Abstract
Background
Prion diseases, also known as transmissible spongiform encephalopathies (TSEs) remain one of the deleterious disorders, which have affected several animal species. Polymorphism of the prion protein (PRNP) gene majorly determines the susceptibility of animals to TSEs. However, only limited studies have examined the variation in PRNP gene in different Nigerian livestock species. Thus, this study aimed to identify the polymorphism of PRNP gene in Nigerian livestock species (including camel, dog, horse, goat, and sheep). We sequenced the open reading frame (ORF) of 65 camels, 31 village dogs and 12 horses from Nigeria and compared with PRNP sequences of 886 individuals retrieved from public databases.
Results
All the 994 individuals were assigned into 162 haplotypes. The sheep had the highest number of haplotypes (n = 54), and the camel had the lowest (n = 7). Phylogenetic tree further confirmed clustering of Nigerian individuals into their various species. We detected five non-synonymous SNPs of PRNP comprising of G9A, G10A, C11G, G12C, and T669C shared by all Nigerian livestock species and were in Hardy-Weinberg Equilibrium (HWE). The amino acid changes in these five non-synonymous SNP were all “benign” via Polyphen-2 program. Three SNPs G34C, T699C, and C738G occurred only in Nigerian dogs while C16G, G502A, G503A, and C681A in Nigerian horse. In addition, C50T was detected only in goats and sheep.
Conclusion
Our study serves as the first to simultaneously investigate the polymorphism of PRNP gene in Nigerian livestock species and provides relevant information that could be adopted in programs targeted at breeding for prion diseases resistance.
Similar content being viewed by others
Background
Prion diseases also known as Transmissible spongiform encephalopathies (TSEs) remain one of the deleterious disorders [1], which have affected several animal species [2, 3]. The unique characterization of Prion diseases is the accumulation of an “infectious” abnormal protease-resistant isoform (PrPSc) of cellular prion proteins (PrPC) encrypted by the prion protein (PRNP) gene [4]. The prion gene family consist of four members namely prion protein gene (PRNP), the prion-like protein gene (PRND), the shadow of the prion protein gene (SPRN), and the prion-related gene (PRNT) [5]. Although only Shadoo (Sho) protein is enclosed by the SPRN gene, and its structure is similar to the PrP protein. The PRND is nearly situated in 20 kb downstream of the PRNP gene, and PRND possess a similar structure with PrP [6]. Prion protein genes are highly maintained among mammals [7] and predominantly synthesized in cells of the central nervous system [8]. Although, it is also expressed in different peripheral tissues [9, 10]. Interestingly, in the central nervous system and lymphoid tissues, TSE diseases encompass a neuronal glycoprotein (i.e. Prion protein) PrPC (encoded by PRNP gene), which is regenerated into an abnormal protease-resistant protein [11, 12]. Prion diseases are grouped as sporadic, familial, and infectious forms and contains two exons with second one carrying the whole open reading frame (ORF) in humans [13]. It was reported that about 85% of prion diseases in humans are Creutzfeldt–Jakob disease (CJD) while 15% of the prion diseases include familial CJD, Gerstmann–Straussler–Scheinker syndrome (GSS), and fatal familial insomnia (FFI) [14,15,16]. Another study has reported the nature of the infectious agents- PrP models of resistant species including dog, rabbit and horses to prion diseases [17]. The β2-α2 loop contributes to their protein structural stabilities while salt bridge contributed to structural stability of horse prion protein [18].
Prion diseases are the sole human neurodegenerative disorders with true associates with mammals thereby enabling rodent suitable models to comprehend the mechanisms of disease transmission and pathogenesis. Scrapie is a detrimental neurodegenerative prion malady and has spread across almost all regions worldwide [19, 20] leading to spongiform brain pathology, brain deposition of misfolded among others. Known for over 250 years, scrapie is one of the TSE and encompasses zoonotic bovine spongiform encephalopathy (BSE) in cattle and Creutzfeldt–Jakob disease (CJD) in humans, which are regulated by the prion protein-encoding gene (PRNP) [21,22,23]. It has been reported that the resistance to scrapie is intently regulated by SNPs of the PRNP gene and controlled by the prion disease agent [7, 24], and the distribution of SNPs at the ORF of PRNP gene in various species was presented [25]. In 1986, classical BSE was first reported in United Kingdom (UK) and has spread through PrPSc affected meat and bone meal. However, different surveillance approaches have been adopted to prevent utilization of contaminated feed and this has drastically reduced the number of classical BSE cases [26]. It was reported that the insertion of G allele at codon 46 of SPRN gene in humans with variant CJD causes a frameshift of this gene and it displays a significant disparity in its distribution between healthy controls and vCJD patients [27]. Moreover, it was reported that somatic mutation of humans’ PRNP was predicted to be one of the factors responsible for prion disease [16]. Also, scrapie could affect small ruminants including sheep and goats [28]. In addition, most forms of TSEs affect different mammalian species but display high dominance in ruminants such as scrapie in goats and sheep [29].
