Abstract
Background
Plant basic leucine zipper (bZIP) transcription factors play crucial roles in plant growth, development, and abiotic stress responses. However, systematic investigation and analyses of the bZIP gene family in peanut are lacking in spite of the availability of the peanut genome sequence.
Results
In this study, we identified 50 and 45 bZIP genes from Arachis duranensis and A. ipaensis genomes, respectively. Phylogenetic analysis showed that Arachis bZIP genes were classified into nine groups, and these clusters were supported by several group-specific features, including exon/intron structure, intron phases, MEME motifs, and predicted binding site structure. We also identified possible variations in DNA-binding-site specificity and dimerization properties among different Arachis bZIPs by inspecting the amino acid residues at some key sites. Our analysis of the evolutionary history analysis indicated that segmental duplication, rather than tandem duplication, contributed greatly to the expansion of this gene family, and that most Arachis bZIPs underwent strong purifying selection. Through RNA-seq and quantitative real-time PCR (qRT-PCR) analyses, the co-expressed, differentially expressed and several well-studied homologous bZIPs were identified during seed development stages in peanut. We also used qRT-PCR to explore changes in bZIP gene expression in response to salt-treatment, and many candidate bZIPs in groups A, B, and S were proven to be associated with the salt-stress response.
Conclusions
This study have conducted a genome-wide identification, characterization and expression analysis of bZIP genes in Arachis genomes. Our results provide insights into the evolutionary history of the bZIP gene family in peanut and the funcntion of Arachis bZIP genes during seed development and in response to salt stress.
Similar content being viewed by others
Background
In plants, transcription factors (TFs) possess specific domains that bind upstream of target genes to regulate gene expression [1, 2]. Of these plant TFs, the basic leucine zipper (bZIP) transcription factor family is one of the largest, and was named and characterized based on the conserved bZIP domain [3, 4]. The domain is 60–80 amino acids in length and is composed of two parts: a basic region and a leucine zipper motif. The basic region is highly conserved and includes 16 amino acid residues with an invariant motif N-× 7-R/K-× 9, independently determining nuclear localization and DNA binding specificity [5, 6]. The leucine zipper motif is less conserved, and contains heptad repeats of leucine (Leu) or other bulky hydrophobic amino acids which is responsible for specific recognition and homo- and/or heterodimerization [4, 7]. The bZIP gene family has been systematically investigated and characterized based on the whole genome sequences of several plants, including Arabidopsis [4], rice [8], sorghum [9], maize [7], grapevine [10], Brachypodium distachyon [11], tomato [12], apple [13], cassava [14] and banana [15].
bZIP genes play important roles in many essential biological processes, including organ differentiation, flower and vascular development, embryogenesis, seed maturation and storage protein gene regulation [16,17,18,19,20]. Considerable evidence also indicates that bZIP genes are important regulators of signaling and the response to abiotic/biotic stress [4, 7]. The phytohormone abscisic acid (ABA) is associated with seed development as well as abiotic stress responses [21]. The ABA-responsive element binding proteins (AREB) or ABRE binding factors (ABFs), which are group A bZIP proteins, have an important role in ABA and stress signaling [22, 23]. For instance, ABI5 is involved in ABA or stress signaling to regulate seed size and development, seed germination and early seedling growth as well as response to abiotic stress [24,25,26,27]. Group B bZIP proteins, which have a transmembrane domain and a specific domain at the C-terminus, also are important to the salt stress response via endoplasmic reticulum stress signaling [28]. For example, slbZIP38, a group G bZIP gene identified in tomato, have proven to be a negative regulator of salt stress tolerance [29]. For Group S bZIP proteins, AtbZIP1, MtbZIP2, and MtbZIP26 from Arabidopsis thaliana and Medicago truncatula, were transcriptionally induced by salt treatment, leading to an increase in salt stress tolerance [30,31,32]. In addition, bZIPs from groups C and S could cooperate with several TFs to form heterodimers and be responsible for the salt stress and seed development crosstalk network [33]. Together, these evidences indicate that bZIP genes have an essential role in both seed development and salt stress.
The peanut (Arachis hypogaea) is an important economical oilseed crop primarily grown in the tropics and semi-arid tropics and provide an important global source of vegetable oil and protein (http://faostat.fao.org/). Despite the economic and nutritional importance of peanuts, and the critical role of bZIP transcription factors in plant development and stress responses, only one AhbZIP gene has been reported that the over-expression of this gene (AREB1) is related to increase abiotic tolerance [34]. In 2016, the genomes of the two diploid ancestors (A. duranensis and A. ipaensis) of cultivated peanut have become available [35], allowing the genome-wide identification and systematic analysis of the bZIP gene family in Arachis genomes. In this study, we identified bZIP genes and analyzed their bZIP domain sequences, gene structure and additional MEME motifs, the DNA-binding-site specificity and dimerization properties of the bZIP proteins. We also investigated the impact of segmental and tandem duplication on the expansion of Arachis bZIP gene family. Using the RNA-seq and quantitative real-time PCR (qRT-PCR) methods, we analyzed their expression profiles in seed developmental stages and salt stress, and identified several candidate Arachis bZIPs responsive to seed development and salt stress.
Methods
Identification of bZIP genes in A. duranensis and A. ipaensis genomes
The genomic sequences of A. duranensis and A. ipaensis and their annotated gene models were downloaded from peanutbase (http://www.peanutbase.org/). BLAST were firstly conducted to search homologous bZIP genes using known bZIP proteins from Arabidopsis [4], rice [8] and maize [7] as queries. The targeting genes with similarity of E-value less than 1e-5 were retained for the following analysis. Subsequently, Hidden Markov Model (HMM) search (http://hmmer.org/) of the bZIP domain profiles (PF00170, PF07716 and PF03131) were performed to identify bZIP domain in these candidate proteins. Finally, Interpro (http://prosite.expasy.org/) and ExPASy Proteomics Server (http://prosite.expasy.org/) were used to confirm the integrity of bZIP domain in candidate genes. Each bZIP gene was given a unique name based on the exact position on chromosome/scaffold (from top to bottom) (Additional file 1).
