Abstract
Background
Prion diseases are zoonotic diseases with a broad infection spectrum among mammalian hosts and are caused by the misfolded prion protein (PrPSc) derived from the normal prion protein (PrPC), which encodes the prion protein gene (PRNP). Currently, although several prion disease-resistant animals have been reported, a high dose of prion agent inoculation triggers prion disease infection in these disease-resistant animals. However, in chickens, natural prion disease-infected cases have not been reported, and experimental challenges with prion agents have failed to cause infection. Unlike other prion disease-resistant animals, chickens have shown perfect resistance to prion disease thus far. Thus, investigation of the chicken PRNP gene could improve for understanding the mechanism of perfect prion-disease resistance. Here, we investigated the genetic characteristics of the open reading frame (ORF) of the chicken PRNP gene in a large sampling of various chicken breeds.
Results
We found only tandem repeat deletion polymorphisms of the chicken PRNP ORF in the 4 chicken breeds including 106 Dekalb White, 100 Ross, 98 Ogolgye and 100 Korean native chickens. In addition, the distribution of chicken insertion/deletion polymorphisms was significantly different among the 4 chicken breeds. Finally, we found significant differences in the number of PRNP SNPs between prion disease-susceptible species and prion disease-resistant species. Notably, chickens lack SNPs in the ORF of the prion protein.
Conclusion
In this study, we found that the absence of SNPs in the chicken PRNP ORF is a notable feature of animals with perfect resistant to prion disease.
Similar content being viewed by others
Background
Prion diseases are zoonotic diseases caused by the misfolded prion protein (PrPSc) derived from the normal prion protein (PrPC) and have a broad infection range in mammalian hosts, including ruminants and humans [1,2,3,4,5,6,7,8,9]. To date, prion disease-affected cases have not been reported in various bird species; however, normal functions of PrPC and tandem repeat domains (octapeptide in mammals and hexapeptide in birds) are well known and conserved in both mammals and birds, respectively [10, 11]. The function of PrPC may be related to regulation of stress protection, myelin maintenance, circadian rhythm, mitochondrial homeostasis and metal-ion homeostasis [12]. Among these functions, metal-ion homeostasis was strictly linked to the tandem repeat domain of prion protein [10].
In mammals, numerous single nucleotide polymorphisms (SNPs) in the open reading frame (ORF) of the prion protein gene (PRNP) have been identified thus far. Among these SNPs, several prion disease-associated SNPs have been reported in prion disease-susceptible species, including humans, sheep and goats. In particular, the PRNP codons 129 and 219 in humans are strongly associated with CJD susceptibility [13, 14]. In addition, the PRNP codons 136, 154 and 171 in sheep [15, 16] and the PRNP codons 127, 142, 143, 146, 154, 171, 211 and 222 in goats are also correlated with the incidence of scrapie [17,18,19,20,21]. Approximately 40 SNPs of PRNP ORF have been reported in cattle. However, prion disease-related polymorphisms have not been reported thus far [22,23,24,25]. In addition, dog, which is known as prion disease resistant animal, showed very little polymorphisms in the PRNP ORF [26]. Although real challenge study has not been performed to confirm the transmission of prion disease to horse, horse prion protein transgenic mouse showed the resistance to infection of several agents of prion diseases, including RML, scrapie, chronic wasting disease (CWD), transmissible mink encephalopathy (TME) and bovine spongiform encephalopathy (BSE) [27]. In addition, horse prion protein had high structural stability and its horse specific amino acids showed the protective effect against prion disease [28, 29]. These results suggest that horse is a prion disease-resistant animal. Interestingly, horse has only one SNP in the ORF of the PRNP gene [5].
Our previous study indicated that genetic polymorphisms, which are considered genetic susceptibility factors in mammals, were significantly different in the chicken prion PRNP gene. Chickens lack SNPs and have only one insertion/deletion polymorphism located on the hexapeptide repeat domain in the prion protein. However, in a study performed with only one chicken breed, Dekalb White, it was unclear whether this is a genetic characteristic of chickens or a characteristic of a chicken breed, Dekalb White [30].
