Liu Y, Tang B, et al. Genome-wide Identification and capsaicinoid biosynthesis-related expression analysis of the R2R3-MYB gene family in Capsicum annuum L. Front Genet. 2020;11:598183.
Wang L, Tang W, Hu Y, et al. A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. 2019;99:359–78.
Wang N, Ma Q, Ma J, et al. A comparative genome-wide analysis of the R2R3-MYB gene family among four gossypium species and their sequence variation and association with fiber quality traits in an interspecific G. hirsutum × G. barbadense population. Front Genet. 2019;10:741.
Wang Z, Tang J, Hu R, et al. Genome-wide analysis of the R2R3-MYB transcription factor genes in Chinese cabbage (Brassica rapa ssp. pekinensis) reveals their stress and hormone responsive patterns. BMC Genomics. 2015;16:17.
Wang Y, Tang H, Debarry JD, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49.
Wong M, Henry R, Ma B, et al. The large subunit of basal transcription factor SNAPc is a Myb domain protein that interacts with Oct-1. Mol Cell Biol. 1998;18:368–77.
Xie Q, Hu Z, Zhang Y, et al. Accumulation and molecular regulation of anthocyanin in purple tumorous stem mustard (Brassica juncea var. tumida Tsen et Lee). J Agric Food Chem. 2014;62:7813–21.
Xu W, Grain D, Bobet S, et al. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014;202:132–44.
Yan C, An G, Zhu T, et al. Independent activation of the BoMYB2 gene leading to purple traits in Brassica oleracea. Theor Appl Genet. 2019;132:895–906.
Zhang Y, Hu Z, Chu G, et al. Anthocyanin accumulation and molecular analysis of anthocyanin biosynthesis-associated genes in eggplant (Solanum melongena L.). J Agric Food Chem. 2014;62:2906–12.
Zhai Y, Yu K, Cai S, et al. Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol J. 2020;18:1153–68.
Zhao L, Gao L, Wang H, et al. The R2R3-MYB, bHLH, WD40, and related transcription factors in flavonoid biosynthesis. Funct Integr Genomics. 2013;13:75–98.
Zhou L, Li Y, Hussain N, et al. Allelic variation of BnaC.TT2.a and its association with seed coat color and fatty acids in rapeseed (Brassica napus L.). PLoS One. 2016;11:e0146661.
Acknowledgements
Thanks to Professor Li Zaiyun for his help in the writing process of the thesis.
Statement on plant guidelines and permission
All materials in this study are comply with relevant institutional, national, and international guidelines, legislation, and sub-section ethical approval and consent to participate.
Gannan Normal University agreed to the sampling of various experimental materials in this study.
Conflict of interest
The authors declare that the study was conducted in the absence of any commercial or financial relationships that could be envisaged and/or construed as a conflict of interest.
Funding
This research was funded by the Natural Science Foundation of Jiangxi Province, grant number 20212BAB215002; the National Natural Science Foundation of China, grant number: 32160454; the Science and Technology Plan Project of Ganzhou, grant number [2020]60.
Author information
Authors and Affiliations
Contributions
CT, BZ and CC conceived and designed the experiments. DC, HC, GD, HZ, YL and WS analyzed the data. DC, CT, BZ and CC wrote the manuscript. All authors approved the manuscript and consent to publication this manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study including the collection on plants material complies with relevant institutional, national, and international guidelines and legislation.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary Fig. 1.
The distribution of R2R3-MYB on the three subgenomic chromosomes of A, B, and C. Different subgenomic chromosomes represented by different colors, and the same crops are marked with the same color.
Additional file 2: Supplementary Fig. 2.
Predicted cis-elements in 7 co-differentially expressed R2R3-MYBs promoters and gene structure. Promoter sequences (-2,000 bp) of 7 co-differentially expressed R2R3-MYBs were analyzed using PlantCARE. Different shapes and colors represent different elements.
Additional file 3: Supplementary Fig. 3.
The domains of R2R3-MYB family genes and protein 3D structural models of R2 and R3 MYB repeats.
Additional file 4: Supplementary Fig. 4.
Six Brassica species with 25 co-differentially expressed R2R3-MYBs correspond to Arabidopsis.
Additional file 5: Supplementary Fig. 5.
The expression pattern of MYBL2 of five Brassica species green and purple leaves.
Additional file 6: Supplementary Table S1.
R2R3-MYB in six Brassica species.
Additional file 7: Supplementary Table S2.
The subfamilies of R2R3-MYB.
Additional file 8: Supplementary Table S3.
Correspondence between 12 cluster tree subfamilies of Brassica R2R3_MYB family and 25 cluster tree subfamilies of Arabidopsis.
Additional file 9: Supplementary Table S4.
Correspondence between the DE R2R3_MYBs of Brassica species and Arabidopsis.
Additional file 10: Supplementary Table S5.
Differentially expressed genes and expression levels related to anthocyanin synthesis in seed coat.
Additional file 11: Supplementary Table S6.
Differentially expressed genes and expression levels of R2R3-MYB in seed coat.
Additional file 12: Supplementary Table S7.
The details of six Brassica species with 25 co-differentially expressed R2R3-MYBs in seed coat correspond to Arabidopsis.
Additional file 13: Supplementary Table S8.
The expression data of R2R3-MYB genes in green and purple leaves of five Brassica species.
Additional file 14: Supplementary Table S9.
Comparison of the identification results of Arabidopsis R2R3-MYB genes.
Additional file 15: Supplementary Table S10.
The detail expression pattern of PAP2 and MYBL2 of five Brassica species green and purple leaves.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Chen, D., Chen, H., Dai, G. et al. Genome-wide identification of R2R3-MYB gene family and association with anthocyanin biosynthesis in Brassica species. BMC Genomics 23, 441 (2022). https://doi.org/10.1186/s12864-022-08666-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-022-08666-7