Sequence alignment and phylogenetic analysis
ClustalX 2.0 [36] were used to align the bZIP sequences of coding DNA and proteins from A. thaliana, A. duranensis and A. ipaensis. The penalties for a gap open and gap extension were 10 and 0.1, respectively. PhyML 3.0 software [37] was used for the reconstruction of the maximum likelihood (ML) phylogenetic tree. The JTT + G model were determined to be the best model for phylogenetic tree construction according to the akaike information criterion implemented in ProtTest 3.0 [38]. 100 replicates were used to produce bootstrap values. MEGA7 [39] was used to edit and show the phylogenetic tree.
Gene structure of bZIP genes
The exon/intron structure of bZIP genes was analyzed and displayed using the GSDS platform (http://gsds.cbi.pku.edu.cn/) [40]. Genewise [41] was used to determine the correspondence on coordinates between DNA (containing exon and intron together) and protein sequences. Then, the coordinates of bZIP domain in protein sequence were transformed to that in gene sequence using in-house perl scripts. The intron splicing phase within the basic and hinge regions of bZIP domains from all bZIP genes were characterized and divided into different types.
Detection of additional conserved motifs of bZIP genes
The MEME tool (http://meme.nbcr.net/meme/) [42] was employed to detect the additional motifs outside the bZIP domain of protein sequences. The motifs with 10–50 amino acids in length and E-value less than 1e - 40 were characterized. All the motifs were compared among bZIP genes to identify the group-conserved or group-specific signatures. These motifs were numbered according to their order in the protein sequences.
Detecting duplicated genes and estimation of nonsynonymous (Ka) and synonymous (Ks) substitutions per site and their ratios
MCScan (http://chibba.agtec.uga.edu/duplication/mcscan) was used to detect the duplicated genomic segments in two Arachis genomes. Tandem duplication cluster was defined to contain at least two consecutive genes with sequence similarity (threshold of e < 10− 20), and one unrelated gene among cluster members was tolerated. The amino acid sequences of duplicated gene pairs were firstly aligned and guide the alignment of cDNA sequences in-house perl-scripts. KaKs_Calculator was used to compute Ka and Ks values of each duplicated gene pair using the YN model [43].
Expression analysis of Arachis bZIP genes during seed development and under salt stress
For investigating the expression of bZIP genes during peanut seed development, we downloaded the previously reported RNA-seq data of peanut seeds at 20, 40 and 60 days after flowering (DAF) [44]. Trimmomatic [45] was used to check, filter or trim RNA-seq reads with low-quality. RNA-seq reads were mapped to reference genome using Hisat2 [46], and the gene expression value were estimated using RSEM [47]. DESeq2 package [48] was used for differential expression (DE) analysis.
For qRT-PCR experiment, the elite peanut cultivar ‘Zhonghua16’ was planted to collect seeds at DAF20, DAF40, and DAF60 according to the previous method [44]. For preparing salt-stress plants, 2-week-old peanut seedlings (at the four-leaf stage) were removed from the soil and hydroponically grown in a 300 mM NaCl solution (Treatment) or deionized water (Control). The time points for salt treatment were setted to be 0, 1, 5, and 10 h, and the seedling roots were collected and frozen immediately in liquid nitrogen for RNA extraction.
Total RNA was extracted with RNAprep Pure Plant Kit (TIANGEN, China) and reverse transcribed into cDNA with cDNA Synthesis Kit (Thermo Fisher Scientific, USA) following the manufacturer’s instructions. qRT-PCR were performed in a 20 μL reaction volume using a CFX connect Real-Time System (Bio-Rad, Hercules, CA, USA) and Hieff qRCR SYBR Green Master Mix (YEASEN, Shanghai, China). The peanut Actin gene (Aradu.W2Y55) was used as the internal control, and the difference in relative target gene expression among the different experimental conditions was calculated using the 2-∆∆Ct method. Standard error was calculated among the three biological replicates of each experiment. Student’s t test was used to test the statistical significance of differences in relative target gene expression.
Results and discussion
Identification, phylogenetic analysis and group classification of bZIP genes in A. duranensis and A. ipaensis
Based on homology searches and domain verification, a total number of 50 and 45 unique bZIP genes were identified in A. duranensis and A. ipaensis genomes, respectively. The details for these genes, including gene ID, genomic position, domain composition, and group classification are given in Additional file 1. According to the existing nomenclature system, we assigned unique names to each of these novel bZIP genes: AdbZIP1–50 and AibZIP1–45. After checking bZIP domains, 93 genes had a typical bZIP domain, including an invariant N-× 7-R/K motif in the basic region and a heptad repeat of Leu positioned exactly nine amino acids upstream of R/K toward the C terminus (Additional file 2). The remaining two bZIP genes, AdbZIP28 and AibZIP22, had an unusual substitution in the basic region: a replacement of the conserved Arg/Lys (R/K) with IIe (I). This replacement has also been reported in other species [8, 49].
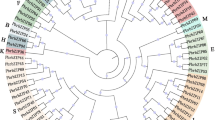
A systematic investigation of the bZIP gene family was first carried out in Arabidopsis [4]. In this analysis, different groups of bZIP genes were distinguished and named based on their phylogenetic relationships and functional divergences. This classification system has since been adopted for other species based on the clustering of bZIP genes from their own and Arabidopsis genomes [7,8,9,10,11,12,13,14,15, 50,51,52,53]. Here, based on a maximum likelihood (ML) analysis of bZIP proteins from Arachis and Arabidopsis genomes, we identified 11 distinct bZIP gene clades (groups A–I, S, and U), all with high bootstrap support (Fig. 1). The subgroup classification of Arachis bZIPs was further confirmed by phylogenetic tree reconstruction after adding bZIPs from soybean (Additional file 3). Most bZIP clades include closely related Arachis bZIPs and their Arabidopsis orthologs; clades E and F have no corresponding members in A. duranensis or A. ipaensis. Notably, bZIP genes within the same clade shared similar group-specific sequence characteristics, including exon/intron structure, intron phases, MEME motifs, and prediction of binding site structure (further analyzed below). This pattern of interspecific group clustering suggested that the group-specific features emerged prior to the divergence of Arachis and Arabidopsis. However, several differences have also accumulated in the bZIP genes of the different plant species over evolutionary time.