Here, we performed direct sequencing of the chicken PRNP gene and confirmed the genetic polymorphism of the chicken PRNP gene in 4 chicken breeds, including Dekalb White, Ross, Ogolgye and Korean native chickens. We also compared the genotype and allele distribution of the chicken PRNP polymorphisms among 4 chicken breeds. Lastly, we investigated and compared SNPs of prion disease-susceptible species (humans, sheep, goats and cattle) and prion disease-resistant species (horses and chickens).
Results
To identify genetic polymorphisms of the chicken PRNP gene, we performed direct sequencing in 298 chickens including 3 chicken breeds (100 Ross, 100 Korean native chickens and 98 Ogolgye). Interestingly, we found only the c.163_180delAACCCAGGGTACCCCCAT (p.55_60delNPGYPH) polymorphism in the 3 chicken breeds (Ross, Ogolgye, and Korean native chickens). The c.268_269insC polymorphism, which was found in the Dekalb White breed, was not found in the 3 studied chicken breeds (Tables 1 and 2 and Additional file 1).
Next, we compared the genotype and allele frequencies of two chicken PRNP polymorphisms in 4 chicken breeds, Dekalb White, Ross, Ogolgye and Korean native chickens. Detailed values of genotype and allele frequencies of the c.163_180delAACCCAGGGTACCCCCAT polymorphism of the chicken PRNP gene are described in Table 1. Except for Dekalb White, all chicken breeds were in HWE. Interestingly, significant differences in genotype and allele distributions were found among Dekalb White and Ross (p < 0.001), Ogolgye (p < 0.001) and Korean native chickens (p < 0.001) (Table 1). In addition, we compared the genotype and allele distributions of the c.268_269insC polymorphism among 4 chicken breeds. Notably, the WT/INS genotype and insertion allele were only identified in 4 out of 106 Dekalb White chickens (3.8%). There were similar distributions in genotype (p = 0.122) and allele (p = 0.1238) frequencies of the c.268_269insC polymorphism between the Dekalb White and 3 chicken breeds (Table 2).
Furthermore, we analyzed haplotypes of the two insertion/deletion polymorphisms among the 4 chicken breeds. Detailed degrees of haplotype distribution in the 4 chicken breeds are described in Table 3. Three major haplotypes were found, and the ht3 haplotype was only detected in the Dekalb White breed. Statistically different distributions of the haplotypes were found between Dekalb White and Ross (p < 0.0001), Ogolgye (p < 0.0001) and Korean native chickens (p < 0.0001).
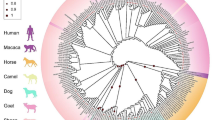
Lastly, we surveyed SNPs of the PRNP gene in prion disease-susceptible species (humans, sheep, goats, and cattle) and prion disease-resistant species (horses and chickens). Over 20 SNPs were found in prion disease-susceptible species. However, in prion disease-resistant animals, only one SNP (N175K) and zero SNPs have been reported in horses and chickens, respectively (Table 4, Fig. 1).