Our previous studies revealed that the SNP sites at codons 139 S, 146 S, 154 H, and 193I were presence in Nigerian goats [30] which have been reported to be susceptible to scrapie in goats [28, 31], and also codons 154 H and 171Q susceptible to classical scrapie in sheep [32, 33] detected in Nigerian sheep [34].
Camelus dromedarius are vastly found in the semiarid northern part of Nigeria and their estimates is about 289,794 heads [35]. They are basically reared for meat, milk, wool, source of transportation, beauty spectacle, and recreational activities [35]. Nigerian village dogs are one of the major sources of the transmission of infectious diseases [36], horses from Nigeria are useful for entertainment, polo games, ceremonies, research, riding etc. [37]. Nigerian sheep are reared in the drier agro-climatic zones of the country with an estimated population of 27 million [38]. There are four major breeds of Nigerian sheep: Yankasa, Uda, Balami, and West Africa Dwarf [39]. Nigerian goats are hardy, tolerant to trypanosomiasis, and adapt easily to the local ecosystem [40]. There are three main indigenous breeds of Nigerian goats: West African Dwarf (WAD), Sokoto Red and Sahelian [41,42,43].
Therefore, this study was designed to understand the PRNP gene sequence variation in different Nigerian livestock species and provide insight into their resistance to prion diseases. Herein, we combined the PRNP sequences of five Nigerian livestock species (camel, dog, horse, sheep, and goat) and analyzed the prion genes. In addition, we retrieved the nucleotide sequences of PRNPs from other mammalians for the SNP analyses.
Results
Haplotype analysis of the 994 sequences of PRNP gene
A total of 994 PRNP gene sequences were analyzed, including 108 de novo and 886 downloaded from GenBank. All the PRNP sequences were assigned into 162 haplotypes (Additional File 1). The sheep had the highest number of haplotypes (n = 54), and the camel had the lowest (n = 7).
Phylogenetic tree based on number of haplotypes
Based on the 162 haplotypes, 180 individuals including Nigerian species and those retrieved sequences were selected to construct the phylogenetic tree. Figure 1 showed the phylogenetic tree from the analysis of PRNP sequences of five Nigerian species together with reported PRNP sequences of Homo sapiens and Macaca mulatta as outgroups. The phylogenetic tree further confirmed clustering of Nigerian individuals into their various species.
Single nucleotide polymorphism (SNPs) of PRNP gene in the five Nigerian species
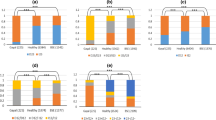
We detected five non-synonymous SNPs of PRNP namely G9A, G10A, C11G, G12C, and T669C in all Nigerian species considered when combined with nucleotide sequences retrieved from public database (Table 1). Further, we determined the genotype and allele frequencies of the five non-synonymous SNPs detected in Nigerian livestock species and were in Hardy-Weinberg Equilibrium (HWE) (Table 2). Based G34C, T699C, and C738G occurred only in PRNP of Nigerian dog while C16G, G502A, G503A, and C681A were identified in Nigerian horse only. In addition, C50T was detected in goat and sheep only. Table 1 shows the non-synonymous SNPs detected in Nigerian livestock species. All SNPs identified in Nigerian livestock species are listed in Additional Table 1.
Assessment of the effects of the non-synonymous SNPs
PolyPhen-2 is an online tool used to predict the possible impact of an amino acid replacement caused by nonsynonymous SNPs on the structure and function of proteins [44]. Based on the polymorphism results, effects of the five non-synonymous SNPs common in the five Nigerian livestock species considered were assessed via PolyPhen-2. It was predicted that the amino acid substitution in the five non-synonymous SNPs was benign (Table 3).