Phylogenetic analysis of peanut and Arabidopsis bZIP genes. Genes at branch ends from different species are denoted by different colored triangles. The peanut bZIP proteins are grouped into nine distinct clades (A–D, G–I, S, and U). bZIP protein sequences were aligned with ClustalX, and the phylogenetic tree was constructed in PhyML using the maximum likelihood method. Bootstrap values are based on 100 replicates
Gene structure of Arachis bZIP genes
As intron and exon organization might indicate the evolutionary trajectory of bZIP genes [8], we examined the structure of Arachis bZIP genes, including intron number, length, and splicing phase (Additional file 4). We found that overall gene structures were identical or similar for Arachis bZIPs within the same phylogenetic group. Considering the number of introns of peanut bZIPs, 24% of AdbZIPs and 22% of AibZIPs were intronless, occurring exclusively in groups S and B. Among the intron-containing genes, the number of introns varied from 1 to 13 in AdbZIP and AibZIP genes. bZIP genes in group G had the most introns, consistent with observations in other legume genomes [32].
The splicing phases were designated as three splicing phases: phase 0 (P0), splicing occurred after the third nucleotide of the codon; phase 1 (P1), splicing occurred after the first nucleotide of the codon; and phase 2 (P2), splicing occurred after the second nucleotide. The phases of splicing sites within the open reading frames (ORFs) were diverse, but were highly conserved in the basic and hinge regions of bZIP domain, because any changes in these regions would affect their code and function. Based on intron position and presence or number of splicing phases in the bZIP domain, four intron patterns (a to d) in Arachis bZIP genes were identified (Fig. 2 and Additional file 2). Pattern a had just one intron inserted at the − 5 position of the hinge region, between the amino acids Gln and Ala; this pattern was identified in all Arachis bZIP genes in groups A and G. Pattern b had two intron insertions with phase 0, one in the basic region and the other in the hinge region; this pattern was identified in all bZIP genes in group D. Pattern c had a single intron inserted at the − 20 position in the basic region in phase 2 (P2), and contains all bZIP genes in groups C and H. Pattern d lacked introns in the basic and hinge regions, and includes all bZIP genes in groups B and S. In addition, most Arachis bZIPs exhibiting pattern d were intronless, except for AdbZIP45 and AibZIP40. Each of these genes had one intron outside the basic and hinge regions. The patterns of splicing phase in Arachis bZIP domain observed here were consistent with those observed in other species [7, 8, 32]. The high conservation of gene structure and intron phases within phylogenetic clades supported the accepted group classification, and suggested that these different patterns of exon splicing may play an important role in functional evolution.
Intron patterns within the basic and hinge regions of the Arachis bZIP domain. The primary structure of the bZIP domain is shown at the top of the image. P0 indicates that the intron splicing site is between codons, and P2 indicates that the intron splicing site is located between the second and third nucleotides of the codon. Based on the intron incidence, intron position, and splicing phase, the Arachis bZIP genes exhibited four different types of patterns (a–d). Details of the intron positions within the bZIP domain of the peanut bZIP proteins are shown in Additional file 2
The motif compositions for different groups of Arachis bZIPs
In addition to the bZIP domain, many additional conserved motifs were detected in bZIP genes by the MEME analysis tool. As shown in Fig. 3, a total of 18 conserved motifs outside the bZIP domain were identified, and the consensus motif compositions for each subgroup were constructed (Additional file 5). These consensus motifs indicated that the overall compositions of the motifs were similar within the same subgroup but different among different groups. This suggested that functional divergence of bZIP genes may be determined by group-specific motifs. Individual examination of these motifs indicated that many were group-specific. For example, motifs 1, 2, 3, and 10 were only identified in group D; motifs 5, 14, and 15 were only identified in group G; motif 6 was only identified in group I; and motif 9 was only identified in in group H. Several motifs may be associated with specific biological functions. For example, Motif 1 is the DELAY OF GERMINATION (DOG) 1 domain, which is required for the induction of dormancy and multiple aspects of seed maturation, in part by interfering with ABA signaling components [54]. Motif 3 contains potential casein kinase II (CK II) phosphorylation sites (S/TxxD/E), which play a key role in cell division and expansion and affect diverse developmental and stress responsive pathways [55, 56]. Interestingly, these group-specific motifs have also been identified in bZIPs from the same group in other legume genomes [32], suggesting that motif composition is conserved across legume plants.
Distribution of additional conserved motifs, as identified by MEME. Motif compositions for each group of peanut bZIP proteins are shown, based on the position of the bZIP domain and additional conserved motifs outside the bZIP domain. The bZIP domains are shown in red, while other motifs are highlighted with colored boxes numbered 1 to 18. Details of the predicted conserved motifs are given in Additional file 6
Arachis bZIP DNA-binding-site structure and dimerization properties
The core basic region and the hinge region of the bZIP domain independently determine DNA-binding specificity, as demonstrated by several experiments [5, 6]. The unusual replacement of the two invariant sites, asparagine (Asn/N; position: − 18) and arginine (Arg/R; position: − 10), altered DNA-binding specificities [5]. We aligned the amino acids sequences of the basic and hinge regions of peanut bZIP proteins to identify conserved and polymorphic amino acid residues within each group (Additional file 6). No replacements of Asn/N at the − 18 position were observed in any peanut bZIPs. However, all members of group I had lysine (Lys/K) instead of arginine (R) at the − 10 position, consistent with the group I bZIPs from other legume species [32]. In addition, AdbZIP28 and AibZIP22 (group U) had a hydrophobic isoleucine (Ile/I) residue instead of an arginine (Arg/R), and such a replacement was demonstrated to completely inhibit the affinity of bZIP for AP1 in yeast [5] and does not recognize G-boxes in rice [49].
The Leu zipper sequence mediates the homo- and/or heterodimerization of bZIP proteins, which are known to bind to DNA as dimers [57, 58]. The Leu zipper region consists of heptad repeats, the amino acids are referred to a, b, c, d, e, f, and g within each heptad [59]. As the amino acids at the a, d, e and g positions are near the Leu zipper interface, these amino acids are the ones that primarily determine Leu zipper oligomerization, dimerization stability, and dimer specificity. We analyzed the compositions of the amino acids found at the a, d, e and g positions of peanut bZIPs (Fig. 4a).