Distribution of the single nucleotide polymorphisms (SNPs) at coding region of the prion protein gene (PRNP) in various species. Previously reported SNPs at coding region of PRNP gene in human, sheep, goats, cattle, horse and chickens. Edged horizontal bar indicates the length of amino acids. Gray boxes indicate tandem repeat domains
Discussion
In the present study, we confirmed that the chicken PRNP gene has only an insertion/deletion polymorphism at the tandem repeat domain in 4 chicken breeds. Because numerous case-control studies have found that SNPs of the PRNP gene play a pivotal role in prion pathogenesis, the absence of SNPs in the chicken PRNP gene is noteworthy. In addition, we identified significant differences in the genetic distribution of tandem repeat polymorphisms among Korean indigenous chickens (Korean native chicken and Ogolgye) and nonindigenous chickens (Dekalb White and Ross). According to previous studies, the tandem repeat domain is a direct binding site of metal ions, especially copper [10, 39,40,41]. Because the tandem repeat domain is a functional domain of the prion protein, further analysis of the highly polymorphic state of the chicken PRNP gene is needed in the future. Indeed, in a recent study, prion protein was expressed in lung epithelial cells and protected apoptosis induced by influenza A viruses. The lung in the prion protein knockout mouse model showed severe damage, higher levels of reactive oxygen species and cleaved caspase-3. In addition, animals with transgene for prion protein to a tandem repeat deleted model were also highly susceptible to influenza A virus infection. The results indicate that prion protein is also important in the susceptibility of influenza A viruses [42]. A previous study reported that chicken prion mRNA was overexpressed in Marek’s disease-infected chicken embryo fibroblasts and that knockdown of chicken prion protein in Marek’s disease virus-infected avian T cells reduced cell viability. This finding implied that Marek’s disease-induced tumors can be inhibited by reduced expression levels of the prion protein [43]. In a recent study, because the number of tandem repeat domains has been directly connected with the functional capacity of the prion protein, our findings in this study can also be related to the malignancy of Marek’s disease-induced tumor [43]. Thus, according to the genotype distribution of the functional domain of the chicken PRNP gene found in this study, investigation of the susceptibility to influenza A and the malignancy of Marek’s viruses is highly desirable in the future.
In addition, we confirmed the absence of SNPs in the ORF of the chicken PRNP gene in a large sampling of various chicken breeds. Notably, although the origins of the evaluated chickens are geographically divided (Korean native chicken: Korea; Ogolgye: China; Ross: Scotland; and Dekalb White: U.S.A), all chicken breeds showed common genetic characteristics of the PRNP gene. Since chicken showed high rate of SNPs in other genes except for PRNP gene, the absence of SNPs of the chicken PRNP gene is remarkable [44, 45]. In addition, natural prion disease-infected cases have not been reported in chickens, and parenteral and oral challenge of prion agent showed the failure of infection [46]. In other words, unlike other prion disease-resistant animals, including horses and dogs, chickens demonstrate perfect resistance to prion disease thus far [26]. Thus, the genetic characteristic of the absence of SNPs in the chicken PRNP gene is a notable features of prion disease-resistant animals. A good approach to understanding the pathogenesis of prion disease may be through the analysis of chicken-specific amino acids in interspecies-conserved domains of prion proteins. Although insertion/deletion polymorphism in the octapeptide repeat domain of bovine prion protein has been reported in cattle, this polymorphism has never been associated with the susceptibility or resistance of prion disease [22,23,24,25, 47,48,49]. However, the variations of octapeptide repeat domain of human prion protein confer the susceptibility to prion disease in human [6, 31]. Amino acid sequences in hexapeptide repeat domain of chicken prion protein showed low homology with those of prion protein in the mammals. In addition, the length and composition of hexapeptide repeat domain of chicken prion protein also showed significant differences in comparison of those of mammalian prion protein [30]. Thus, further study is needed to investigate the association between prion disease and hexapeptide repeat polymorphism of chicken prion protein in the future.
Conclusion
In conclusion, the present study surveyed chicken PRNP polymorphisms in large samples of 4 chicken breeds, Dekalb white, Ross, Ogolgye and Korean native chickens. We found only tandem repeat deletion polymorphisms and compared the genotype and allele distribution of tandem repeat polymorphisms among 4 chicken breeds. We confirmed that chickens have significantly different genotypes and allele distributions of tandem repeat polymorphisms among 4 chicken breeds. Finally, we found significant differences in the number of PRNP SNPs between prion disease-susceptible species and prion disease-resistant species. To the best of our knowledge, no SNP has been identified in the chicken PRNP gene thus far.
