Abstract
Background
A carbapenem-resistant hypermucoviscous Klebsiella pneumoniae isolate was recovered from human sputum.
Methods
Whole genome sequencing of this isolate was carried out to reveal its clonal background, antimicrobial resistance determinants and virulence factors. Virulence assays were performed using wax moth larvae. The transfer of blaNDM-5 between bacterial strains was tested using conjugation. 59 genome assemblies of ST29 K. pneumoniae and 230 IncX3 plasmids regardless of the carriage of resistance gene were employed for phylogenetic analysis, respectively.
Results
The strain carried a virulence plasmid pVir-SCNJ1 bearing the virulence gene rmpA and exhibited a high virulence in wax moth. This hypervirulent strain belongs to sequence type 29 and carries blaNDM-5, which is located on a conjugative plasmid, designated pNDM5-SCNJ1, belonging to type IncX3. pNDM5-SCNJ1 was fully sequenced and shows high similarity with pNDM_MGR194, except some deletion inside the ISAba125 region. Phylogenetic analysis of IncX3 plasmids revealed that although blaNDM-5 can be evolved from blaNDM-1 via point mutations within some IncX3 plasmids, most of blaNDM-5-carrying IncX3 plasmids probably have acquired blaNDM-5 in multiple events.
Conclusions
In this study, we characterized a blaNDM-5-positive hypervirulent K. pneumoniae of sequence type 29 in China. Our results highlight the need for active surveillance on this lineage of carbapenem-resistant K. pneumoniae.
Similar content being viewed by others
Introduction
Hypervirulent Klebsiella pneumoniae (hvKP) is a worldwide concern due to its capacity to cause life-threatening, community-acquired infections in healthy individuals with high morbidity and mortality [1, 2]. hvKP strains are usually less resistant to most antimicrobials than classic K. pneumoniae [3], but the increasing emergence of carbapenemase-producing hypervirulent K. pneumoniae (CP-hvKP) compromises options of antimicrobial agents for infection control and drives a global crisis [2, 4]. These CP-hvKP strains are thought to be the result of acquiring plasmid-mediated resistance and virulence markers, either by transferring of resistance plasmids into hvKP strains or virulence plasmids into carbapenem-resistant strains [5]. There have now been several reports of infections caused by carbapenemase-producing hypervirulent K. pneumoniae strains [2, 4, 6,7,8,9]. These CP-hvKP isolates mainly produce KPC (a group of serine-lactamases), followed by IMP (a group of metallo-β-lactamases), and they belong to the widely distributed sequence type (ST) 11 [2, 8] and several other STs, e.g., ST25, ST65 [6] and ST36 [4]. Here, we identified an ST29 CR-hvKP clinical strain with K54 serotype carrying blaNDM-5 gene and reported on its characterization.
Materials and methods
Bacterial identification and PCR analysis
The strain SCNJ1 was recovered from the sputum of a patient with an acute bronchiolitis in a hospital of Sichuan Province in November 2018. The initial species identification was performed using the Vitek-2 compact system (bioMérieux, Marcy-l’Étoile, France). A further species confirmation was performed by PCR amplifying of the 16S rRNA gene using the primer pair 27F/1492R [10]. PCR products were purified and then sequenced by Sanger sequencing. The resulting 16S rRNA gene sequences were compared with sequences in GenBank (NCBI) database using BLAST software. The presence of the acquired carbapenemase genes blaKPC, blaNDM, blaGES, blaIMP, blaOXA-48, and blaVIM in this isolate was screened via PCR using primers as previously described [11,12,13,14].
Antimicrobial susceptibility tests
In vitro susceptibility tests of cefepime, piperacillin-tazobactam, ampicillin, ampicillin/sulbactam, cefotetan, ceftriaxone, cefazolin, nitrofurantoin, ceftazidime, tobramycin, ciprofloxacin, trimethoprim/sulfamethoxazole, aztreonam, amikacin, gentamicin and levofloxacin were performed using Vitek-2 system. The minimum inhibitory concentrations (MICs) of imipenem, meropenem and colistin against the isolate were determined using the microdilution broth method following recommendations of the Clinical Laboratory Standards Institute (CLSI) [15]. Breakpoints of colistin was defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org/), otherwise, we applied those defined by the CLSI.