Prediction of dimerization properties of the Arachis bZIP proteins. a Pie charts indicating the frequency of various amino acids in each of the four positions (a, d, e, and g) in the Leu zipper of the Arachis bZIP domains. b Histogram of the frequency of Asn (N) in the a position of the Leu zipper across all Arachis bZIP proteins. c Histogram showing the frequency of attractive or repulsive g↔e’ pairs per heptad across all Arachis bZIP proteins. The g↔e′ pairs are classified into four groups according to the electrostatic charges at the g and e positions. The +/− attractive, showed by orange box, indicates that the g position is basic and the following e position is acidic. The −/+ attractive, showed by skyblue box, indicates that the g position is acidic and the following e position is basic. The basic repulsive (pink box) and acidic repulsive (green box) indicate that the g and the following e positions have a similar charge, either both basic or both acidic
At the a position, about 20% of the residues were asparagine (Asn/N), which can form a polar pocket in the hydrophobic interface, allowing for more stable N-N interactions at a↔a′ (the corresponding position in the opposite helix), as compared to other amino acids [60]. Across the different heptads, the second and the fifth heptads had the highest frequency of Asn/N residues in the a position (61.46 and 60.22%, respectively; Fig. 4b). At the d position (Fig. 4a), the Leu was found in 45% of all peanut bZIPs and is one of the most dimer-stabilizing aliphatic amino acids [61]. At the e position, 37% of all peanut bZIPs had acidic amino acids D or E, while at the g position, 44% of all peanut bZIPs had the basic amino acids R or K (Fig. 4a). These charged amino acids are thought to form salt bridges between helices in electrostatic interactions [62]. The attractive or repulsive g↔e′ electrostatic interactions can also form interhelical salt bridges that affect dimerization specificity and stability [62]. For investigating the contribution of charged residues at the e and g positions in governing dimerization properties of Arachis bZIP proteins, the frequencies of attractive and repulsive g↔e′ pairs in each heptad was calculated (Fig. 4c). Across all heptads, the attractive g↔e′ pairs were concentrated in the second (15.6%), fifth (35%) and sixth (30%) heptads, indicating they can form complete attractive g↔e′ interactions and contribute to stability through complementation in a heterodimer. Three groups comprising 28 subfamilies (BZ1–BZ28) were further divided based on homo- and heterodimerization properties, particularly dimerization specificity [60, 63] (Additional file 7).
The impact of whole genome duplication and tandem duplication on the expansion of Arachis bZIP gene family
We identified the genome-wide collinear duplicated blocks in the A. duranensis and A. ipaensis genomes and the orthologous collinear blocks between two genomes. The pairwise synonymous distances (Ks values) between the paralogs and orthologs within collinear blocks were calculated, and their frequency distributions were plotted (Fig. 5a; Ks bin = 0.05). The peak Ks frequency between A. duranensis and A. ipaensis, representing average sequence variation, was 0.035. This represented the sequence divergence between these two closely related Arachis species, which was estimated to have diverged ~ 2.16 million years ago [35]. Further, the Ks peaks for A. duranensis and A. ipaensis paralogs were 0.90 and 0.95, respectively, corresponding to the sequence divergence of early papilionoid whole genome duplication (WGD) event occurred ~ 58 million years ago [35].
Whole genome duplication (WGD)-derived Arachis bZIP genes. a The Ks distribution of paralogs from WGD-derived duplicated genomic blocks in A. duranensis and A. ipaensis. b The duplicated bZIP paralogs derived from WGD were linked by blue (in A. duranensis) and green (in A. ipaensis) lines. The bZIP orthologs between A. duranensis and A. ipaensis were linked by read lines
We detected 35 AdbZIPs and 32 AibZIPs involved in duplicated genomic blocks, accounting for around 70% (35/50) and 71% (32/45) of the bZIP genes in each species (Fig. 5b and Additional file 8). Moreover, the duplicated bZIP gene pairs occurred either within a chromosome or between chromosomes, and some of these pairs were segmentally duplicated once, twice, or three times. This result indicated preferential gene retention and frequent chromosomal arrangements after WGD. Tandem duplications were detected for only two gene pairs (AdbZIP33/AdbZIP34 and AdbZIP41/AdbZIP42) in A. duranensis and only one gene pair (AibZIP28/AibZIP29) in A. ipaensis. This suggested that tandem duplication occurred rarely and was not more important than segmental duplication in the expansion of the bZIP gene family. We also used phylogenetic and syntenic analyses to identify 35 orthologous bZIP gene pairs between A. duranensis and A. ipaensis. These genes were also homeologs between the two subgenomes of the tetraploid peanut.
To understand the evolutionary constraints acting on the Arachis bZIP genes, we calculated Ka/Ks values for each duplicated bZIP gene pair in two Arachis species (Additional file 9). For most of these pairwise comparisons, the Ka/Ks values were less than 0.5 (only one pairwise comparison between duplicated AdbZIPs and only two between duplicated AibZIPs were larger than 0.5). This suggested that strong purifying selection acted on the Arachis duplicated bZIPs to remove deleterious mutations at the protein level.
Expression analysis of Arachis bZIP genes during peanut seed development
To profile bZIP gene expression, we used our previously published RNA-seq data [44], which documents gene expression in peanut seeds at different developmental stages: 20, 40, and 60 days after flowering (DAF). Using this data, we identified the FPKM values for all Arachis bZIPs and all differentially expressed bZIPs across the three developmental stages. With the exception of 24 bZIPs, which were not expressed at any developmental stage, four groups including corresponding bZIP genes with specific expression profile were recognized(Fig. 6a and Additional file 10). The first group comprised 37 bZIPs that were up-regulated during early development (20 DAF), but down-regulated thereafter (at 40 and 60 DAF). The second group comprised 15 bZIPs that were up-regulated at 40 DAF, while the third group comprised 17 bZIPs that were down-regulated at 40 DAF. The fourth group comprised 22 bZIPs that were highly expressed across all three developmental stages. The highly expressed bZIPs in group four were mainly distributed in clades A, C, and S. Several of these bZIPs were homologous to genes that have been implicated in seed development in other plants, such as Arabidopsis [4], rice [8] and maize [7]. Here, 12 bZIPs, which were highly expressed and homologous to previous well-studied genes in seed development, were selected for qRT-PCR confirmation, and found that the expression patterns determined by RNA-seq were consistent with those found using qRT-PCR (Fig. 6b).
