Abstract
Background
Carbapenem-resistant hypervirulent K. pneumoniae (CR-hvKP) causes serious infections with significant morbidity and mortality. However, the epidemiology and transmission mechanisms of CR-hvKP and the corresponding carbapenem-resistant plasmids require further investigation. Herein, we have characterized an ST11 K. pneumoniae strain EBSI041 from the blood sample encoding both hypervirulence and carbapenem resistance phenotypes from a patient in Egypt.
Results
K. pneumoniae strain EBSI041 showed multidrug-resistance phenotypes, where it was highly resistant to almost all tested antibiotics including carbapenems. And hypervirulence phenotypes of EBSI041 was confirmed by the model of Galleria mellonella infection. Whole-genome sequencing analysis showed that the hybrid plasmid pEBSI041-1 carried a set of virulence factors rmpA, rmpA2, iucABCD and iutA, and six resistance genes aph(3′)-VI, armA, msr(E), mph(E), qnrS, and sul2. Besides, blaOXA-48 and blaSHV-12 were harboured in a novel conjugative IncL-type plasmid pEBSI041-2. The blaKPC-2-carrying plasmid pEBSI041-3, a non-conjugative plasmid lacking the conjugative transfer genes, could be transferred with the help of pEBSI041-2, and the two plasmids could fuse into a new plasmid during co-transfer. Moreover, the emergence of the p16HN-263_KPC-like plasmids is likely due to the integration of pEBSI041-3 and pEBSI041-4 via IS26-mediated rearrangement.
Conclusion
To the best of our knowledge, this is the first report on the complete genome sequence of KPC-2- and OXA-48-coproducing hypervirulent K. pneumoniae from Egypt. These results give new insights into the adaptation and evolution of K. pneumoniae during nosocomial infections.
Similar content being viewed by others
Background
Carbapenem-resistant Klebsiella pneumoniae (CRKP) is one of the most critical threats to global public health associated with significant morbidity and mortality [1,2,3]. K. pneumoniae producing KPCs, NDMs, and OXA-48-like carbapenemases have become rapidly disseminated worldwide [2]. The blaKPC and blaNDM genes in K. pneumoniae have been reported on multiple plasmid types, including IncF, IncA/C, IncR, IncX and IncL/M. Among them, the IncF and IncA/C type plasmids are predominantly responsible for the transfer of blaKPC and blaNDM, respectively [4, 5]. Besides, the blaKPC gene is strongly associated with Tn4401 flanking by ISKpn6 and ISKpn7, and the blaNDM gene is closely related to Tn125 structure with two ISAba125 elements [6, 7]. Unlike KPCs and NDMs, IncL group plasmid has been shown to be the major genetic carrier for blaOXA in K. pneumoniae and the composite transposon Tn1999 is mainly responsible for integration of the blaOXA gene [4]. Furthermore, dissemination of KPC-producing K. pneumoniae in worldwide is largely caused by expansion of the dominant ST258 clones [8]. Differently, the blaOXA and blaNDM genes are detected in various K. pneumoniae clones, in which ST11 is a major high-risk sequence type in many countries, such as China [9, 10], Australia [11], Poland [12], Spain [13] and Turkey [14]. Co-carriage of different carbapenems resistance genes is not common but renders clinical K. pneumoniae strains extremely highly resistant to different carbapenems, which leads to more difficult infection treatment [15, 16].
Seriously, carbapenem-resistant hypervirulent K. pneumoniae (CR-hvKP) has been increasingly reported in nosocomial infection and can cause higher mortality [17, 18]. The plasmid-mediated genetic factors conferring the hypervirulent phenotype including rmpA and rmpA2 (regulators that increase capsule production), and several siderophore gene clusters [19]. Notably, a multinational prospective cohort study warns of the severity of carbapenem resistance in low-income and middle-income countries, including Egypt [3]. In the previous studies, the high prevalence of carbapenemase-mediated resistance in K. pneumoniae isolates in the clinical setting from Egypt was reported [20,21,22,23], but few studies have analyzed the genome characteristic of carbapenem-resistant K. pneumoniae by whole-genome sequencing. In this study, we report the in-depth characterization of co-producing blaKPC-2 and blaOXA-48 hypervirulent ST11 K. pneumoniae strain from Egypt. The dissemination of two plasmids carrying blaOXA-48 and blaKPC-2 and novel plasmid structures were identified.