String test
String test was performed by stretching a mucoviscous string from the colony using a standard bacteriologic loop as described previously [16]. Strains that formed viscous strings > 5 mm in length were defined as hypermucoviscous.
Conjugation
Conjugation experiments were carried out using broth-based methods with the azide-resistant Escherichia coli strain J53 as the recipient and transconjugants were selected using 2 μg/ml meropenem plus 150 μg/ml sodium azide. The presence of blaNDM-5 in transconjugants was confirmed by PCR and sequencing.
Virulence assay
The virulence potential of the SCNJ1 strain was assessed using wax moth (Galleria mellonella) larvae weighing 250 to 350 mg (Tianjin Huiyude Biotech Company, Tianjin, China) with method described previously [17]. Overnight cultures of K. pneumoniae strains were adjusted with phosphate-buffered saline (PBS) to concentrations of 1 × 104 CFU/ml, 1 × 105 CFU/ml, 1 × 106 CFU/ml, 1 × 107 CFU/ml after being washed with PBS. 10 μl of inoculum was injected into the hemocoel of sixteen larvae using a 25-μl Hamilton syringe via the last left proleg. The larvae were then incubated at 37 °C and the number of live larvae was counted at 12 h intervals for 3 days. Two blaKPC-2-carrying carbapenem-resistant K. pneumoniae clinical isolates of ST11:K47, KPNJ2 and KPLZ1050, without rmpA and rmpA2, were used as the control.
Genome sequencing and analysis
The strain was subjected to whole genomic sequencing using an Illumina HiSeq 2000 system with the 150-bp paired-end approach and 150 × coverage. Reads were trimmed using Trimmomatic [18]. Draft genome was then assembled using the SPAdes program [19]. Annotation was carried out using Prokka [20]. Sequence type and capsular type of this strain were determined using the assembled contigs to query the Multi-Locus Sequence Typing (MLST) v 2.0 (https://cge.cbs.dtu.dk/services/MLST/) and wzc genotyping system as previously described [21], respectively. In addition, the wzi genotyping system [22] and KLeborate (https://github.com/katholt/Kleborate/) were employed to confirm the sequence type and capsular type. Clonal complexes (CCs) were determined by using eBURST v3 based on K. pneumoniae MLST data (https://eburst.mlst.net). Virulence genes were identified using the Virulence Factors Database (VFDB) available at http://www.mgc.ac.cn/VFs/main.htm. Antimicrobial resistance genes were identified using the ResFinder v3.1 software of the Center for Genomic Epidemiology (CGE, http://genomicepidemiology.org/).
Plasmids pVir-SCNJ1 and pNDM5-SCNJ1 were completely circularized with gaps between the contigs closed by PCR and respective amplicons sequenced using Sanger sequencing, respectively. Identification of plasmid incompatibility types were performed on complete sequences of plasmids via the online service PlasmidFinder v2.0 at CGE (https://cge.cbs.dtu.dk/services/PlasmidFinder/). The annotations of the plasmid sequences were conducted using the RAST tools and edited manually [23]. Sequence alignment of blaNDM-5-carrying plasmids was performed using BLAST and visualized with Easyfig v 2.2.3 [24]. Alignments with highly homologous complete plasmid sequences of pVir-SCNJ1 available in NCBI were performed by using the BRIG tool [25]. The circular map of pNDM5-SCNJ1 was also generated using BRIG [25].
Phylogenetic analysis
All assembled K. pneumoniae genomes (n = 6823; accessed by March 1, 2019) were retrieved from GenBank. MLST typing was performed using the script (https://github.com/tseemann/mlst). A total of 59 assemblies of ST29 K. pneumoniae were included and aligned with that of strain SCNJ1 using CSI Phylogeny 1.4 (https://cge.cbs.dtu.dk/services/CSIPhylogeny/) (Additional file 1 :Table S2). Gubbins (version 2.3.4) was used to remove single nucleotide polymorphisms (SNPs) on recombination sites [26]. The filtered SNPs were then used as input for inferring a phylogenetic tree using RAxML with the GTRGAMMA model and 1000 bootstraps [27]. ABRicate (https://github.com/tseemann/abricate) was used to identify antimicrobial resistance genes in these genomes and the capsular typing of K. pneumoniae was performed with the wzc genotyping system.