Arachis bZIP gene expression during peanut seed development. a Four groups (groups I - IV) including corresponding bZIP genes with specific expression profile were recognized. In each subgroup, the gray lines indicated the expression values of bZIPs at DAF20, DAF40 and DAF60. The red line show the average FPKM of all bZIP genes. b qRT-PCR verification of 12 bZIP genes expressed during seed development. The relative gene expression levels as measured by qRT-PCR (orange histograms) and by RNA-seq (blue lines) are shown. Results are based on three biological replicates; error bars represent SE. c Expression pattern of bZIP A. duranensis and A. ipaensis orthologs during seed development. The similar (denoted as Y) or diverged (denoted as N) expression pattern between orthologs were indicated
In group A, AdbZIP33 and AibZIP28 were orthologous to Arabidopsis ABA insensitive 5 (ABI5), which is associated with ABA-signaling as well as the regulation of seed development and longevity in Arabidopsis [64] and legumes [27]. Our RNA-seq and qRT-PCR results showed that both orthologous ABI5 copies from the two subgenomes of the tetraploid peanut were highly expressed during development, suggesting the function of these genes may be similar in peanut and Arabidopsis. Our qRT-PCR results also indicated that the group A genes AdbZIP42, AdbZIP48 and AibZIP31 were stably expressed during development (Fig. 6b and Additional file 11). These genes are homologous to ABFs and AREB, which are involved in ABA-mediated seed development, germination, and embryo maturation [65]. Three genes in group C (AdbZIP23, AdbZIP37, and AibZIP30) were also highly expressed, and are homologous to the maize bZIP factor Opaque2. Opaque2 regulates protein accumulation and amino acid and sugar metabolism in maize seeds [66,67,68,69]. In addition, the group S genes AibZIP10, AdbZIP12, AdbZIP24, AdbZIP26, and AdbZIP36 were extremely highly expressed in peanut seeds (Fig. 6b and Additional file 11). Interestingly, the group S genes AdbZIP24 and AdbZIP36 had a similar expression pattern to the group C genes AdbZIP37 and AibZIP30: a decrease in expression level as seed development progressed.
We then further investigated the divergences in gene expression between homeologous genes from the AA and BB genomes of the tetraploid peanut. The heatmap analysis indicated that the overall expression patterns across seed development were similar for 31 pairs of homeologous/orthologous genes from the AA and BB genomes. We used the differential expression analysis method in combination with statistical methods to calculate differences in gene expression between these gene pairs for each sample. We found that 3 pairs of genes (AdbZIP5 and AibZIP5, AdbZIP17 and AibZIP15, AdbZIP46 and AibZIP41) were differentially expressed at 20 DAF, 3 pairs (AdbZIP3 and AibZIP1, AdbZIP4 and AibZIP4, AdbZIP49 and AibZIP45) at 40 DAF, and 5 pairs (AdbZIP3 and AibZIP1, AdbZIP33 and AibZIP28, AdbZIP37 and AibZIP30, AdbZIP10 and AibZIP10, AdbZIP1 and AibZIP3) at 60 DAF. These results indicated the overall expression conservation between two genomes, but suggested that 20% of the genes had diverged in expression during the parallel evolution and polyploidization of two genomes (Fig. 6c).
qRT-PCR expression profiles of Arachis bZIP genes under salt stress
We used qRT-PCR to explore changes in bZIP gene expression in response to salt-treatment (Fig. 7 and Additional file 12). We were unable to clearly amplify 4 bZIPs with PCR. After peanut roots were treated with salt for 1 h, 20 genes were significantly differentially expressed; after 5 h, 27 genes were significantly differentially expressed; and after 10 h, 41 genes were significantly differentially expressed (Fig. 7j; Student’s t test: P < 0.05). At each time point, many more genes were up-regulated than were down-regulated (14 vs. 6 at 1 h; 21 vs. 6 at 5 h; and 34 vs. 7 at 10 h). Among these differentially expressed bZIPs after salt treatment, many of them were distributed in groups A and S (Fig. 7k), indicating bZIPs in these groups play important roles in sugar signaling and abiotic stress regulation [4, 70, 71].
Arachis bZIP gene expression levels in peanut roots after 0, 1, 5, and 10 h of salt treatment. a–i bZIP gene expression levels in different groups. *: P < 0.05. j The number of significantly differently expressed bZIP genes in each group. k The number of significant differently expressed bZIP genes after 1, 5, and 10 h of salt treatment
Group A bZIPs possess the CKII and Ca2 + −dependent protein kinase phosphorylation site motifs involved in stress and/or ABA signaling, and these motifs are important for plant adaptation to various abiotic environmental stressors [72]. Indeed, many group A genes are associated with the salt stress response. In Arabidopsis, ABI5 and ABFs/AREB are key ABA-dependent signal transduction factors involved in abiotic stress tolerance [22, 73]. The over-expression of GhABF2 significantly improved salt stress tolerance both in Arabidopsis and cotton [74]. In tomato, slAREB1 and slbZIP1 knockout increased salt stress tolerance, while slAREB1 and slbZIP1 over-expression reduced salt stress tolerance [75, 76]. Here, genes AdbZIP42 and AibZIP35 were significantly up-regulated in response to salt stress, and these genes are homologous to ABFs, GhABF2, slAREB1, and slbZIP1. In addition, these genes have been reported to be phosphorylated by the ABA-activated SnRK2 protein kinases [77,78,79,80], suggesting phosphorylating ABA response element-binding factors may be critical for the ABA-mediated salt stress response.
The group B genes AdbZIP45 and AibZIP40 were up-regulated after 10 h of salt stress, and these genes are homologous to AtbZIP17, which could improve the expression of several salt stress response genes in Arabidopsis [28]. Seven group G bZIP genes (AdbZIP7, AdbZIP15, AdbZIP19, AdbZIP50, AibZIP17, AibZIP21, and AibZIP38) were homologous to Arabidopsis AtbZIP41 and tomato slbZIP38, and these genes have both been shown to negatively regulate salt stress [29]. Of these seven genes, AdbZIP15 was significantly down-regulated after 1 h and 5 h of salt stress treatment, while AdbZIP19 and AibZIP17 were significantly up-regulated after 10 h of salt stress. Thus, AdbZIP15, AdbZIP19 and AibZIP17 might confer resistance to salt stress. AdbZIP15 might be a negative regulator of salt stress, as its expression pattern was similar to that of slbZIP38 in response to salt stress.