Results
Strains characteristic
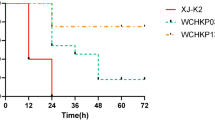
K. pneumoniae EBSI041 was collected from the blood sample of a male emergency ICU patient in March 2012 in Egypt. This strain showed high resistance to almost all tested antibiotics, including imipenem, meropenem, ertapenem, piperacillin-tazobactam, cefotaxime, ceftazidime, cefepime, aztreonam, gentamicin, amikacin, ciprofloxacin, fosfomycin, chloramphenicol, but sensitivity to colistin, tigecycline, tetracycline, and trimethoprim-sulfamethoxazole (Table S1). EBSI041 exhibited extensive resistance to carbapenems with high resistance levels (MIC > 32 mg/L). In addition, it was also highly resistant to piperacillin-tazobactam, a β-lactam combination agent, with MIC > 512 mg/L, and cephems cefotaxime (MIC > 256 mg/L), ceftazidime (MIC > 256 mg/L), cefepime (MIC = 256 mg/L). The virulence level of strain EBSI041 was tested in wax moth (G. mellonella) larvae. The larval mortality rate increased dramatically to 60% within 12 h post-infection with EBSI041. On being infected for 48 h at an inoculum of 1 × 104 colony-forming units (CFU), the survival of G. mellonella were 10 and 0% for EBSI041 and the hypervirulence control strains HvKP4, respectively (Fig. S1).
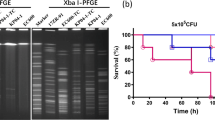
In vitro conjugation demonstrated that carbapenem resistance genetic factors in EBSI041 can be transferred to the recipient strains E. coli J53 and EC600. Further S1-PFGE and PCR assay revealed OXA-48-carrying plasmid (~ 90 kb) can be self-transferred and KPC-2-producing plasmid has no transferability. However, KPC-2-producing plasmid can be transferred to the recipient strains with the help of OXA-48-carrying plasmid, and the two plasmids may be fused a larger plasmid (~ 180 kb) carring both blaOXA-48 and blaKPC-2 according to results of S1-PFGE (Fig. S2).
Genomic features of the carbapenem-resistant hypervirulent K. pneumoniae EBSI041
Genomic analysis showed EBSI041 included a 5,516,355 bp chromosome and seven plasmids: pEBSI041-1 (299,522 bp), pEBSI041-2 (97,179 bp), pEBSI041-3 (85,594 bp), pEBSI041-4 (45,103 bp), pEBSI041-5 (10,060 bp, pEBSI041-6 (5596 bp), pEBSI041-7 (1780 bp) (Table 1). EBSI041 carried 19 antibiotic resistance genes (ARGs), four of them were located on the chromosomes including the blaSHV-11, oqxB, oqxA, and fosA6 genes. The two carbapenemase genes, blaOXA-48 and blaKPC-2, were identified in plasmids pEBSI041-2 and pEBSI041-3, respectively. In addition to resistance genes, eighty-six putative virulence genes were annotated in the genome of EBSI041, including genes coding for fimbriae, capsule, yersiniabactin, iron-enterobactin, mucoid and aerobactin. Most virulence factors were found on chromosome, except for seven genes (rmpA, rmpA2, iucABCD and iutA) in plasmid pEBSI041-1. The virulence of EBSI041 was demonstrated using the Galleria mellonella larvae model. EBSI041 was identified as resistant and virulent ST11 clone.
For source tracking bacterial pathogens, EBSI041 was found to be similar to a ST11 K. pneumoniae strain (WJTB01) isolated from bronchoalveolar lavage of patient in China, with 317 SNPs, based on SNP (sequence-based) strategy using BacWGSTdb 2.0 [24]. The core-genome-based MLST (cgMLST) analysis showed that 67 strains were less than or equal to 50 alleles different from EBSI041. All strains were isolated from China, except one from USA. EBSI041 and HA_74 (PJOU01) were on the same branch of the phylogenetic tree (Fig. S3). ST11 K. pneumoniae strain HA_74 was isolated from China in 2015. However, because of limited sources from Egypt or Africa, we were not able to clarify the domestic transmission route.
The MDR pEBSI041-1 co-harbouring virulence genes via recombination
The plasmid pEBSI041-1 is 299,522 bp in size and belongs to an IncFIB:IncHI1B type hybrid plasmid. A 27,568-bp multidrug-resistance (MDR) module in pEBSI041-1 harboured six resistance genes armA, msr(E), mph(E), qnrS, aph(3′)-VI and sul2. The armA gene was mediated by an intact IS26 (IS6 family, 820 bp) element upstream. With a 2300-bp space to armA downstream, a locus of msr(E) and mph(E) was flanked by an intact ISEc29 (IS4 family, 1325 bp) (Fig. 1). For the rest of ARGs, each of them was flanked independently by different IS elements. This module was almost identical to the sequences in plasmid p51015_NDM_1 (CP050380) which was identified in a human K. pneumoniae isolate in the Czech Republic. The only difference between them was an inversion of a 7710-bp sequence bounded by two IS26 occurred in p51015_NDM_1. Besides this MDR module, pEBSI041-1 harbours the plasmid-located virulence factors, including regulator of the mucoid phenotype (rmpA), the regulator of mucoid phenotype 2 (rmpA2), aerobactin (iucABCD, iutA) (Fig. 1). A BLASTn search showed that a 36,929-bp sequence containing the virulence genes in pEBSI041-1 was identical to the sequences in pF16KP0084-1 (CP052159.1; South Korea). Furthermore, close to this sequence, a 37,030-bp sequence was also found to be identical to the sequence in pF16KP0084-1 with a reversion order, while the sequence harbours the virulence gene cluster iroBCD and iroN in pF16KP0084-1 was lacking in our plasmid (Fig. 1). Also, these two sequence fragments in pEBSI041-1 caused the main difference to plasmid pKpvST101_5 (CP031372.2; United Kingdom), indicating that the emergence of the MDR-virulent pEBSI041-1 was due to the transfer of virulence determinants into a pKpvST101_5-like MDR plasmid. Sequence alignments showed that pEBSI041-1 shared > 99% identity with plasmid pKpvST147B_virulence (CP040726.1; United Kingdom), pKpvST383L (CP034201.2; United Kingdom), and p51015_NDM_1 (CP050380.1) with query coverages of 96-99.5%, all of which are MDR-virulent hybrid plasmids (Fig. 1).