Methods
Genetic analysis
Genomic DNA was purified from 20 mg of brain tissue sample using a Hiyield genomic DNA mini kit (Real Biotech Corporation, Taiwan). Polymerase chain reaction (PCR) was performed with chicken PRNP gene-specific primers: forward primer: 5′-TGGGATGATGCTTGATTTCGGT-3′ and reverse primer: 5′ ATCCCTGTCACGCTCCAGAA-3′. These primers were designed based on the chicken PRNP gene sequence from GenBank (Gene ID: 396452) and amplified the entire ORF of the chicken PRNP gene. The length of the PCR products was 978 bp. A 25 μl reaction mixture containing 2.5 μl of 10 X Taq DNA polymerase buffer, 1 μl of genomic DNA, 10 pmol each primer, 0.5 μl of 0.2 μM dNTP mixture, 0.2 μl of 0.04 units of Taq DNA polymerase and sterile deionized water to a total volume of 25 μl. The PCR conditions were as follows: denaturing at 95 °C for 2 min, followed by 34 cycles of 95 °C for 20 s, 65 °C for 30 s, and 72 °C for 1 min 30 s and 1 cycle of 72 °C for 5 min. Purified PCR products were directly sequenced using an ABI 3730 sequencer (ABI, Foster City, California, USA), and sequencing electropherograms were analyzed using Finch TV software (Geospiza Inc., Seattle, USA).
Statistical analysis
Genotype, allele and haplotype frequencies of the chicken PRNP gene was compared among 4 chicken breeds by chi-square test using SAS 9.4 Software (SAS Institute Inc., Cary, NC, USA). The Hardy-Weinberg Equilibrium (HWE) test and haplotype analysis were performed using Haploview version 4.2 (Broad Institute, Cambridge, MA, USA).
Literature search
A literature search was conducted to search for SNPs of prion protein in humans, sheep, goats, cattle, horses and chickens using PubMed. The search terms were “prion”, “SNP” combined with “human,” “sheep,” “goats,” “cattle,” “horse” or “chickens”. Moreover, we supplemented our search by screening the reference lists of the relevant studies, including original articles and reviews. References for all identified publications are indicated in Table 4.
Availability of data and materials
All data generated or analysed during this study are included in supplementary information file and available from the corresponding author on reasonable request.
Abbreviations
- BSE:
-
Bovine spongiform encephalopathy
- CWD:
-
Chronic wasting disease
- HWE:
-
Hardy-Weinberg Equilibrium
- ORF:
-
Open reading frame
- PCR:
-
Polymerase chain reaction
- PRNP :
-
Prion protein gene
- PrPC :
-
Normal prion protein
- PrPSc :
-
Misfolded prion protein
- SNPs:
-
Single nucleotide polymorphisms
- TME:
-
Transmissible mink encephalopathy
References
Kim YC, Jeong BH. The first report of prion-related protein gene (PRNT) polymorphisms in goat. Acta Vet Hung. 2017;65:291–300.
Jeong MJ, Kim YC, Jeong BH. Prion-like protein gene (PRND) polymorphisms associated with scrapie susceptibility in Korean native black goats. PLoS One. 2018;13:e0206209.
Kim YC, Jeong BH. Bovine spongiform encephalopathy (BSE) associated polymorphisms of the prion-like protein gene (PRND) in Korean dairy cattle and Hanwoo. J Dairy Res. 2018;85:7–11.
Kim YC, Jeong BH. First report of prion-related protein gene (PRNT) polymorphisms in cattle. Vet Rec. 2018;182:717.
Kim YC, Jeong BH. The first report of polymorphisms and genetic characteristics of the prion protein gene (PRNP) in horses. Prion. 2018;12:245–52.
Jeong BH, Kim YS. Genetic studies in human prion diseases. J Korean Med Sci. 2014;29:623–32.
Hunter N, Goldmann W, Foster JD, Cairns D, Smith G. Natural scrapie and PrP genotype: case-control studies in British sheep. Vet Rec. 1997;141:137–40.
Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–83.
Prusiner SB. The prion diseases. Brain Pathol. 1998;8:499–513.
Di Natale G, Pappalardo G, Milardi D, Sciacca MF, Attanasio F, La Mendola D, Rizzarelli E. Membrane interactions and conformational preferences of human and avian prion N-terminal tandem repeats: the role of copper (II) ions, pH, and membrane mimicking environments. J Phys Chem B. 2010;114:13830–8.
Stanczak P, Luczkowski M, Juszczyk P, Grzonka Z, Kozlowski H. Interactions of Cu2+ ions with chicken prion tandem repeats. Dalton Trans. 2004;21:2102–7.
Wulf MA, Senatore A, Aguzzi A. The biological function of the cellular prion protein: an update. BMC Biol. 2017;15:34.
Jeong BH, Lee KH, Kim NH, Jin JK, Kim JI, Carp RI, Kim YS. Association of sporadic Creutzfeldt-Jakob disease with homozygous genotypes at PRNP codons 129 and 219 in the Korean population. Neurogenetics. 2005;6:229–32.
Brandel JP, Preece M, Brown P, Croes E, Laplanche JL, Agid Y, Will R, Alperovitch A. Distribution of codon 129 genotype in human growth hormone-treated CJD patients in France and the UK. Lancet. 2003;362:128–30.
Groschup MH, Lacroux C, Buschmann A, Luhken G, Mathey J, Eiden M, Lugan S, Hoffmann C, Espinosa JC, Baron T, Torres JM, Erhardt G, Andreoletti O. Classic scrapie in sheep with the ARR/ARR prion genotype in Germany and France. Emerg Infect Dis. 2007;13:1201–7.
Laplanche JL, Chatelain J, Westaway D, Thomas S, Dussaucy M, Brugere-Picoux J, Launay JM. PrP polymorphisms associated with natural scrapie discovered by denaturing gradient gel electrophoresis. Genomics. 1993;15:30–7.
Bouzalas IG, Dovas CI, Banos G, Papanastasopoulou M, Kritas S, Oevermann A, Papakostaki D, Evangelia C, Papadopoulos O, Seuberlich T, Koptopoulos G. Caprine PRNP polymorphisms at codons 171, 211, 222 and 240 in a Greek herd and their association with classical scrapie. J Gen Virol. 2010;91:1629–34.
Colussi S, Vaccari G, Maurella C, Bona C, Lorenzetti R, Troiano P, Casalinuovo F, Di Sarno A, Maniaci MG, Zuccon F, Nonno R, Casalone C, Mazza M, Ru G, Caramelli M, Agrimi U, Acutis PL. Histidine at codon 154 of the prion protein gene is a risk factor for Nor98 scrapie in goats. J Gen Virol. 2008;89:3173–6.
Goldmann W, Martin T, Foster J, Hughes S, Smith G, Hughes K, Dawson M, Hunter N. Novel polymorphisms in the caprine PrP gene: a codon 142 mutation associated with scrapie incubation period. J Gen Virol. 1996;77:2885–91.
Ortiz-Pelaez A, Georgiadou S, Simmons MM, Windl O, Dawson M, Arnold ME, Neocleous P, Papasavva-Stylianou P. Allelic variants at codon 146 in the PRNP gene show significant differences in the risk for natural scrapie in Cypriot goats. Epidemiol Infect. 2015;143:1304–10.
Ricci A, Bolton D, Chemaly M, Davies R. Genetic resistance to transmissible spongiform encephalopathies (TSE) in goats. EFSA J. 2017;15:4962.
Choi S, Woo HJ, Lee J. Sequence variations of the bovine prion protein gene (PRNP) in native Korean Hanwoo cattle. J Vet Sci. 2012;13:127–37.