The group S genes AdbZIP24 and AdbZIP36 were homologous to AtbZIP1, AtbZIP53, MtbZIP2, and MtbZIP26, and the expression patterns of these genes in response to salt stress were similar (Fig. 7). In particular, AdbZIP36 was significantly up-regulated after 10 h of salt stress. Two homologous genes in Arabidopsis, AtbZIP1 and AtbZIP53, were shown to reprogram the primary carbohydrate and amino acid metabolism to help roots adapt to salt stress [30]. The homologs MtbZIP2 and MtbZIP26 are also transcriptionally induced by salt treatment, and improve plant tolerance to salt stress [32]. Notably, the expression pattern of AdbZIP36 was similar to those of AtbZIP1, MtbZIP2, and MtbZIP26 in Arabidopsis and M. truncatula [30, 32], suggesting that AdbZIP36 might be a positive regulator of tolerance to salt stress in the peanut. In summary, our study of expression analysis has identified several candidate peanut bZIPs, which may be associated with the salt-stress response, as targets for future research.
Conclusions
Despite the importance of bZIP transcription factors for plant growth, development, and abiotic stress responses, little is known about the bZIP gene family in peanut. Here, we used the previously published peanut reference genome to perform a comprehensive analysis of peanut bZIPs, including sequence identification, phylogenetic construction, motif composition characterization, gene structure analysis, and determination of DNA-binding-site specificity and dimerization properties. We also investigated evolutionary expansion of the bZIP gene family. bZIP genes were clearly divided into phylogenetic clades. These clades were supported by various group-specific sequence characteristics, including exon/intron structure, intron phases in domain, MEME motif composition, DNA-binding specificity, and dimerization properties. By analyzing changes in bZIP gene expression during seed development and in response to salt stress, we characterized the overall expression patterns for different groups of bZIPs. We also identified several candidate bZIP proteins that may be important for seed development and the salt stress response. The information generated in this study could facilitate further research on bZIP gene family and other gene families in peanut.
Abbreviations
- ABA:
-
Abscisic acid
- ABF:
-
ABRE binding factors
- ABI5:
-
A insensitive 5
- AREB:
-
ABA-responsive element binding proteins
- bZIP:
-
Basic leucine zipper
- DAF:
-
Days after flowering
- DE:
-
Differential expression
- DOG:
-
Delay of germination
- ER:
-
Endoplasmic reticulum
- HMM:
-
Hidden markov model
- Leu:
-
Leucine
- MEME:
-
Multiple Em for Motif Elicitation
- ML:
-
Maximum likelihood
- ORFs:
-
Open reading frames
- qRT-PCR:
-
Quantitative real-time PCR
- TF:
-
Transcription factor
- WGD:
-
Whole genome duplication
References
Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245(4916):371–8.
Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386(6625):569–77.
Hurst HC. Transcription factors 1: bZIP proteins. Protein Profile. 1995;2(2):101–68.
Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7(3):106–11.
Suckow M, Schwamborn K, Kisters-Woike B, von Wilcken-Bergmann B, Muller-Hill B. Replacement of invariant bZip residues within the basic region of the yeast transcriptional activator GCN4 can change its DNA binding specificity. Nucleic Acids Res. 1994;22(21):4395–404.
Niu X, Renshaw-Gegg L, Miller L, Guiltinan MJ. Bipartite determinants of DNA-binding specificity of plant basic leucine zipper proteins. Plant Mol Biol. 1999;41(1):1–13.
Wei K, Chen J, Wang Y, Chen Y, Chen S, Lin Y, Pan S, Zhong X, Xie D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012;19(6):463–76.
Nijhawan A, Jain M, Tyagi AK, Khurana JP. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008;146(2):333–50.
Wang J, Zhou J, Zhang B, Vanitha J, Ramachandran S, Jiang SY. Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum. J Integr Plant Biol. 2011;53(3):212–31.
Liu J, Chen N, Chen F, Cai B, Dal Santo S, Tornielli GB, Pezzotti M, Cheng ZM. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genomics. 2014;15:281.
Liu X, Chu Z. Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium distachyon. BMC Genomics. 2015;16:227.
Li D, Fu F, Zhang H, Song F. Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (Solanum lycopersicum L.). BMC Genomics. 2015;16:771.
Zhao J, Guo R, Guo C, Hou H, Wang X, Gao H. Evolutionary and expression analyses of the apple basic leucine zipper transcription factor family. Front Plant Sci. 2016;7:376.
Hu W, Yang H, Yan Y, Wei Y, Tie W, Ding Z, Zuo J, Peng M, Li K. Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci Rep. 2016;6:22783.
Hu W, Wang L, Tie W, Yan Y, Ding Z, Liu J, Li M, Peng M, Xu B, Jin Z. Genome-wide analyses of the bZIP family reveal their involvement in the development, ripening and abiotic stress response in banana. Sci Rep. 2016;6:30203.
Walsh J, Waters CA, Freeling M. The maize gene liguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade-sheath boundary. Genes Dev. 1998;12(2):208–18.
Chuang CF, Running MP, Williams RW, Meyerowitz EM. The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev. 1999;13(3):334–44.
Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309(5737):1052–6.
Shen H, Cao K, Wang X. A conserved proline residue in the leucine zipper region of AtbZIP34 and AtbZIP61 in Arabidopsis thaliana interferes with the formation of homodimer. Biochem Biophys Res Commun. 2007;362(2):425–30.
Lara P, Onate-Sanchez L, Abraham Z, Ferrandiz C, Diaz I, Carbonero P, Vicente-Carbajosa J. Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J Biol Chem. 2003;278(23):21003–11.
Romagosa I, Prada D, Moralejo MA, Sopena A, Munoz P, Casas AM, Swanston JS, Molina-Cano JL. Dormancy, ABA content and sensitivity of a barley mutant to ABA application during seed development and after ripening. J Exp Bot. 2001;52(360):1499–506.