Discussion
The polymorphism of the PRNP gene plays a great role in the susceptibility of animals to prion protein diseases. In horses, the stability of prion protein is associated with disease progression.
Previous studies have identified single nucleotide mutations at codons 136 (A > V), 154 (R > H), and 171 (R > Q/H) of PRNP gene [45, 46]. Interestingly, the variation of amino acids at codons 141 and 154 were reported to be related to various forms of classical scrapie by altering the configuration of prion protein [47, 48]. In addition, the changes in amino acids A to V and Q to R at codons 136 and 171, respectively were reported to increase resistance to scrapie in sheep [20, 45].
The susceptibility of small ruminants (i.e. goat and sheep) to scrapie are affected by the genetic variation of the PRNP gene. Goats and sheep share about 99% protein sequence homology for their prion proteins. Although, the fragments of their amino acids associated with scrapie vulnerability are not similar [5, 21]. PRNP genes are highly polymorphic in goats [49,50,51,52], and the distributions of genotype and haplotype frequencies at codons 139, 146, and 154 were highly associated with vulnerability to scrapie in goats [50, 52].
We combined the PRNP sequences of five Nigerian livestock species (camel, dog, goat, horse, and sheep) and retrieved sequences of human, monkey, camel, dog, goat, sheep, goat, mule deer, Rocky Mountain elk and fallow deer from available databases. We detected 162 haplotypes using the 994 sequences (Additional File 3). Based on the phylogenetic tree when Homo sapiens and Macaca mulatta are outgroups shows the clustering of Nigerian individuals into their various species (Fig. 2). The sheep had the highest number of haplotypes (n = 54), and the camel had the lowest (n = 7). We assumed that the Nigerian sheep might be more susceptible to prion related disease than the other four Nigerian livestock species (goat, dog, horse, and camel).
Further, based on the SNP analysis, we detected five non-synonymous SNPs of PRNP namely G9A, G10A, C11G, G12C, and T669C in all Nigerian species considered as shown in Table 1. The result shows that the five Nigerian livestock species might be susceptible to prion related diseases. These SNP sites are unique to the Nigerian livestock species considered in this study. Contrarily, previous studies on polymorphism of PRNP gene in Nigerian small ruminants, 29 SNPs (14 non-synonymous and 23 novel SNPs) and 19 SNPs (14 non-synonymous SNPs with T718C as a novel SNP) were revealed in Nigerian goats and sheep, respectively [30, 34]. Recent studies have reported low variation in dromedary PRNP gene in Egypt and Iran [53, 54]. Two non-synonymous SNPs (G205A and G401A) were identified in PRNP gene in Algerian dromedary [55] but not detected in the present study for Nigerian camel. It has been reported that dog are resistance to prion infection due to change of asparagine at codon 163 [56]. In a previous study, the substitutions of amino acids at canine shadow of prion protein (Sho) were all neutral except 70_71DelAA that was deleterious [57]. Previous study on prion protein gene identified only one non-synonymous SNP at c.525 A (N175K) in Thoroughbred horse [58]. Based on PolyPhen-2, it was predicted that the amino acid substitution in the five non-synonymous SNPs common to all the Nigerian livestock species was benign (tolerant).
Conclusion
This preliminary study aims to examine the single nucleotide polymorphism (SNP) in the open frame region (ORF) of PRNP in Nigerian livestock species. Based on our results, we detected five non-synonymous SNPs of PRNP namely G9A, G10A, C11G, G12C, and T669C in all Nigerian species. We assumed that Nigerian livestock species might be susceptible to prion related diseases based on these codons identified in our current study. Our preliminary study provides baseline information on prion gene polymorphism in Nigerian livestock species and subsequent studies will examine the functional relationship between clinical signals with prion SNPs from our genomic studies in connection with genotypes of prion protein. In addition, future studies will incorporate large sample size, utilize different coat colors, detect the prevalence of pion protein disease, and functional analyses in PRNP gene in Nigerian animals.