The sequence of all available IncX3 plasmids regardless of the carriage of resistance gene (n = 230; accessed by March 24, 2019) were retrieved from the GenBank (Additional file 1 :Table S4). Orthogroups were identified using OrthoFinder [28] and used for multiple sequence alignments (MSA) with MAFFT [29]. The species tree was inferred from the concatenated MSA using FastTree [30]. The STRIDE algorithm (Specie Tree Root Inference from Duplication Events) was used to root the species tree in OrthoFinder.
Results and discussion
Antimicrobial susceptibility test showed that the K. pneumoniae SCNJ1 strain was resistant to imipenem (MIC, > 256 μg/ml) and meropenem (MIC, > 256 μg/ml) cefepime (MIC, 16 μg/ml), piperacillin-tazobactam (MIC, ≥ 128 μg/ml), ampicillin (MIC, ≥ 32 μg/ml), ampicillin/sulbactam (MIC, ≥ 32 μg/ml), cefotetan (MIC, ≥ 64 μg/ml), ceftriaxone (MIC, ≥ 64 μg/ml), cefazolin (MIC, ≥ 64 μg/ml), nitrofurantoin (MIC,128 μg/ml) and ceftazidime (MIC, ≥ 64 μg/ml), but was susceptible to colistin (MIC, 2 μg/ml), tobramycin (MIC, ≤ 1 μg/ml), ciprofloxacin (MIC, ≤ 0.25 μg/ml), trimethoprim/sulfamethoxazole (MIC, ≤ 20 μg/ml), aztreonam (MIC, ≤ 1 μg/ml), amikacin (MIC, ≤2 μg/ml), gentamicin (MIC, ≤1 μg/ml) and levofloxacin (MIC, ≤ 0.25 μg/ml). Strain SCNJ1 showed hypermucoviscosity phenotype as evidenced by forming a viscous string about 35 mm, which is beyond the > 5 mm to define hypermucoviscous.
PCR and sequencing showed that blaNDM-5 was the only carbapenemase-encoding gene carried by the strain SCNJ1. NDM-5, a variant of NDM (New Delhi Metallo-β-lactamase), was first identified in an E. coli ST648 isolate (EC045) in the UK in 2011 from a patient with a recent hospitalization history in India [31]. Although blaNDM-5 has been widely found in K. pneumoniae strains since its first discovery (Additional file 1 :Table S1), blaNDM-5-carrying hypermucoviscous K. pneumoniae remains uncommon. We found one publication that described a blaNDM-5-positive K. pneumoniae isolate (K2/ST14) in China in 2015 [32], which was speculated to be hypermucoviscous on the basis of genome analysis. However, no experimental data was included.
Draft genome sequences of SCNJ1 was assembled into 29 contigs (28 were > 1000 bp in length), which comprises 5,474,953 bp, with a 57.29% GC content. SCNJ1 was assigned to capsular type wzi115-K54 and sequence type ST29 (gapA-infB-mdh-pgi-phoE-rpoB-tonB allele number 2–3–2-3-6-4-4). K54 is a hypervirulent member of K. pneumoniae [16] and has been described in several previous publications as linked to ST29 [33,34,35,36,37]. To date, K. pneumoniae strains with the ST29 group has a worldwide distribution and has been found carrying a variety of carbapenem genes, including blaNDM-1 [38,39,40], blaKPC [41], blaOXA-48 [42] and blaOXA-181 [38], as well as several extended-spectrum β-lactamases (ESBLs) genes [43, 44] in various countries. However, the currently available evidence is insufficient to demonstrate whether ST29, a member of CC29, is an epidemic clone mediating the spread of specific and clinically relevant antibiotic resistance genes.