Structure analysis of pEBSI041-1. Major structural features of plasmid pEBSI041-1 were compared with plasmids pF16KP0084-1 (CP052159.1), pKpvST101_5 (CP031372.2), pKpvST147B_virulence (CP040726.1), p51015_NDM_1 (CP050380.1) and pKpvST383L (CP034201.2). Blue shading indicates shared regions with a high degree of homology. Red and purple represent the antibiotic resistance and virulence genes, respectively, and yellow is the insertion sequences and transposons
The bla OXA-48-carrying pEBSI041-2 harbours an exogenetic chromosome-located fragment
The IncL/M-type plasmid pEBSI041-2 was 97,179 bp in size with an average GC content of 52.45% and contained 134 open reading frames (ORFs). It possessed a complete array of genes involved in replication (repA gene), stabilization (stbAB genes), toxin-antitoxin system (pemIK genes) and conjugation (traHIJKLMNOPQR and traUWXYX genes). The blaOXA-48 gene, encoding the class D carbapenemase OXA-48, was found in this plasmid. The blaOXA-48 gene was surrounded by multiple IS elements, namely ΔIS10A (IS4 family, 1329 bp), IS1R (IS1 family, 768 bp) and IS10A (IS4 family, 1329 bp) (Fig. 2). pEBSI041-2 exhibited a high similarity (80% coverage, 99.58% identity) with plasmid pOXA-48_1639 (CP025105.1), which also encodes OXA-48 and identified from E. coli. Besides, pEBSI041-2 was also similar to plasmids pOXA-48_920 (LR025095.1) and pUR17313-1 (KP061858.1), which are from K. pneumoniae and Enterobacter cloacae respectively. This indicates that pOXA-48_1639-like plasmids have been widely spread among bacteria of different species. However, compared with pOXA-48_1639, an 18,779-bp sequence harbouring blaSHV-12 was missing in pEBSI036-2. The blaSHV-12 gene was surrounded by two IS26 (IS6 family, 820 bp) elements, and this 6100-bp fragment was similar to plasmid p680_1 (CP038659.1) from Citrobacter freundii. A BLASTn search did not find the homologous plasmid-located sequence to the remaining region (47,727-55,495 bp) while exhibited limited similarities (~ 50% coverage, > 99% identity) to the chromosome of some K. michiganensis strains (CP023185.1; CP022348.1; CP003683.1) (Fig. 2). The mobilization of blaSHV-12 can be attributed to the gene exchange by recombination, while the acquisition of exogenous chromosome sequence needs to be further considered. Indeed, the conjugation experiment demonstrated that pEBSI041-2 transferred from the donor strain K. pneumoniae EBSI041 to the recipients E. coli J53 and EC600.
The comparative schematic diagram of plasmid pEBSI041-2. pEBSI041-2 was compared with three OXA-48-carrying plasmids pOXA-48_1639 (CP025105.1), pOXA-48_920 (LR025095.1) and pUR17313-1 (KP061858.1). Besides, plasmid p680_1 (CP038659.1) and the chromosome of K. michiganensis strains (CP023185.1) were analyzed. Red represent the antibiotic resistance, and yellow is the insertion sequences and transposons. Plasmid transferability genes were shown in green
The transfer of non-conjugative bla KPC-2-carrying pEBSI041-3 needs the help of pEBSI041-2
The pEBSI041-3 was an IncR-type plasmid. In pEBSI041-3, two antibiotic resistance genes blaKPC-2 and blaSHV-12 were found, which were separated by a fragment of ISKpn27-ΔTn3-IS26-ΔTnAs1 (Fig. 3). Besides, an ΔISKpn6 (IS1182 family, 1540 bp) was located in upstream of blaKPC-2. The pEBSI041-3 acquired blaKPC-2 by a transposon unit with the core structure of ΔISKpn6-blaKPC-2-ISKpn27 as pKPC-LK30 (KC405622.1) [25] and pKPC-L111 (CP030134.1) [15] and captured blaSHV-12 with an IS26-interrupted TnAs1 (Tn3 family, 6694 bp) element. The complete transfer operon (locus tra-trb) was not detected in pEBSI041-3, except traA and traM, which may explain why pEBSI041-3 does not transfer conjugatively to the recipients E. coli J53 and EC600. However, the results of vitro conjugation and PFGE experiments showed pEBSI041-3 can be transferred with the help of conjugative plasmid pEBSI041-2. Moreover, pEBSI041-2 and pEBSI041-3 may fuse into a larger plasmid (~ 180 kb) carried both blaOXA-48 and blaKPC-2 genes during co-transfer (Fig. S2).