Materials and methods
Samplings and DNA extraction
We collected about 10 ml of blood samples from 65 camels (25 males and 40 females) from four states in Nigeria including: Kaduna (n = 19 males; n = 26 females), Sokoto (n = 5 males; n = 5 females), Kebbi (n = 1 male; n = 7 females), and Katsina (n = 2 females), 31 village dogs from Oyo (n = 10 males; n = 8 females) and Taraba (n = 6 males; n = 7 females) and 12 horses from Oyo (n = 5) and Taraba (n = 7) states (Fig. 2, Additional File 2). During sample collection, we avoided individuals from clustered populations. The whole blood samples were stored at -20 ◦C prior to DNA extraction. Genomic DNA was extracted at Kunming Institute of Zoology, Chinese Academy of Sciences (CAS), using the phenol-chloroform method [59]. We quantified the genomic DNA using the Thermo Scientific™ NanoDrop 2000 spectrophotometer to evaluate its purity. In addition, to check for molecular quality, we ran gel electrophoresis of the genomic DNA using a 2% agarose gel against a 2 Kilobase (kb) DNA ladder marker. In addition, we retrieved nucleotide sequences of PRNP of 126 sheep [34] and 132 goats [30] from Nigeria, human, monkey, camel, dog, sheep, goat and horse individuals from database (Additional File 2).
Polymerase chain reaction (PCR) and DNA sequencing
We amplified the base pairs of the PRNP gene in the animals to reveal its variable sites. Primers from each animal were designed based on the nucleotide sequence of the PRNP gene retrieved from the NCBI website (Additional Table 2). The 25 µl PCR mixture and sequencing reactions contained 1 µl of genomic DNA,10 pmol of each primer, 2.5mM dNTPs and 5 units of Takara Taq DNA polymerase in a 10 pmol reaction buffer containing 1.5 mM MgCl2.
The PCR was carried out in a thermocycler (detailed PCR reactions of each species is presented in Additional File 3. PCR products were purified for sequencing analysis with a QIAquick Gel Extraction Kit (Qiagen, Valencia, California, USA). The PCR products were bidirectionally sequenced using an ABI 3730XL sequencer (Applied Biosystems, Foster City, California, USA).
Sequences alignment, haplotype, phylogenetic and statistical analyses
The sequences were aligned with MEGA (v.11.0.8) [60]. Nucleotide and amino acid alignments were produced using ClustalW and adjusted manually. We computed genetic distances using MEGA (v.11.0.8). The number of haplotypes in the 994 sequences was analyzed using DnaSP 6 [61]. Further, we determined the distance matrices under the assumptions of Kimura’s two-parameter model and were adopted to infer dendrograms by the neighbor-joining method [62]. The confidence values for individual branches of the resulting tree were determined by bootstrap analysis with 1000 replicates [63]. The allelic and genotypic frequencies of the non-synonymous SNPs common to the five Nigerian species were tested by chi-square test (χ2) or Fisher’s exact test using SPSS version 21.0 (IBM Corp., Armonk, NY).
Assessment of the effects of the non-synonymous SNPs
The effects of the three (3) nonsynonymous SNPs of PRNP gene common to the five Nigerian livestock species were evaluated using PolyPhen- 2 (https://genetics.bwh.harvard.edu/pph2/).
Data availability
The nucleotide sequences are available on NCBI with accession numbers: MZ463488 - MZ463499 for horse, MZ463325 - MZ463355 for dog, and OK041226 - OK041290 for camel.
Abbreviations
- PRNP :
-
Prion protein
- TSEs:
-
Transmissible spongiform encephalopathies
- PrPSc:
-
Protease-resistant isoform
- PrPC:
-
Cellular prion proteins
- SPRN :
-
Shadow of the prion protein gene
- PRNT :
-
The prion-related gene
- CJD:
-
Creutzfeldt–Jakob disease
- SNPs:
-
Single Nucleotide Polymorphism
- WAD:
-
West African Dwarf
- ORF:
-
Open reading frame
References
Kim Y-C, Won S-Y, Jeong B-H. Altered expression of glymphatic system-related proteins in prion diseases: implications for the role of the glymphatic system in prion diseases. Cell Mol Immunol. 2021;18:2281–3.
Wan J, Bai X, Liu W, Xu J, Xu M, Gao H. Polymorphism of prion protein gene in Arctic fox (Vulpes lagopus). Mol Biol Rep. 2009;36:1299–303.
Kim YC, Kim SK, Jeong BH. Scrapie susceptibility-associated indel polymorphism of shadow of prion protein gene (SPRN) in Korean native black goats. Sci Rep. 2019;9.
Prusiner SB. Molecular biology of prion diseases. Science (80-). 1991;252.