Kerr TCC, Abdel-Mageed H, Aleman L, Lee J, Payton P, Cryer D, Allen RD. Ectopic expression of two AREB/ABF orthologs increases drought tolerance in cotton (Gossypium hirsutum). Plant Cell Environ. 2018;41(5):898-07.
Fujita Y, Yoshida T, Yamaguchi-Shinozaki K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol Plant. 2013;147(1):15–27.
Cheng ZJ, Zhao XY, Shao XX, Wang F, Zhou C, Liu YG, Zhang Y, Zhang XS. Abscisic acid regulates early seed development in Arabidopsis by ABI5-mediated transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell. 2014;26(3):1053–68.
Albertos P, Romero-Puertas MC, Tatematsu K, Mateos I, Sanchez-Vicente I, Nambara E, Lorenzo O. S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat Commun. 2015;6:8669.
Skubacz A, Daszkowska-Golec A, Szarejko I. The role and regulation of ABI5 (ABA-insensitive 5) in plant development, Abiotic Stress Responses and Phytohormone Crosstalk. Front Plant Sci. 2016;7:1884.
Zinsmeister J, Lalanne D, Terrasson E, Chatelain E, Vandecasteele C, Vu BL, Dubois-Laurent C, Geoffriau E, Le Signor A, Dalmais M, et al. ABI5 is a regulator of seed maturation and longevity in legumes. Plant Cell. 2016;28(11):2735–54.
Liu JX, Srivastava R, Che P, Howell SH. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 2007;51(5):897–909.
Pan Y, Hu X, Li C, Xu X, Su C, Li J, Song H, Zhang X, Pan Y. SlbZIP38, a Tomato bZIP Family Gene Downregulated by Abscisic Acid, Is a Negative Regulator of Drought and Salt Stress Tolerance. Genes. 2017;8(12):402.
Hartmann L, Pedrotti L, Weiste C, Fekete A, Schierstaedt J, Gottler J, Kempa S, Krischke M, Dietrich K, Mueller MJ, et al. Crosstalk between two bZIP signaling pathways orchestrates salt-induced metabolic reprogramming in Arabidopsis roots. Plant Cell. 2015;27(8):2244–60.
Sun X, Li Y, Cai H, Bai X, Ji W, Ding X, Zhu Y. The Arabidopsis AtbZIP1 transcription factor is a positive regulator of plant tolerance to salt, osmotic and drought stresses. J Plant Res. 2012;125(3):429–38.
Wang Z, Cheng K, Wan L, Yan L, Jiang H, Liu S, Lei Y, Liao B. Genome-wide analysis of the basic leucine zipper (bZIP) transcription factor gene family in six legume genomes. BMC Genomics. 2015;16:1053.
Weltmeier F, Rahmani F, Ehlert A, Dietrich K, Schutze K, Wang X, Chaban C, Hanson J, Teige M, Harter K, et al. Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: availability of heterodimerization partners controls gene expression during stress response and development. Plant Mol Biol. 2009;69(1–2):107–19.
Li XY, Liu X, Yao Y, Li YH, Liu S, He CY, Li JM, Lin YY, Li L. Overexpression of Arachis hypogaea AREB1 gene enhances drought tolerance by modulating ROS scavenging and maintaining endogenous ABA content. Int J Mol Sci. 2013;14(6):12827–42.
Bertioli DJ, Cannon SB, Froenicke L, Huang G, Farmer AD, Cannon EK, Liu X, Gao D, Clevenger J, Dash S, et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat Genet. 2016;48(4):438–46.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–82.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–21.
Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27(8):1164–5.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4.
Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Yi Chuan. 2007;29(8):1023–6.
Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14(5):988–95.
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–8.
Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17(1):32–43.
Wan L, Li B, Lei Y, Yan L, Ren X, Chen Y, Dai X, Jiang H, Zhang J, Guo W, et al. Mutant Transcriptome Sequencing Provides Insights into Pod Development in Peanut (Arachis hypogaea L.). Front Plant Sci. 2017;8:1900.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60.
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106.
Nantel A, Quatrano RS. Characterization of three rice basic/leucine zipper factors, including two inhibitors of EmBP-1 DNA binding activity. J Biol Chem. 1996;271(49):31296–305.
Baloglu MC, Eldem V, Hajyzadeh M, Unver T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS One. 2014;9(4):e96014.
Jin Z, Xu W, Liu A. Genomic surveys and expression analysis of bZIP gene family in castor bean (Ricinus communis L.). Planta. 2014;239(2):299–312.
Pourabed E, Ghane Golmohamadi F, Soleymani Monfared P, Razavi SM, Shobbar ZS. Basic leucine zipper family in barley: genome-wide characterization of members and expression analysis. Mol Biotechnol. 2015;57(1):12–26.
Zhou Y, Xu D, Jia L, Huang X, Ma G, Wang S, Zhu M, Zhang A, Guan M, Lu K, et al. Genome-Wide Identification and Structural Analysis of bZIP Transcription Factor Genes in Brassica napus. Genes. 2017;8(10):288.
Nishimura N, Tsuchiya W, Moresco JJ, Hayashi Y, Satoh K, Kaiwa N, Irisa T, Kinoshita T, Schroeder JI, Yates JR, et al. Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nat Commun. 2018;9(1):2132.
Mulekar JJ, Bu Q, Chen F, Huq E. Casein kinase II alpha subunits affect multiple developmental and stress-responsive pathways in Arabidopsis. Plant J. 2012;69(2):343–54.
Moreno-Romero J, Espunya MC, Platara M, Arino J, Martinez MC. A role for protein kinase CK2 in plant development: evidence obtained using a dominant-negative mutant. Plant J. 2008;55(1):118–30.
Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240(4860):1759–64.
Ellenberger TE, Brandl CJ, Struhl K, Harrison SC. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992;71(7):1223–37.
McLachlan AD, Stewart M. Tropomyosin coiled-coil interactions: evidence for an unstaggered structure. J Mol Biol. 1975;98(2):293–304.
Deppmann CD, Acharya A, Rishi V, Wobbes B, Smeekens S, Taparowsky EJ, Vinson C. Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: a comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Res. 2004;32(11):3435–45.