Potential recombination of plasmids pEBSI041-4 and pEBSI041-3
Plasmid pEBSI041-4 was identified as an IncFII type. A 20,894 bp multi-drug resistance region in pEBSI041-4 harboured five resistance genes, including blaCTX-M-65, blaTEM-1B, catA2, fosA3 and rmtB. Those genes were separated by eight IS26 elements, one IS903B (IS5 family, 1057 bp) element, one ISKpn26 (IS5 family, 1196 bp) element, one ISCfr3 (ISNCY family, 1081 bp) element and a few truncated insertion sequences and transposons (Fig. 3).
The segment contained genes blaCTX-M-65, blaTEM-1B, fosA3, and rmtB was similar to F33:A-:B-type plasmid pHN7A8, which isolated from an E. coli in China [26]. The pEBSI041-4 shared 99.98% identity with pHN7A8 (JN232517.1) with query coverages of 71%. Compared to pHN7A8, pEBSI041-4 has an additional resistance gene catA2 flanked by two IS26 elements. Besides, the downstream region of catA2 has ISKpn26, ISCfr3 and IS26 elements. The presence of multiple insertion sequences, especially IS26, indicates that recombination may repeatedly have occurred in this multi-drug resistance region.
Moreover, the combination of pEBSI041-3 and pEBSI041-4 was almost identical to p16HN-263_KPC (CP045264.1) (Fig. 3). The p16HN-263_KPC was collected from Klebsiella pneumoniae of bloodstream infection in China and shared high similarity with pKP1034 (KP893385.1) and p69-2(CP025458.1) [27, 28]. Those KPC-2-producing plasmids belonged to IncR-F33:A-:B- type and carried all resistance genes in pEBSI041-3 and pEBSI041-4, except p69-2 without fosA3. The comparison suggested that pEBSI041-3 and pEBSI041-4 might have undergone rearrangement by recombination to form p16HN-263_KPC-like plasmids.
Discussion
ST11 is the dominant clone of carbapenemases-producing K. pneumoniae in Asia, especially in China, and ST11 clone was reported to account for up to 60% of carbapenem-resistant K. pneumoniae [9, 29, 30]. Recently, ST11 has been reported in several clinical infection cases from African countries, such as Egypt [31, 32] and Tunisian [33]. And in ST11 carbapenem-resistant K. pneumoniae isolates, the most predominant carbapenemase genes are blaKPC-2, blaNDM-1 and blaOXA-48 [10, 34]. The emerging threat of carbapenem resistance of K. pneumoniae in Egyptian hospitals has been highlighted over recent years [3, 35]. To the best of our knowledge, this is the first report on the complete genome sequence of KPC-2-and OXA-48-coproducing virulent K. pneumoniae from Egypt.
The uncommon co-carriage of genes encoding different classes of carbapenemases endowed EBSI041 with high carbapenems resistance. Not restricted in this study, the co-harboring blaKPC-2 and blaOXA-48 in K. pneumoniae isolates were also found from other clinical setting [15]. The presence of carbapenemase genes on mobile elements greatly promotes the spread and stacking of carbapenems resistance. The blaOXA-48-carrying pEBSI041-2 was identified as an IncL/M-type plasmids, which are commonly self-conjugative among Enterobacteriaceae according to the previous studies [36]. It’s worth noting that non-conjugative pEBSI041-3 carried blaKPC-2 gene, was successfully transferred with the help of pEBSI041-2. The results warn that the mechanisms of blaOXA-48 or blaKPC-2 -carrying plasmid transfer need to be further studied to better control the spread of carbapenemase-producing K. pneumoniae.
The genetic structure of blaOXA-48 or blaKPC-2 -carrying plasmid in this study is different from that of reported plasmids due to the presence of multiple transposons and insertion sequences. The blaOXA-48-carrying pEBSI041-2 was similar to other IncL/M plasmids previously sequenced, the majority of which only carry blaOXA-48 [15]. However, pEBSI041-2 harboured an exogenetic chromosome-located fragment and acquired additional resistance gene blaSHV-12 due to the recombination of the IS26-like elements. In pEBSI041-3, the blaKPC-2 gene was located on a transposon unit with the core structure of ΔISKpn6-blaKPC-2-ISKpn27 as that reported [15, 25]. Further, pEBSI041-3 and MDR plasmid pEBSI041-4, which carried eight IS26 elements, almost constitutes another KPC-2-producing plasmids [27, 28]. Therefore, the novel genetic structure of these plasmids are likely to be created by IS-mediated recombination.