Kim YC, Jeong BH. The first report of prion-related protein gene (PRNT) polymorphisms in goat. Acta Vet Hung. 2017;65.
Jeong MJ, Jeong BH. NO polymorphisms in the coding region of the prion-like protein gene in Thoroughbred racehorses. Acta Vet Hung. 2019;67:174–82.
Myers R, Cembran A, Fernandez-Funez P. Insight from animals resistant to Prion diseases: deciphering the genotype– morphotype– phenotype code for the prion protein. Front Cell Neurosci. 2020;14:1–15.
Wulf MA, Senatore A, Aguzzi A. The biological function of the cellular prion protein: an update. BMC Biol. 2017;15:1–13.
Salvesen Ø, Espenes A, Reiten MR, Vuong TT, Malachin G, Tran L, et al. Goats naturally devoid of PrPC are resistant to scrapie. Vet Res. 2020;51:1–14.
Douet JY, Huor A, Cassard H, Lugan S, Aron N, Arnold M, et al. Wide distribution of prion infectivity in the peripheral tissues of vCJD and sCJD patients. Acta Neuropathol. 2021;141:383–97.
Tranulis MA, Gavier-Widén D, Våge J, Nöremark M, Korpenfelt SL, Hautaniemi M, et al. Chronic wasting disease in Europe: new strains on the horizon. Acta Vet Scand. 2021;63:1–15.
Pérez DR, Damberger FF, Wüthrich K. Horse prion protein NMR structure and comparisons with related variants of the mouse prion protein. J Mol Biol. 2010;400:121–8.
Imran M, Mahmood S. An overview of human prion diseases. Virol J. 2011;8.
Kovács GG, Puopolo M, Ladogana A, Pocchiari M, Budka H, van Duijn C et al. Genetic prion disease: the EUROCJD experience. Hum Genet. 2005;118.
Lloyd SE, Mead S, Collinge J. Genetics of prion diseases. Curr Opin Genet Dev. 2013;23.
Won SY, Kim YC, Jeong BH. First report of the potential bovine spongiform encephalopathy (Bse)-related somatic mutation e211k of the prion protein gene (prnp) in cattle. Int J Mol Sci. 2020;21.
Zhang J. The nature of the infectious agents: Prp models of resistant species to prion diseases (dog, rabbit and horses). In: Prions and Prion Diseases: New Developments. 2012.
Zhang J. The structural stability of wild-type horse prion protein. J Biomol Struct Dyn. 2011;29.
Detwiler LA, Baylis M. The epidemiology of scrapie. OIE Revue Scientifique et Technique. 2003;22.
Acín C, Bolea R, Monzón M, Monleón E, Moreno B, Filali H, et al. Classical and atypical scrapie in sheep and goats: review on the etiology, genetic factors, pathogenesis, diagnosis, and control measures of both diseases. Animals. 2021;11:1–20.
Baylis M, Goldmann W. The Genetics of Scrapie in Sheep and Goats. Curr Mol Med. 2005;4.
Agrimi U, Conte M, Morelli L, Di Bari MA, Di Guardo G, Ligios C et al. Animal transmissible spongiform encephalopathies and genetics. Vet Res Commun. 2003;27 SUPPL. 1.
Lee J, Kim SY, Hwang KJ, Ju YR, Woo HJ. Prion diseases as Transmissible Zoonotic diseases. Osong Public Health Res Perspect. 2013;4.
Gelasakis AI, Boukouvala E, Babetsa M, Katharopoulos E, Palaska V, Papakostaki D et al. Polymorphisms of codons 110, 146, 211 and 222 at the goat prnp locus and their association with scrapie in Greece. Animals. 2021;11.
Kim YC, Won SY, Jeong BH. Absence of single nucleotide polymorphisms (SNPs) in the open reading frame (ORF) of the prion protein gene (PRNP) in a large sampling of various chicken breeds. BMC Genomics. 2019;20:1–7.
Dudas S, Czub S, Atypical BSE. Current knowledge and knowledge gaps. Food Saf. 2017;5.
Beck JA, Campbell TA, Adamson G, Poulter M, Uphill JB, Molou E et al. Association of a null allele of SPRN with variant Creutzfeldt-Jakob disease. J Med Genet. 2008;45.
Fragkiadaki EG, Vaccari G, Ekateriniadou LV, Agrimi U, Giadinis ND, Chiappini B, et al. PRNP genetic variability and molecular typing of natural goat scrapie isolates in a high number of infected flocks. Vet Res. 2011;42:2–7.