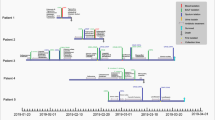
The gene blaNDM-5 is described for the first time in a strain of K. pneumoniae ST29 in our work, as demonstrated by the phylogenetic tree based on filtered SNPs of all available ST29 K. pneumoniae strains (Fig. 1). The phylogenetic analysis also showed that strain SCNJ1 was clustered with four isolates recovered in China and was closest to strain SCLZ15–011 (GCA_001630805, carrying no carbapenemase gene, recovered in 2016 in China) with 198 SNPs difference (Fig. 1). It should be noted that sporadic cases due to ST29 K. pneumoniae were frequently detected, mainly from liver abscess patients [37, 43, 45, 46], and multidrug-resistant ST29 hvKP strains have been reported in different locations of China [5, 33, 47]. This highlights the need to monitor the epidemiology of the ST29 clones of K. pneumoniae isolates in China.
We found that strain SCNJ1 harbored genes encoding regulators of the mucoid phenotype (rmpA), aerobactin (iucABCD and iutA), ent siderophore (entABCDEFS, fepABCDG), salmochelin (iroBCDEN), yersiniabactin (fyuA, irp1, irp2 and ybtAEPQSTUX) and type 3 fimbriae (mrkABCDFHIJ) etc. These genes are frequently associated with hypervirulence phenotype of K. pneumoniae [16, 48]. The rmpA, iutA, iucABCD and iroBCDN genes were carried by a 211,807-bp plasmid, designated pVir-SCNJ1. The rmpA2 (another regulator of mucoid phenotype) gene on the pVir-SCNJ1 plasmid was truncated, due to a frameshift mutation introducing an internal stop codon. pVir-SCNJ1 was an IncHI1/IncFIB-type plasmid and was 99.71% identical to the known virulence plasmid pLVPK (219,385 bp, GenBank accession no. NC_005249) at 93% coverage [49] (Fig. 2). It was notable that pVir-SCNJ1 was highly similar (99% coverage and 99.99% identity, Fig. 2) to the recently-identified plasmid pL22–1 (212, 635 bp, GenBank accession no. NZ_CP031258) that recovered from a Klebsiella quasipneumoniae strain L22, which suggests that the pLVPK-like plasmid has the potential to mediate inter- and intra-species transfer of virulence genes [50]. Our virulence assays showed that survival of G. mellonella was 0% with strains SCNJ1, while survival was 56.2 and 50.0% with the control strains KPLZ1050 and KPNJ2, at an inoculum of 1 × 105 cfu/mL at 72 h after infection, respectively (Fig. 3, Additional file 1 :Table S3). These findings suggest that strain SCNJ1 was hypervirulent.
Alignment of pVir-SCNJ1 with 3 hypervirulence-encoding plasmids. The alignment was performed using BRIG and pLVPK was used as a reference. Accession numbers for the plasmids are NZ_CP031258 (pL22–1), NC_006625 (pK2044), NC_005249 (pLVPK). The locations of virulence genes rmpA2, iutA, iucDCBA, rmpA and iroNDCB are indicated
Virulence potential of K. pneumoniae strains in a G. mellonella infection model. The effect of 1 × 105 CFU/ml of each isolate on survival of G. mellonella is shown. The results for other doses of each K pneumoniae strain are shown in Supplementary Table S3. KPLZ1050 and KPNJ2, two blaKPC − 2-carrying K pneumoniae clinical isolates of ST11 that did not harbour a virulence plasmid, were used as the control
Conjugation assays showed that strain SCNJ1 transferred a plasmid carrying blaNDM-5 to E. coli J53 at a frequency of 10− 6 (transconjugant/recipient) by mating, suggesting that blaNDM-5 was carried on a self-transmissible plasmid, which was assigned pNDM5-SCNJ1. In addition to the blaNDM-5, strain SCNJ1 had a few chromosomal resistance genes, including the ESBL gene blaSHV-187, fluoroquinolone-resistance genes oqxA and oqxB, and fosfomycin-resistance gene fosA.