Conclusion
This study reported the co-carriage of distinct blaKPC-2 and blaOXA-48 plasmids in a single ST11 hypervirulent Klebsiella pneumoniae isolate in Egypt. The recombination and rearrangement of MDR plasmids and virulent plasmids have occurred during evolution. These results give new insights into the adaptation and evolution of K. pneumoniae plasmids during nosocomial infections in Egypt.
Materials and methods
Bacterial strain
K. pneumoniae EBSI041 was collected from the blood sample of a patient in Egypt. The clinical strain was initially isolated on MacConkey agar (Oxoid, UK). Species identification was determined primarily with an automated VITEK®2 AST-16 Gram-negative susceptibility card (bioMérieux, Marcy-l’Étoile, France) and confirmed by matrix-assisted laser desorption ionisation-time of flight mass spectrometry (MALDI-TOF MS). Investigation of carbapenemase production in the routine hospital laboratory procedure using the Modified Hodge Test (MHT) [37] and Carba NP test [38] showed that EBSI041 was resistant to carbapenems. We tested EBSI041 for carbapenems-resistant genes and confirmed that it carried blaOXA-48 and blaKPC-2. Then, EBSI041 was selected for whole-genome sequencing for further identification. Ethical approval for this study was given by Zhongshan School of Medicine of Sun Yat-sen University under approval number 068. All methods involved in this study were carried out in accordance with relevant guidelines and regulations.
Antimicrobial susceptibility testing
Minimum inhibitory concentrations (MICs) were determined for the following 17 different antibiotics: cefotaxime (CTX), ceftazidime (CAZ), cefepime (FEP), colistin (CT), tigecycline (TGC), imipenem (IMP), ertapenem (ETP), meropenem (MEM), ciprofloxacin (CIP), fosfomycin (FOS), trimethoprim-sulfamethoxazole (SXT), piperacillin-tazobactam (PTZ), amikacin (AMK), gentamicin (GEN), chloramphenicol (CHL), tetracycline (TET), and aztreonam (ATM) for EBSI041 using the agar microdilution method excepted for colistin using the broth microdilution method. Escherichia coli ATCC 25922 strain was used as the reference control. MICs were interpreted following the Clinical and Laboratory Standards Institute (CLSI 2018) [39] guidelines, except for tigecycline and colistin, which were interpreted using the EUCAST 2019 guidelines (ECOFFs; http://www.eucast.org/).
In vitro conjugation and S1-PFGE
The horizontal transfer of plasmids was examined by in vitro conjugation using K. pneumoniae EBSI041 as a donor and E. coli strain EC600 and J53 as a recipient, respectively. Briefly, the EBSI041, EC600 and J53 strains were cultured to OD600 0.4-0.6, mixed in a 1: 1 donor-to-recipient ratio, platted onto Luria-Bertani (LB) agar plates and incubated at 37 °C overnight. One ml of sterile saline was used to remove the conjugation mix from the LB agar plates. Transconjugants were then selected by plating LB agar plates containing rifampicin (Rif; 500 μg/ml) and imipenem (Imp; 2 μg/ml) for EC600, and sodium azide (NaN3; 100 μg/ml) and imipenem (Imp; 2 μg/ml) for J53. The transfer of the plasmid was checked by PCR analysis and MICs. The blaKPC-2 and blaOXA-48 genes were confirmed by PCR and sequencing with primers KPC-A (TGTAAGTTACCGCGCTGAGG), KPC-B (CCAGACGACGGCATAGTCATF) [40], and OXA-A (TTGGTGGCATCGATTATCGG), OXA-B (GAGCACTTCTTTTGTGATGGC) [41]. And S1-PFGE was used to determine the sizes and numbers of plasmids harboured by the isolate EBSI041 and transconjugants [42].
Galleria mellonella virulence assay
The virulence of strain EBSI041 was tested using the wax moth (Galleria mellonella) larvae model. Briefly, 30 larvae weighing about 300 mg were randomly selected for each isolate and maintained on woodchips in the dark at 15 °C until being used. Overnight cultures of K. pneumoniae strains were washed with phosphate-buffered saline (PBS) and further adjusted with PBS to concentrations of 1×106 CFU/mL (10 ul for injection). Colony counts were conducted by serial dilution with final plating on LB agar. The G. mellonella were infected with the tested bacteria, as previously described [43]. PBS injection controls and the negative controls (receiving no injection) were used to evaluate trauma and attrition, respectively. EC600 strain was used as non-virulent control, while, HvKP4 strain was used as the hypervirulent control [18]. HvKP4, a ST11 carbapenem-resistant hypervirulent K. pneumoniae outbreak strain, was isolated from China. The larvae were incubated at 37 °C in the dark and observed every 12 h for 7 days. We recorded the survival rate of the G. mellonella over 48 h post-infection. Results were not included if greater than or equal to two larvae died in either of the control groups. All experiments were done in triplicate.