Jeong BH, Sohn HJ, Lee JO, Kim NH, Kim JI, Lee SY, Cho IS, Joo YS, Carp RI, Kim YS. Polymorphisms of the prion protein gene (PRNP) in Hanwoo (Bos taurus coreanae) and Holstein cattle. Genes Genet Syst. 2005;80:303–8.
Msalya G, Shimogiri T, Okamoto S, Kawabe K, Minezawa M, Namikawa T, Maeda Y. Gene and haplotype polymorphisms of the prion gene (PRNP) in Japanese Brown, Japanese native and Holstein cattle. Anim Sci J. 2009;80:520–7.
Zhao H, Wang XY, Zou W, Zhang YP. Prion protein gene (PRNP) polymorphisms in native Chinese cattle. Genome. 2010;53:138–45.
Won SY, Kim YC, Kim K, Kim AD, Jeong BH. The first report of polymorphisms and genetic features of the prion-like protein gene (PRND) in a prion disease-resistant animal, Dog. Int J Mol Sci. 2019;20:E1404.
Bian J, Khaychuk V, Angers RC, Fernandez-Borges N, Vidal E, Meyerett-Reid C, Kim S, Calvi CL, Bartz JC, Hoover EA, Agrimi U, Richt JA, Castilla J, Telling GC. Prion replication without host adaptation during interspecies transmissions. Proc Natl Acad Sci U S A. 2017;114:1141–6.
Sanchez-Garcia J, Fernandez-Funez P. D159 and S167 are protective residues in the prion protein from dog and horse, two prion-resistant animals. Neurobiol Dis. 2018;119:1–12.
Zhang J. The structural stability of wild-type horse prion protein. J Biomol Struct Dyn. 2011;29:369–77.
Kim YC, Jeong MJ, Jeong BH. The first report of genetic variations in the chicken prion protein gene. Prion. 2018;12(3–4):197–203.
Lloyd S, Mead S, Collinge J. Genetics of prion disease. Top Curr Chem. 2011;305:1–22.
Jeong BH, Nam JH, Lee YJ, Lee KH, Jang MK, Carp RI, Lee HD, Ju YR, Ahn Jo S, Park KY, Kim YS. Polymorphisms of the prion protein gene (PRNP) in a Korean population. J Hum Genet. 2004;49:319–24.
Guan F, Pan L, Li J, Tang H, Zhu C, Shi G. Polymorphisms of the prion protein gene and their effects on litter size and risk evaluation for scrapie in Chinese Hu sheepm. Virus Genes. 2011;43:147–52.
Frootan F, Nikbakht G, Ozgenturk NO, Un C. Prion protein coding gene (PRNP) variability in sheep from Turkey and Iran. Biochem Genet. 2012;50:277–84.
Seabury CM, Derr JN. Identification of a novel ovine PrP polymorphism and scrapie-resistant genotypes for St. Croix White and a related composite breed, Cytogenet Genome Res. 2004;102:85–8.
Ekateriniadou LV, Panagiotidis CH, Terzis A, Ploumi K, Triantafyllidis A, Deligiannidis P, Triantaphyllidis C, Sklaviadis T. Genotyping for PrP gene polymorphisms in rare Greek breeds of sheep. Vet Rec. 2007;160:194–5.
Vaccari G, Panagiotidis CH, Acin C, Peletto S, Barillet F, Acutis P, Bossers A, Langeveld J, van Keulen L, Sklaviadis T, Badiola JJ, Andreeoletti O, Groschup MH, Agrimi U, Foster J, Goldmann W. State-of-the-art review of goat TSE in the European Union, with special emphasis on PRNP genetics and epidemiology. Vet Res. 2009;40:48.
Kim SK, Kim YC, Won SY, Jeong BH. Potential scrapie-associated polymorphisms of the prion protein gene (PRNP) in Korean native black goats. Sci Rep. 2019;9:15293.