Pitarch JL, Raksa HC, Arnal MC, Revilla M, Martínez D, De Fernández D et al. Low sequence diversity of the prion protein gene (PRNP) in wild deer and goat species from Spain. Vet Res. 2018;49.
Adeola AC, Bello SF, Abdussamad AM, Mark AI, Sanke OJ, Onoja AB, et al. Scrapie-associated polymorphisms of the prion protein gene (PRNP) in Nigerian native goats. Gene. 2022;855:147121.
Vaccari G, Di Bari MA, Morelli L, Nonno R, Chiappini B, Antonucci G et al. Identification of an allelic variant of the goat PrP gene associated with resistance to scrapie. J Gen Virol. 2006;87.
Clouscard C, Beaudry P, Elsen JM, Milan ~ D, Dussaucy ~ M, Bounneau C et al. Different allelic effects of the codons 136 and 171 of the prion protein gene in sheep with natural scrapie. 1995.
Baylis M, Goldmann W, Houston F, Cairns D, Chong A, Ross A et al. Scrapie epidemic in a fully PrP-genotyped sheep flock. J Gen Virol. 2002;83.
Adeola AC, Bello SF, Abdussamad AM, Mark AI, Sanke OJ, Onoja AB, et al. Polymorphism of prion protein gene (PRNP) in Nigerian sheep. Prion. 2023;17:44–54.
Mohammed I, Hoffmann I. Management of draught camels (Camelus dromedarius) in crop-livestock production systems in Northwest Nigeria. Livest Res Rural Dev. 2006;18.
Eke C, Omotowo B, Ukoha M, Ibe B. Human rabies: still a neglected preventable disease in Nigeria. Niger J Clin Pract. 2015;18:268–72.
Ehizibolo DO, Gusi AM, Ehizibolo PO, Mbuk EU, Ocholi RA. Serologic prevalence of brucellosis in horse stables in two northern states of Nigeria. J Equine Sci. 2011;22.
Lombin LH. Report on the internet. 2007. http://www.africanagricultureblog.com/2007/12/nigeria-has-16-million-cattle.html 2007.
Yunusa AJ, Salako AE, Oladejo OA. Morphometric characterization of Nigerian indigenous sheep using multifactorial discriminant analysis. 2013;5 October:661–5.
Serranito B, Taurisson-Mouret D, Harkat S, Laoun A, Ouchene-Khelifi NA, Pompanon F, et al. Search for selection signatures related to Trypanosomosis Tolerance in African goats. Front Genet. 2021;12:1–15.
Wheto M, Ilori BM, Sanda AJ, Adeleke MA, Durosaro SO, Adenaike AS et al. Morphological characterization and evaluation of heat tolerance traits in Nigerian goat breeds. Niger J Anim Prod. 2021;42.
Yakubu A, Salako AE, Imumorin IG. Comparative multivariate analysis of biometric traits of west African dwarf and Red Sokoto goats. Trop Anim Health Prod. 2011;43.
Murital I, Afolayan O, Bemji MN, Dadi O, Landi V, Martínez A et al. Genetic diversity and population structure of Nigerian indigenous goat using DNA microsatellite markers. Arch Zootec. 2015;64.
Goldmann W, Hunter N, Smith G, Foster J, Hope J. PrP genotype and agent effects in scrapie: change in allelic interaction with different isolates of agent in sheep, a natural host of scrapie. J Gen Virol. 1994;75.
Greenlee JJ, Review. Update on classical and atypical Scrapie in Sheep and Goats. Vet Pathol. 2019;56:6–16.
Cassmann ED, Greenlee JJ. Pathogenesis, detection, and control of scrapie in sheep. Am J Vet Res. 2020;81:600–14.
Yang S, Thackray AM, Hopkins L, Monie TP, Burke DF, Bujdoso R. Polymorphisms at amino acid residues 141 and 154 influence conformational variation in ovine PrP. Biomed Res Int. 2014;2014.
Andrade CP, Neto JDB, Driemeier D. Identification of single nucleotide polymorphisms in the prion protein gene in Santa Ines and Dorset sheep. Pesqui Vet Bras. 2018;38:624–8.