Whole-genome sequencing (WGS)
The long-read MinION sequencer (Oxford Nanopore Technologies, Oxford, UK) was performed to sequencing the EBSI041 strain with a mean read length of 24 Kbp. De novo hybrid assembly was performed using Unicycler v0.4.3 [44]. Complete circular contigs were then corrected using Pilon v1.22 with Illumina reads (Illumina, USA). In silico multilocus sequence typing (MLST) was performed by MLST 1.8 (https://cge.cbs.dtu.dk/services/MLST/). The antibiotic resistance genes and virulence genes were identified using ABRicate version 0.5 (https://github.com/tseemann/abricate). Insertion sequence (IS) elements were determined with ISFinder (https://www-isfinder.biotoul.fr). The SNP (sequence-based) and core-genome-based MLST (cgMLST) strategies on BacWGSTdb 2.0 were used for source tracking bacterial pathogens, and the phylogenetic tree was generated and visualized by Grapetree [24, 45].
Nucleotide accession numbers
The annotated sequences of all four plasmids have been deposited in the GenBank nucleotide sequence database under accession numbers MW245019 (pEBSI041-1), MW245020 (pEBSI041-2), MW245021 (pEBSI041-3) and MW245022 (pEBSI041-4).
Availability of data and materials
The newly sequenced plasmid sequences are available in GenBank under accession numbers MW245019 (pEBSI041-1) (https://www.ncbi.nlm.nih.gov/nuccore/MW245019), MW245020 (pEBSI041-2) (https://www.ncbi.nlm.nih.gov/nuccore/MW245020), MW245021 (pEBSI041-3) (https://www.ncbi.nlm.nih.gov/nuccore/MW245021) and MW245022 (pEBSI041-4) (https://www.ncbi.nlm.nih.gov/nuccore/MW245022). The data in the present study are available from the corresponding author upon reasonable request.
Change history
06 April 2022
A Correction to this paper has been published: https://doi.org/10.1186/s12864-022-08477-w
Abbreviations
- CRKP:
-
Carbapenem-resistant Klebsiella pneumonia
- CR-hvKP:
-
Carbapenem-resistant hypervirulent K. pneumoniae
- ICU:
-
Intensive Care Unit
- S1-PFGE:
-
S1 nuclease pulsed field gel electrophoresis
- ARGs:
-
Antibiotic resistance genes
- MDR:
-
Multidrug-resistance
- ORFs:
-
Open reading frames
- MALDI-TOF MS:
-
matrix-assisted laser desorption ionisation-time of flight mass spectrometry
- MICs:
-
Minimum inhibitory concentrations
- WGS:
-
Whole-genome sequencing
- MLST:
-
Multilocus sequence typing
References
Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13:785–96.
Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895.
Stewardson AJ, Marimuthu K, Sengupta S, Allignol A, El-Bouseary M, Carvalho MJ, et al. Effect of carbapenem resistance on outcomes of bloodstream infection caused by Enterobacteriaceae in low-income and middle-income countries (PANORAMA): a multinational prospective cohort study. Lancet Infect Dis. 2019;19:601–10.
Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59:5873–84.
Poirel L, Dortet L, Bernabeu S, Nordmann P. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother. 2011;55:5403–7.
Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22:686–96.
Wailan AM, Paterson DL, Kennedy K, Ingram PR, Bursle E, Sidjabat HE. Genomic characteristics of NDM-producing Enterobacteriaceae isolates in Australia and their blaNDM genetic contexts. Antimicrob Agents Chemother. 2016;60:136–41.
Bowers JR, Kitchel B, Driebe EM, MacCannell DR, Roe C, Lemmer D, et al. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS One. 2015;10:e0133727.
Wang Q, Wang X, Wang J, Ouyang P, Jin C, Wang R, et al. Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012-2016). Clin Infect Dis. 2018;67:S196–205.
Liao W, Liu Y, Zhang W. Virulence evolution, molecular mechanisms of resistance and prevalence of ST11 carbapenem-resistant Klebsiella pneumoniae in China: a review over the last 10 years. J Glob Antimicrob Resist. 2020;23:174–80.
Shoma S, Kamruzzaman M, Ginn AN, Iredell JR, Partridge SR. Characterization of multidrug-resistant Klebsiella pneumoniae from Australia carrying blaNDM-1. Diagn Microbiol Infect Dis. 2014;78:93–7.
Baraniak A, Izdebski R, Fiett J, Gawryszewska I, Bojarska K, Herda M, et al. NDM-producing Enterobacteriaceae in Poland, 2012-14: inter-regional outbreak of Klebsiella pneumoniae ST11 and sporadic cases. J Antimicrob Chemother. 2016;71:85–91.
Branas P, Villa J, Viedma E, Mingorance J, Orellana MA, Chaves F. Molecular epidemiology of carbapenemase-producing Klebsiella pneumoniae in a hospital in Madrid: successful establishment of an OXA-48 ST11 clone. Int J Antimicrob Agents. 2015;46:111–6.