Moitra J, Szilak L, Krylov D, Vinson C. Leucine is the most stabilizing aliphatic amino acid in the d position of a dimeric leucine zipper coiled coil. Biochemistry. 1997;36(41):12567–73.
Vinson CR, Hai T, Boyd SM. Dimerization specificity of the leucine zipper-containing bZIP motif on DNA binding: prediction and rational design. Genes Dev. 1993;7(6):1047–58.
Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M. Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol. 2002;22(18):6321–35.
Dekkers BJ, He H, Hanson J, Willems LA, Jamar DC, Cueff G, Rajjou L, Hilhorst HW, Bentsink L. The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. Plant J. 2016;85(4):451–65.
Bensmihen S, Giraudat J, Parcy F. Characterization of three homologous basic leucine zipper transcription factors (bZIP) of the ABI5 family during Arabidopsis thaliana embryo maturation. J Exp Bot. 2005;56(412):597–603.
Schmidt RJ, Burr FA, Aukerman MJ, Burr B. Maize regulatory gene opaque-2 encodes a protein with a “leucine-zipper” motif that binds to zein DNA. Proc Natl Acad Sci U S A. 1990;87(1):46–50.
Lohmer S, Maddaloni M, Motto M, Di Fonzo N, Hartings H, Salamini F, Thompson RD. The maize regulatory locus Opaque-2 encodes a DNA-binding protein which activates the transcription of the b-32 gene. EMBO J. 1991;10(3):617–24.
Brochetto-Braga MR, Leite A, Arruda P. Partial purification and characterization of lysine-ketoglutarate reductase in normal and opaque-2 maize endosperms. Plant Physiol. 1992;98(3):1139–47.
Yunes JA, Cord Neto G, Leite A, Ottoboni LM, Arruda P. The role of the Opaque2 transcriptional factor in the regulation of protein accumulation and amino acid metabolism in maize seeds. Anais da Academia Brasileira de Ciencias. 1994;66(Su 1 ( Pt 2)):227–37 quiz 237-228.
Hanson J, Smeekens S. Sugar perception and signaling--an update. Curr Opin Plant Biol. 2009;12(5):562–7.
Smeekens S, Ma J, Hanson J, Rolland F. Sugar signals and molecular networks controlling plant growth. Curr Opin Plant Biol. 2010;13(3):274–9.
Hossain MA, Cho JI, Han M, Ahn CH, Jeon JS, An G, Park PB. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J Plant Physiol. 2010;167(17):1512–20.
Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci U S A. 2000;97(21):11632–7.
Liang C, Meng Z, Meng Z, Malik W, Yan R, Lwin KM, Lin F, Wang Y, Sun G, Zhou T, et al. GhABF2, a bZIP transcription factor, confers drought and salinity tolerance in cotton (Gossypium hirsutum L.). Scientific Rep. 2016;6:35040.
Orellana S, Yanez M, Espinoza A, Verdugo I, Gonzalez E, Ruiz-Lara S, Casaretto JA. The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ. 2010;33(12):2191–208.
Zhu M, Meng X, Cai J, Li G, Dong T, Li Z. Basic leucine zipper transcription factor SlbZIP1 mediates salt and drought stress tolerance in tomato. BMC Plant Biol. 2018;18(1):83.
Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, Yamamoto A, Hattori T. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005;44(6):939–49.
Kulik A, Wawer I, Krzywinska E, Bucholc M, Dobrowolska G. SnRK2 protein kinases--key regulators of plant response to abiotic stresses. OMICS. 2011;15(12):859–72.
Yoshida T, Fujita Y, Maruyama K, Mogami J, Todaka D, Shinozaki K, Yamaguchi-Shinozaki K. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015;38(1):35–49.
Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61(4):672–85.
Acknowledgements
Not applicable.
Funding
This research was supported by the National Key Research and Development Program of China (2018YFD1000900), the National Natural Science Foundation of China (nos. 31671734, 31461143022, 31770250 and 31371662), the Knowledge Innovation Program of Chinese Academy of Agricultural Sciences, Central Public-interest Scientific Institution Basal Research Fund, National High Technology Research and Development Program of China (863 Program, no. 2013AA102602), Agriculture Research System of China (CARS-14).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Authors' contributions
ZW, YL and BL conceived the project and research plans, designed the experiments, and wrote the manuscript with contributions from all authors. LY, LW, DH, YK and LS performed the experiments. ZW analyzed the data and prepared figures, ZW, HJ, YL and BL revised the manuscript critically. All authors have read and approved the final version of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Identified bZIP proteins in peanuts and related information. (XLSX 18 kb)
Additional file 2:
Positions and patterns of introns within the basic and hinge regions of the bZIP domains of the Arachis bZIP transcription factors. (PDF 327 kb)
Additional file 3:
The phylogenetic tree of bZIP genes from Arabidopsis thaliana, Arachis duranensis, Arachis ipaensis, and Glycine max. (PDF 683 kb)
Additional file 4:
Map of intron-exon arrangements for the Arachis bZIP genes. (PDF 980 kb)
Additional file 5:
MEME motif composition of the Arachis bZIP proteins. (PDF 520 kb)
Additional file 6:
Alignment of the basic and hinge regions of 95 Arachis bZIP proteins. (XLSX 18 kb)
Additional file 7:
Amino acid sequence alignment of the leucine zipper region of 95 Arachis bZIP proteins for prediction of dimerization properties. (PDF 894 kb)
Additional file 8:
Chromosomal distributions of the Arachis bZIP genes. (PDF 512 kb)
Additional file 9:
The Ka, Ks and Ka/Ks values for duplicated bZIP gene pairs in two Arachis genomes. (XLSX 14 kb)
Additional file 10:
Four groups including corresponding bZIP genes with specific expression profile were recognized. (XLSX 5042 kb)
Additional file 11:
Phylogenetic analysis of some Arachis bZIP proteins and their homologs in different plant species. (PDF 182 kb)
Additional file 12:
Gene-specific primers used for qRT-PCR. (PDF 463 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wang, Z., Yan, L., Wan, L. et al. Genome-wide systematic characterization of bZIP transcription factors and their expression profiles during seed development and in response to salt stress in peanut. BMC Genomics 20, 51 (2019). https://doi.org/10.1186/s12864-019-5434-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-019-5434-6