Öner Y, Yilmaz O, Eriş C, Ata N, Ünal C, Koncagül S. Genetic diversity and population structure of Turkish native cattle breeds. South Afr J Anim Sci. 2019. https://doi.org/10.4314/sajas.v49i4.4.
Fantazi K, Migliore S, Kdidi S, Racinaro L, Tefiel H, Boukhari R, et al. Analysis of differences in prion protein gene (PRNP) polymorphisms between Algerian and Southern Italy’s goats. Ital J Anim Sci. 2018;17:578–85.
Kim SK, Kim YC, Won SY, Jeong BH. Potential scrapie-associated polymorphisms of the prion protein gene (PRNP) in Korean native black goats. Sci Rep. 2019;9:1–10.
Billinis C, Psychas V, Leontides L, Spyrou V, Argyroudis S, Vlemmas I et al. Prion protein gene polymorphisms in healthy and scrapie-affected sheep in Greece. J Gen Virol. 2004;85.
Tahmoorespur M, Jelokhani Niaraki S. Analysis of sequence variations of prion protein gene in dromedary camels in Iran. J Appl Anim Res. 2014;42:238–43.
Abdel-Aziem SH, Abd El-Kader HAM, Alam SS, Abd El-Moneim OM, Othman OE. Nucleotide structure of prion protein gene in Egyptian camels. J Appl Anim Res. 2019;47:123–8.
Zoubeyda K, Imane M, Youcef C, Baaissa B, Suheil GSB, Michela C et al. Variability of the prion protein gene (PRNP) in Algerian dromedary populations. Anim Gene. 2020;17–8 February.
Vidal E, Fernández-Borges N, Eraña H, Parra B, Pintado B, Sánchez-Martín MA, et al. Dogs are resistant to prion infection, due to the presence of aspartic or glutamic acid at position 163 of their prion protein. FASEB J. 2020;34:3969–82.
Kim YC, Kim HH, Kim AD, Jeong BH. Novel insertion/deletion polymorphisms and genetic features of the shadow of prion protein gene (SPRN) in dogs, a prion-resistant animal. Front Vet Sci. 2022;9.
Kim YC, Jeong BH. The first report of polymorphisms and genetic characteristics of the prion protein gene (PRNP) in horses. Prion. 2018;12.
Sambrook J, Russell DW. Molecular cloning: a laboratory manual (3-volume set). Molecular cloning: a laboratory manual. 2001.
Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5.
Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017. https://doi.org/10.1093/molbev/msx248.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4.
Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17.
Acknowledgements
We recognize all who assisted in the success of this study.
Funding
This work was supported by the Sino-Africa Joint Research Center, Chinese Academy of Sciences (SAJC202103), and the Animal Branch of the Germplasm Bank of Wild Species, Chinese Academy of Sciences (the Large Research Infrastructure Funding). In addition, this work has been successful through the Chinese Academy of Sciences President’s International Fellowship Initiative (CAS-PIFI) who provided grant support to Adeola Charles Adeniyi (2021FYB0006).
Author information
Authors and Affiliations
Contributions
A.C.A., S.F.B., A.M.A., and R.A.M.A led the project, designed, and conceived the study. A.C.A., and S.F.B. performed data analysis, interpreted results, prepared, and developed the manuscript. A.C.A., and S.F.B carried out experiments. A.E.S., A.M.A., N.A., R.A.M.A., A.B.O., A.I.M., O.J.S., S.C.O., G.F.M., J.I., P.M.D., S.K., and M.H.Y. revised the manuscript. A.C.A., R.A.M.A., A.I.M., O.J.S., S.C.O., G.F.M., J.I., A.M.A., N.A., A.E.S., P.M.D., A.B.O., S.K., and M.H.Y. performed sampling. All authors contributed and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All experimental procedures in this present study were performed in accordance with Research Guidelines for the Institutional Review Board of Kunming Institute of Zoology, Chinese Academy of Sciences (SMKX-20160524-119) and current study is approved by the Institutional Review Board of Kunming Institute of Zoology, Chinese Academy of Sciences (SMKX-20160524-119). We have complied with ARRIVE at submission.
Consent for publication
Not Applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Adeola, A.C., Bello, S.F., Abdussamad, A.M. et al. Single nucleotide polymorphisms (SNPs) in the open reading frame (ORF) of prion protein gene (PRNP) in Nigerian livestock species. BMC Genomics 25, 177 (2024). https://doi.org/10.1186/s12864-024-10070-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-024-10070-2