pNDM5-SCNJ1 was a 45,255-bp IncX3 plasmid, with an average GC content of 46.83% and had no other known antimicrobial resistance genes except blaNDM-5. pNDM5-SCNJ1 consists of a 30-kb backbone comprising several sets of genes (pir and bis encoding replication initiation protein, parA for plasmid partitioning, hns and topB for maintenance and a gene cluster responsible for conjugation) and a genetic load region with high GC content between the resolvase and the hns gene, which are typical of IncX3 plasmids (Fig.4a). BLASTn revealed that the sequence of pNDM5-SCNJ1 was highly similar (100% coverage and 99.99% identity) to the plasmid pNDM_MGR194 (GenBank accession no. KF220657) recovered from a K. pneumoniae isolate in India, as well as a number of previously described IncX3 plasmids carrying blaNDM-5 in China.
Plasmid analysis of pNDM5-SCNJ1. (a) Genetic structure of IncX3 plasmid pNDM5-SCNJ1. This map was used to illustrate the backbone and the location of the genetic load region of pNDM5-SCNJ1. Genes are denoted by arrowheads and colored based on gene function classification. (b) Comparative analysis of the genetic load region of pNDM5-SCNJ1. Genes and insertion sequences are indicated by arrows. Light gray shades denote shared regions with a high degree of homology. The accession numbers were: pEC14_35 (JN935899), pBJ01(JX296013), pNDM_MGR194 (KF220657), pP744T (MF547511), pTK1044 (LC000627). The alignment is a pairwise BLASTn alignment performed using Easyfig [24]
In the genetic load region of pNDM5-SCNJ1(Fig. 4b), the umuD gene was split into two fragments at the nucleotide position 336 bp by the blaNDM-5-containing structure (IS26-ΔctuA1-tat-trpF-bleMBL-blaNDM-5-ΔISAba125-IS5-ΔISAba125-IS3000-ΔTn2), resulting in a pair of 3-bp direct repeats (TGT). In such a genetic context, an IncX3 plasmid pEC14_35 (GenBank accession no. JN935899) without any antibiotic-resistance gene, which was isolated from a patient in the USA in 1989, was likely to be the ancestral vector. It is also likely that pNDM5-SCNJ1 has diverged recently from blaNDM-1-positive plasmids pBJ01 (GenBank accession no. JX296013) by sequential mutations (Fig. 4b). In the subsequent genetic variant, the ISAba125 was truncated by the insertion of IS5 element (at 166 bp upstream blaNDM-5 start codon) and a 4-bp flanking direct repeats (CTAA) was identified. Comparisons of the genetic contexts of blaNDM-5 in pNDM_MGR194, pP744T, pTK1044 and pNDM5-SCNJ1 showed that the remnant of ISAba125 (73 bp of 1087 bp) upstream of blaNDM-5 was conserved, but the length of the remnant of ISAba125 between IS3000 and IS5 differed (pNDM_MGR19: 1002 bp, pP744T: 404 bp, pNDM-SCNJ1: 112 bp, pTK1044: 0 bp), suggesting that IS5 has inserted into ISAba125 at the same position in these plasmids and that gene deletions caused by homologous recombination plays a possible role in the formation of diversified ΔISAba125 region.
IncX3 plasmids are narrow-host-range vectors of the Enterobacteriaceae [51, 52]. Searches on IncX3 plasmids in NCBI showed that they were recovered from various species of Enterobacteriaceae (Additional file 1 :Table S4), including E. coli, K. pneumoniae, Citrobacter freundii, Enterobacter cloacae, Klebsiella oxytoca, Enterobacter hormaechei, Salmonella enterica, Kluyvera intermedia, Morganella morganii, Raoultella planticola and Raoultella ornithinolytica from different countries, suggesting a wide distribution of IncX3 plasmids. Three kinds of carbapenemase genes were found to be carried by IncX3 plasmids, including blaNDM, blaOXA-181 and blaKPC. The carriage rate of blaNDM was significantly higher (n = 150, 64.94%) than those of blaKPC (n = 18, 7.79%) and blaOXA-181 (n = 17, 7.35%) (Additional file 1 :Table S4). Of note, IncX3 plasmids were found to carry many different blaNDM alleles, including NDM-1, 4, 5, 6, 7, 13, 19, 20, 21, which were mainly recovered from China, confirming that IncX3 plasmids function as a common vehicle in facilitating the rapid dissemination of NDM-type MBLs among Enterobacteriaceae in China.