Lascols C, Peirano G, Hackel M, Laupland KB, Pitout JD. Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-like enzymes in North America. Antimicrob Agents Chemother. 2013;57:130–6.
Wang YC, Tang HL, Liao YC, Chiou CS, Chen YT, Chiang MK, et al. Cocarriage of distinct blaKPC-2 and blaOXA-48 plasmids in a single sequence type 11 carbapenem-resistant Klebsiella pneumoniae isolate. Antimicrob Agents Chemother. 2019;63:e02282–18.
Liu Y, Long D, Xiang TX, Du FL, Wei DD, Wan LG, et al. Whole genome assembly and functional portrait of hypervirulent extensively drug-resistant NDM-1 and KPC-2 co-producing Klebsiella pneumoniae of capsular serotype K2 and ST86. J Antimicrob Chemother. 2019;74:1233–40.
Huang YH, Chou SH, Liang SW, Ni CE, Lin YT, Huang YW, et al. Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J Antimicrob Chemother. 2018;73:2039–46.
Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18:37–46.
Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32:e00001–19.
Metwally L, Gomaa N, Attallah M, Kamel N. High prevalence of Klebsiella pneumoniae carbapenemase-mediated resistance in K. pneumoniae isolates from Egypt. East Mediterr Health J. 2013;19:947–52.
Makharita RR, El-Kholy I, Hetta HF, Abdelaziz MH, Hagagy FI, Ahmed AA, et al. Antibiogram and genetic characterization of carbapenem-resistant gram-negative pathogens incriminated in healthcare-associated infections. Infect Drug Resist. 2020;13:3991–4002.
Khalifa HO, Soliman AM, Ahmed AM, Shimamoto T, Hara T, Ikeda M, et al. High carbapenem resistance in clinical gram-negative pathogens isolated in Egypt. Microb Drug Resist. 2017;23:838–44.
El Mahallawy H, Zafer MM, Al-Agamy M, Amin MA, Mersal MM, Booq RY, et al. Dissemination of ST101 blaOXA-48 producing Klebsiella pneumoniae at tertiary care setting. J Infect Dev Countries. 2018;12:422–8.
Feng Y, Zou S, Chen H, Yu Y, Ruan Z. BacWGSTdb 2.0: a one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 2021;49:D644–D50.
Chen YT, Lin JC, Fung CP, Lu PL, Chuang YC, Wu TL, et al. KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J Antimicrob Chemother. 2014;69:628–31.
He L, Partridge SR, Yang X, Hou J, Deng Y, Yao Q, et al. Complete nucleotide sequence of pHN7A8, an F33:a-:B- type epidemic plasmid carrying blaCTX-M-65, fosA3 and rmtB from China. J Antimicrob Chemother. 2013;68:46–50.
Nishida S, Ono Y. Genomic analysis of a pan-resistant Klebsiella pneumoniae sequence type 11 identified in Japan in 2016. Int J Antimicrob Agents. 2020;55:105854.
Xiang DR, Li JJ, Sheng ZK, Yu HY, Deng M, Bi S, et al. Complete sequence of a novel IncR-F33:A-:B- plasmid, pKP1034, harboring fosA3, blaKPC-2, blaCTX-M-65, blaSHV-12, and rmtB from an epidemic Klebsiella pneumoniae sequence type 11 strain in China. Antimicrob Agents Chemother. 2015;60:1343–8.
Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215:S28–36.
Zhang R, Liu L, Zhou H, Chan EW, Li J, Fang Y, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106.
Gamal D, Fernandez-Martinez M, Salem D, El-Defrawy I, Montes LA, Ocampo-Sosa AA, et al. Carbapenem-resistant Klebsiella pneumoniae isolates from Egypt containing blaNDM-1 on IncR plasmids and its association with rmtF. Int J Infect Dis. 2016;43:17–20.
Gamal D, Egea P, Elias C, Fernandez-Martinez M, Causse M, Perez-Nadales E, et al. High-risk clones and novel sequence type ST4497 of Klebsiella pneumoniae clinical isolates producing different alleles of NDM-type and other carbapenemases from a single tertiary-care Centre in Egypt. Int J Antimicrob Agents. 2020;56:106164.
Jaidane N, Bonnin RA, Mansour W, Girlich D, Creton E, Cotellon G, et al. Genomic insights into colistin-resistant Klebsiella pneumoniae from a Tunisian teaching hospital. Antimicrob Agents Chemother. 2018;62:e01601–17.
Zhang Y, Jin L, Ouyang P, Wang Q, Wang R, Wang J, et al. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother. 2020;75:327–36.
Barwa R, Shaaban M. Molecular characterization of Klebsiella pneumoniae clinical isolates with elevated resistance to carbapenems. Open Microbiol J. 2017;11:152–9.
Potter RF, D'Souza AW, Dantas G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist Updat. 2016;29:30–46.