Brown DR, Qin K, Herms JW, Madlung A, Manson J, Strome R, Fraser PE, Kruck T, von Bohlen A, Schulz-Schaeffer W, Giese A, Westaway D, Kretzschmar H. The cellular prion protein binds copper in vivo. Nature. 1997;390:684–7.
Miura T, Hori-i A, Mototani H, Takeuchi H. Raman spectroscopic study on the copper (II) binding mode of prion octapeptide and its pH dependence. Biochemistry. 1999;38:11560–9.
Redecke L, Meyer-Klaucke W, Koker M, Clos J, Georgieva D, Genov N, Echner H, Kalbacher H, Perbandt M, Bredehorst R, Voelter W, Betzel C. Comparative analysis of the human and chicken prion protein copper binding regions at pH 6.5. J Biol Chem. 2005;280:13987–92.
Chida J, Hara H, Yano M, Uchiyama K, Das NR, Takahashi E, Miyata H, Tomioka Y, Ito T, Kido H, Sakaguchi S. Prion protein protects mice from lethal infection with influenza a viruses. PLoS Pathog. 2018;14:e1007049.
Wan X, Yang R, Liu G, Zhu M, Zhang T, Liu L, Wu R. Downregulation of cellular prion protein inhibited the proliferation and invasion and induced apoptosis of Marek’s disease virus-transformed avian T cells. J Vet Sci. 2016;17:171–8.
Kim YC, Jeong MJ, Jeong BH. Genetic characteristics and polymorphisms in the chicken interferon-induced transmembrane protein (IFITM3) gene. Vet Res Commun. 2019; https://doi.org/10.1007/s11259-019-09762-y.
Zhong C, Li S, Li J, Li F, Ran M, Qiu L, Li D, Zhu Q, Wang Y, Yin H, Shu G, Yang C, Zhao X. Polymorphisms in the Egl nine homolog 3 (EGLN3) and peroxisome proliferator activated receptor-alpha (PPARα) genes and their correlation with hypoxia adaptation in Tibetan chickens. PLoS One. 2018;13:e0194156.
Moore J, Hawkins SA, Austin AR, Konold T, Green RB, Blamire IW, Dexter I, Stack MJ, Chaplin MJ, Langeveld JP, Simmons MM, Spencer YI, Webb PR, Dawson M, Wells GA. Studies of the transmissibility of the agent of bovine spongiform encephalopathy to the domestic chicken. BMC Res Notes. 2011;4:501.
Collee JG, Bradley R. BSE: a decade on--Part 2. Lancet. 1997;349:715–21.
Collee JG, Bradley R. BSE: a decade on--Part I. Lancet. 1997;349:636–41.
Murdoch BM, Murdoch GK. Genetics of prion disease in cattle. Bioinform Biol Insights. 2015;9:1–10.
Acknowledgements
Not applicable.
Funding
This research was supported by the Basic Science Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2018R1D1A1B07048711) and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2017R1A6A1A03015876). S. Y. Won and Y.C. Kim were supported by the BK21 Plus Program in the Department of Bioactive Material Sciences. The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
YCK, SYW and BHJ conceived and designed the experiment. YCK and SYW performed the experiments. YCK and BHJ analyzed the data. YCK, SYW and BHJ wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Brain tissue samples from 298 chickens, including 3 chicken breeds (100 Ross, 100 Korean native chickens and 98 Ogolgye), were obtained from slaughter houses in the Republic of Korea. All experimental procedures were accredited by the Institute of Animal Care and Use Committee of Chonbuk National University (CBNU 2017–0030).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
Sanger sequencing data of chicken PRNP gene in Dekalb White, Ross, Ogolgye and Korean native chicken.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kim, YC., Won, SY. & Jeong, BH. Absence of single nucleotide polymorphisms (SNPs) in the open reading frame (ORF) of the prion protein gene (PRNP) in a large sampling of various chicken breeds. BMC Genomics 20, 922 (2019). https://doi.org/10.1186/s12864-019-6315-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-019-6315-8