Phylogenetic analysis based on concatenated MSA of IncX3 plasmids revealed that pNDM5-SCNJ1 was closely related (100% coverage, 99.99% identity) to plasmid pQDE2-NDM (GenBank accession no. MH917280), which also carried blaNDM-5 that was recovered from a K. pneumoniae isolate in Shandong, China, in 2015 (Fig. 5). The phylogenetic tree also showed that most of blaNDM-5-carrying IncX3 plasmids are tightly clustered with each other and formed a relatively distinct clade, with only sporadic ones clustered into clades with blaNDM-1-carrying plasmids, suggesting that although blaNDM-5 is likely to evolve from blaNDM-1 via point mutations on some IncX3 plasmids, most of IncX3 plasmids probably have acquired blaNDM-5 in multiple events.
Conclusion
In conclusion, our work identified an ST29 CP-hvKP carrying the carbapenemase gene blaNDM-5 and provided additional evidence of the rapid dissemination of blaNDM-5 by pNDM-MGR194-like plasmid among Enterobacteriaceae in China. The association of the epidemic IncX3 plasmid carrying blaNDM-5 with a hypervirulent K. pneumoniae lineage, ST29/K54 in this case, is quite worrisome and may pose a great threat to humans. More extensive surveillance and effective action to control its further dissemination are urgently required.
Availability of data and materials
Draft whole-genome sequences of the SCNJ1 strain has been deposited into GenBank under the accession no. SPSD00000000. The complete sequences of pVir-SCNJ1 and pNDM5-SCNJ1 have been deposited into GenBank under accession no. MK715436 and MK715437, respectively.
References
Zhang Y, Zhao C, Wang Q, Wang X, Chen H, Li H, Zhang F, Li S, Wang R, Wang H. High prevalence of hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother. 2016;60(10):6115–20.
Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW-C, Shu L, Yu J, Zhang R, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46.
Liu YM, Li BB, Zhang YY, Zhang W, Shen H, Li H, Cao B. Clinical and molecular characteristics of emerging hypervirulent Klebsiella pneumoniae bloodstream infections in mainland China. Antimicrob Agents Chemother. 2014;58(9):5379–85.
Feng Y, Lu Y, Yao Z, Zong Z. Carbapenem-resistant hypervirulent Klebsiella pneumoniae of sequence type 36. Antimicrob Agents Chemother. 2018;62(7):e02644–17.
Chen L, Kreiswirth BN. Convergence of carbapenem-resistance and hypervirulence in Klebsiella pneumoniae. Lancet Infect Dis. 2018;18(1):2–3.
Yao B, Xiao X, Wang F, Zhou L, Zhang X, Zhang J. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int J Infect Dis. 2015;37:107–12.
Zhang R, Lin D, Chan EW, Gu D, Chen GX, Chen S. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother. 2016;60(1):709–11.
Zhan L, Wang S, Guo Y, Jin Y, Duan J, Hao Z, Lv J, Qi X, Hu L, Chen L, et al. Outbreak by Hypermucoviscous Klebsiella pneumoniae ST11 isolates with carbapenem resistance in a tertiary hospital in China. Front Cell Infect Microbiol. 2017;7:182.
Zhang Y, Zeng J, Liu W, Zhao F, Hu Z, Zhao C, Wang Q, Wang X, Chen H, Li H, et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect. 2015;71(5):553–60.
Lane DJ. 16S/23S rRNA sequencing. Nucleic acid techniques in bacterial systematics. 1991:115–75.
Szekely E, Damjanova I, Janvari L, Vas KE, Molnar S, Bilca DV, Lorinczi LK, Toth A. First description of bla (NDM-1), bla (OXA-48), bla (OXA-181) producing Enterobacteriaceae strains in Romania. Int J Med Microbiol. 2013;303(8):697–700.
Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(3):490–5.
Ellington MJ, Kistler J, Livermore DM, Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother. 2007;59(2):321–2.
Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. Biochemical sequence analyses of GES-1, a novel class a extended-spectrum beta-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2000;44(3):622–32.
Clinical and Laboratory Standards Institute. M100S. Performance standards for antimicrobial susceptibility testing. 29th edition. USA: Wayne, PA; 2019.
Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–18.
Lu Y, Feng Y, McNally A, Zong Z. The Occurence of colistin-resistant hypervirulent Klebsiella pneumoniae in China. Front Microbiol. 2018;9:2568.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77.
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–9.
Pan YJ, Lin TL, Chen YH, Hsu CR, Hsieh PF, Wu MC, Wang JT. Capsular types of Klebsiella pneumoniae revisited by wzc sequencing. PLoS One. 2013;8(12):e80670.
Brisse S, Passet V, Haugaard AB, Babosan A, Kassis-Chikhani N, Struve C, Decre D. wzi gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol. 2013;51(12):4073–8.
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75.
Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–10.
Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402.
Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43(3):e15.
Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–3.
Emms DM, Kelly S. OrthoFinder2: fast and accurate phylogenomic orthology analysis from gene sequences. bioRxiv. 2018:466201.
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–80.
Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490.
Hornsey M, Phee L, Wareham DW. A novel variant, NDM-5, of the New Delhi metallo-beta-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother. 2011;55(12):5952–4.
Liu PP, Liu Y, Wang LH, Wei DD, Wan LG. Draft genome sequence of an NDM-5-Producing Klebsiella pneumoniae sequence type 14 strain of serotype K2. Genome Announc. 2016;4(2):e01610–15.
Chuang YC, Lee MF, Yu WL. Mycotic aneurysm caused by hypermucoviscous Klebsiella pneumoniae serotype K54 with sequence type 29: an emerging threat. Infection. 2013;41(5):1041–4.
Iwasaki Y, Inokuchi R, Harada S, Aoki K, Ishii Y, Shinohara K. Bacterial meningitis caused by hypervirulent Klebsiella pneumoniae capsular genotype K54 with development of granuloma-like nodal enhancement in the brain during the subacute phase. Intern Med. 2017;56(3):373–6.
Yan Q, Zhou M, Zou M, Liu WE. Hypervirulent Klebsiella pneumoniae induced ventilator-associated pneumonia in mechanically ventilated patients in China. Eur J Clin Microbiol Infect Dis. 2016;35(3):387–96.
Cubero M, Grau I, Tubau F, Pallares R, Dominguez MA, Linares J, Ardanuy C. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007-2013). Clin Microbiol Infect. 2016;22(2):154–60.
Liao CH, Huang YT, Chang CY, Hsu HS, Hsueh PR. Capsular serotypes and multilocus sequence types of bacteremic Klebsiella pneumoniae isolates associated with different types of infections. Eur J Clin Microbiol Infect Dis. 2014;33(3):365–9.
Balm MN, La MV, Krishnan P, Jureen R, Lin RT, Teo JW. Emergence of Klebsiella pneumoniae co-producing NDM-type and OXA-181 carbapenemases. Clin Microbiol Infect. 2013;19(9):E421–3.
Castanheira M, Deshpande LM, Farrell SE, Shetye S, Shah N, Jones RN. Update on the prevalence and genetic characterization of NDM-1-producing Enterobacteriaceae in Indian hospitals during 2010. Diagn Microbiol Infect Dis. 2013;75(2):210–3.
Gharout-Sait A, Alsharapy SA, Brasme L, Touati A, Kermas R, Bakour S, Guillard T, de Champs C. Enterobacteriaceae isolates carrying the New Delhi metallo-beta-lactamase gene in Yemen. J Med Microbiol. 2014;63(Pt 10):1316–23.
Barria-Loaiza C, Pincheira A, Quezada M, Vera A, Valenzuela P, Dominguez M, Lima CA, Araya I, Araya P, Prat S, et al. Molecular typing and genetic environment of the bla KPC gene in Chilean isolates of Klebsiella pneumoniae. J Glob Antimicrob Resist. 2016;4:28–34.