Mathers AJ, Carroll J, Sifri CD, Hazen KC. Modified Hodge test versus indirect carbapenemase test: prospective evaluation of a phenotypic assay for detection of Klebsiella pneumoniae carbapenemase (KPC) in Enterobacteriaceae. J Clin Microbiol. 2013;51:1291–3.
Campana EH, Chuster SG, da Silva IR, Paschoal RP, Bonelli RR, Moreira BM, et al. Modified Carba NP test for the detection of carbapenemase production in gram-negative rods: optimized handling of multiple samples. Braz J Microbiol. 2017;48:242–5.
CLSI. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Wayne: M100-S28. CLSI; 2018.
Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66:307–12.
Poirel L, Heritier C, Tolun V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:15–22.
Sirichote P, Hasman H, Pulsrikarn C, Schonheyder HC, Samulioniene J, Pornruangmong S, et al. Molecular characterization of extended-spectrum cephalosporinase-producing salmonella enterica serovar Choleraesuis isolates from patients in Thailand and Denmark. J Clin Microbiol. 2010;48:883–8.
McLaughlin MM, Advincula MR, Malczynski M, Barajas G, Qi C, Scheetz MH. Quantifying the clinical virulence of Klebsiella pneumoniae producing carbapenemase Klebsiella pneumoniae with a galleria mellonella model and a pilot study to translate to patient outcomes. BMC Infect Dis. 2014;14:31.
Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595.
Zhou Z, Alikhan NF, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28(9):1395–404.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (grant numbers 81830103, 82061128001, 81902123), National Key Research and Development Program (grant number 2017ZX10302301), Guangdong Natural Science Foundation (grant number 2017A030306012), Guangdong Province Medical Science Research Foundation (grant number A2018413), Project of high-level health teams of Zhuhai at 2018 (The Innovation Team for Antimicrobial Resistance and Clinical Infection), 111 Project (grant number B12003), and Open project of Key Laboratory of Tropical Disease Control (Sun Yat-sen University), Ministry of Education (grant numbers 2020kfkt04, 2020kfkt07), China Postdoctoral Science Foundation (grant number 2019 M653192), Science, Technology & Innovation Commission of Shenzhen Municipality (JCYJ20190807151601699), Guangdong Provincial Bureau of Traditional Chinese Medicine research fund (grant number 20201407).
Author information
Authors and Affiliations
Contributions
Conceptualization, Guo-Bao Tian, Xubin Huang and Lin Xu; Data curation, Yanxian Yang and Yongqiang Yang; Funding acquisition, Guo-Bao Tian, Yongqiang Yang, Lingqing Xu and Lin Xu; Investigation, Yanxian Yang, Yongqiang Yang, Reem Mostafa Hassan and Mohamed Abd El-Gawad El-Sayed Ahmed; Methodology, Yanxian Yang, Mingyang Qin, Ruowen He, Yiping Wu, Xiaoxue Liang; Resources, Guo-Bao Tian, Reem Mostafa Hassan and Mohamed Abd El-Gawad El-Sayed Ahmed; Visualization, Yanxian Yang; Writing-original draft, Yanxian Yang and Yongqiang Yang; Writing-review & editing, Guo-Bao Tian, Xubin Huang, Lin Xu, Mohamed Abd El-Gawad El-Sayed Ahmed, Lan-Lan Zhong, Ping Chen, Baoguo Deng, Weihong Wen and Lingqing Xu; Supervision, Guo-Bao Tian. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Ethical approval for this study was given by the Ethics Committee of Zhongshan School of Medicine of Sun Yat-sen University under approval number 068 and sample collection was approved by the Ethics Committee of Souad-Kafafi teaching hospital in Misr University for Science and Technology. Informed consent was provided by the guardians of the patients in the study. All methods involved in this study were carried out in accordance with relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: there was an error in the correspondence box and a typographical error in the body of the article.
Supplementary Information
Additional file 1: Table S1
Minimum inhibitory concentrations (MICs) of K. pneumoniae EBSI041 strain and transconjugants. Figure S1 Virulence potential of strain EBSI041 as depicted in a Galleria mellonella infection model with an inoculum of 1 × 104 CFU. Figure S2 The S1-PFGE map of K. pneumoniae EBSI041 strain and transconjugants. Transconjugant a and b were E. coli J53 as the recipient strains, transconjugant c and d were E. coli EC600 as the recipient strains. Figure S3 The phylogenetic tree of 67 K. pneumoniae strains based on core-genome-based MLST (cgMLST) analysis using BacWGSTdb 2.0 (threshold 50). The tree was generated and visualized by Grapetree.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yang, Y., Yang, Y., Ahmed, M.A.EG.ES. et al. Carriage of distinct blaKPC-2 and blaOXA-48 plasmids in a single ST11 hypervirulent Klebsiella pneumoniae isolate in Egypt. BMC Genomics 23, 20 (2022). https://doi.org/10.1186/s12864-021-08214-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-021-08214-9