Uz Zaman T, Aldrees M, Al Johani SM, Alrodayyan M, Aldughashem FA, Balkhy HH. Multi-drug carbapenem-resistant Klebsiella pneumoniae infection carrying the OXA-48 gene and showing variations in outer membrane protein 36 causing an outbreak in a tertiary care hospital in Riyadh, Saudi Arabia. Int J Infect Dis. 2014;28:186–92.
Luo Y, Wang Y, Ye L, Yang J. Molecular epidemiology and virulence factors of pyogenic liver abscess causing Klebsiella pneumoniae in China. Clin Microbiol Infect. 2014;20(11):O818–24.
Gorrie CL, Mirceta M, Wick RR, Judd LM, Wyres KL, Thomson NR, Strugnell RA, Pratt NF, Garlick JS, Watson KM, et al. Antimicrobial-resistant Klebsiella pneumoniae carriage and infection in specialized geriatric care wards linked to acquisition in the referring hospital. Clin Infect Dis. 2018;67(2):161–70.
Qu TT, Zhou JC, Jiang Y, Shi KR, Li B, Shen P, Wei ZQ, Yu YS. Clinical and microbiological characteristics of Klebsiella pneumoniae liver abscess in east China. BMC Infect Dis. 2015;15:161.
Xiao SZ, Wang S, Wu WM, Zhao SY, Gu FF, Ni YX, Guo XK, Qu JM, Han LZ. The resistance phenotype and molecular epidemiology of Klebsiella pneumoniae in bloodstream infections in Shanghai, China, 2012-2015. Front Microbiol. 2017;8:250.
Moura Q, Esposito F, Fernandes MR, Espinoza-Munoz M, Souza TA, Santos SR, Cerdeira L, Cassettari V, Lincopan N. Genome sequence analysis of a hypermucoviscous/hypervirulent and MDR CTX-M-15/K19/ST29 Klebsiella pneumoniae isolated from human infection. Pathog Dis. 2017;75(9):ftx121.
Hsieh PF, Lin TL, Lee CZ, Tsai SF, Wang JT. Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis. 2008;197(12):1717–27.
Chen YT, Chang HY, Lai YC, Pan CC, Tsai SF, Peng HL. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene. 2004;337:189–98.
Lu Y, Feng Y, McNally A, Zong ZZ. Occurrence of colistin-resistant hypervirulent Klebsiella variicola. J Antimicrob Chemother. 2018;73(11):3001–4.
Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid. 2012;68(1):43–50.
Wu WJ, Feng Y, Tang GM, Qiao F, McNally A, Zong ZZ. NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin Microbiol Rev. 2019;32(2):e00115–8.
Acknowledgements
Not applicable.
Funding
This work was supported by Project of Education Department in Sichuan, China (18ZB0633), Natural Science Foundation of Southwest Medical University (No. 2017-ZRZD-022 and 2018-ZRZD-011), and National Undergraduate Innovation and Entrepreneurship Project (No.201816032021). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author information
Authors and Affiliations
Contributions
LZ designed the experiments. GW, CL, FZ and WC performed the experiments. SN, YM and JZ analyzed the data. YL analyzed the data and wrote the manuscript. YY wrote the manuscript. YC edited the original draft. All authors read and approved the final manuscript.
Authors’ information
Not applicable
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The current study was approved by the Ethics Committee of Southwest Medical University (No.201903–194) and were carried out in accordance with the approved guidelines. Written informed consent was exempted, since this retrospective study mainly focused on bacteria and patient intervention was not required.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Table S1. Background information on the blaNDM-5-positive K. pneumoniae isolates. Table S2. ST29 K. pneumoniae strains with genome sequences available in the GenBank. Table S3. Survival (number of larvae) of G. mellonella after infection by K. pneumoniae strain SCNJ1. Table S4. The names, host species, accession numbers, carbapenemase genes and locations of IncX3 plasmids. (DOCX 109 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yuan, Y., Li, Y., Wang, G. et al. blaNDM-5 carried by a hypervirulent Klebsiella pneumoniae with sequence type 29. Antimicrob Resist Infect Control 8, 140 (2019). https://doi.org/10.1186/s13756-019-0596-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-019-0596-1